Distribution of Runs of Homozygosity and Their Relationship with Candidate Genes for Productivity in Kazakh Meat–Wool Sheep Breed
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction and SNP Genotyping
2.3. Quality Control of Data
2.4. Calculation of Inbreeding Coefficient
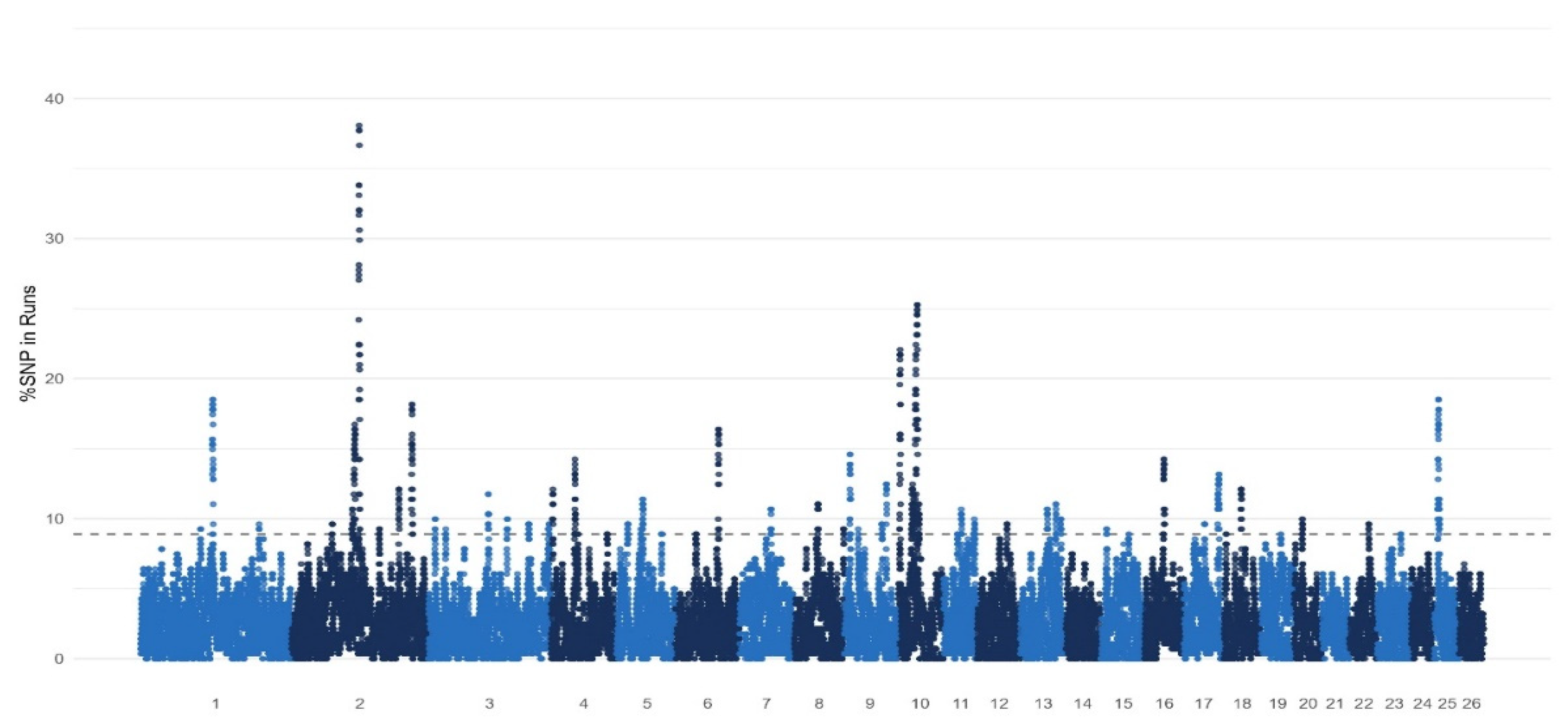
2.5. Detection of ROH Islands
3. Results
3.1. Runs of Homozygosity Analysis
3.2. Genomic Inbreeding Coefficients
3.3. Gene Annotation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tarlykov, P.; Atavliyeva, S.; Auganova, D.; Akhmetollayev, I.; Loshakova, T.; Varfolomeev, V.; Ramankulov, Y. Mitochondrial DNA analysis of ancient sheep from Kazakhstan: Evidence for early sheep introduction. Heliyon 2021, 7, e08011. [Google Scholar] [CrossRef]
- Sabdenov, K.S.; Makhatov, B.M.; Nurzhanova, K.H. Modern Technology of Production of Sheep Breeding Products; Aitumar: Almaty, Kazakhstan, 2015. (In Russian) [Google Scholar]
- Khamzin, K.P. Intensive Growing and Fattening of Young Stock of Multiparous Meat-Wool Sheep. Candidate Dissertation, KazNAU, Almaty, Kazakhstan, 1999. (In Russian). [Google Scholar]
- Li, X.; Yang, J.; Shen, M.; Xie, X.L.; Liu, G.J.; Xu, Y.X.; Lv, F.H.; Yang, H.; Yang, Y.L.; Liu, C.B.; et al. Whole-genome resequencing of wild and domestic sheep identifies genes associated with morphological and agronomic traits. Nat. Commun. 2020, 11, 2815. [Google Scholar] [CrossRef] [PubMed]
- El-Seedy, A.S.; Hashem, N.M.; El-Azrak, K.M.; Nour El-Din, A.; Ramadan, T.A.; Taha, T.A.; Salem, M.H. Genetic screening of FecB, FecXG and FecXI mutations and their linkage with litter size in Barki and Rahmani sheep breeds. Reprod. Domest. Anim. 2017, 52, 1133–1137. [Google Scholar] [CrossRef]
- Granado-Tajada, I.; Rodríguez-Ramilo, S.T.; Legarra, A.; Ugarte, E. Inbreeding, effective population size, and coancestry in the Latxa dairy sheep breed. J. Dairy. Sci. 2020, 103, 5215–5226. [Google Scholar] [CrossRef] [PubMed]
- Rafter, P.; McHugh, N.; Pabiou, T.; Berry, D.P. Inbreeding trends and genetic diversity in purebred sheep populations. Animal 2022, 16, 100604. [Google Scholar] [CrossRef] [PubMed]
- Peripolli, E.; Munari, D.P.; Silva, M.V.G.B.; Lima, A.L.F.; Irgang, R.; Baldi, F. Runs of homozygosity: Current knowledge and applications in livestock. Anim. Genet. 2017, 48, 255–271. [Google Scholar] [CrossRef]
- Martikainen, K.; Koivula, M.; Uimari, P. Identification of runs of homozygosity affecting female fertility and milk production traits in Finnish Ayrshire cattle. Sci. Rep. 2020, 10, 3804. [Google Scholar] [CrossRef]
- Bosse, M.; Megens, H.J.; Madsen, O.; Paudel, Y.; Frantz, L.A.; Schook, L.B.; Crooijmans, R.P.; Groenen, M.A. Regions of homozygosity in the porcine genome: Consequence of demography and the recombination landscape. PLoS Genet. 2012, 8, e1003100. [Google Scholar] [CrossRef]
- Sumreddee, P.; Toghiani, S.; Hay, E.H.; Roberts, A.; Aggrey, S.E.; Rekaya, R. Runs of homozygosity and analysis of inbreeding depression. J. Anim. Sci. 2020, 98, skaa361. [Google Scholar] [CrossRef]
- Forutan, M.; Ansari Mahyari, S.; Baes, C.; Melzer, N.; Schenkel, F.S.; Sargolzaei, M. Inbreeding and runs of homozygosity before and after genomic selection in North American Holstein cattle. BMC Genom. 2018, 19, 98. [Google Scholar] [CrossRef]
- Ku, C.S.; Naidoo, N.; Teo, S.M.; Pawitan, Y. Regions of homozygosity and their impact on complex diseases and traits. Hum. Genet. 2011, 129, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Metzger, J.; Karwath, M.; Tonda, R.; Beltran, S.; Águeda, L.; Gut, M.; Gut, I.G.; Distl, O. Runs of homozygosity reveal signatures of positive selection for reproduction traits in breed and non-breed horses. BMC Genom. 2015, 16, 764. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhuo, Y.; Ning, C.; Zhou, L.; Liu, J.F. Estimate of inbreeding depression on growth and reproductive traits in a Large White pig population. G3 2022, 12, jkac118. [Google Scholar] [CrossRef] [PubMed]
- Deniskova, T.; Dotsev, A.; Lushihina, E.; Shakhin, A.; Kunz, E.; Medugorac, I.; Reyer, H.; Wimmers, K.; Khayatzadeh, N.; Sölkner, J.; et al. Population Structure and Genetic Diversity of Sheep Breeds in the Kyrgyzstan. Front. Genet. 2019, 12, 1311. [Google Scholar] [CrossRef]
- Tao, L.; He, X.; Wang, F.; Gan, S.; Di, R.; Chu, M. Luzhong mutton sheep: Inbreeding and selection signatures. J. Anim. Sci. Technol. 2020, 62, 777–789. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, Z.; Fang, Y.; Cao, C.; Zhang, Z.; Pan, Y.; Wang, Q. Runs of homozygosity revealed reproductive traits of Hu sheep. Genes 2022, 13, 1848. [Google Scholar] [CrossRef]
- Cheng, J.; Zhao, H.; Chen, N.; Cao, X.; Hanif, Q.; Pi, L.; Hu, L.; Chaogetu, B.; Huang, Y.; Lan, X.; et al. Population structure, genetic diversity, and selective signature of Chaka sheep revealed by whole genome sequencing. BMC Genom. 2020, 21, 520. [Google Scholar] [CrossRef]
- Granero, A.; Anaya, G.; Demyda-Peyrás, S.; Alcalde, M.J.; Arrebola, F.; Molina, A. Genomic Population Structure of the Main Historical Genetic Lines of Spanish Merino Sheep. Animals 2022, 12, 1327. [Google Scholar] [CrossRef]
- McQuillan, R.; Leutenegger, A.-L.; Abdel-Rahman, R.; Franklin, C.S.; Pericic, M.; Barac-Lauc, L.; Smolej-Narancic, N.; Janicijevic, B.; Polasek, O.; Tenesa, A.; et al. Runs of homozygosity in European populations. Am. J. Hum. Genet. 2008, 83, 359–372. [Google Scholar] [CrossRef]
- Gorssen, W.; Meyermans, R.; Janssens, S.; Buys, N. A publicly available repository of ROH islands reveals signatures of selection in different livestock and pet species. Genet. Sel. Evol. 2021, 53, 2. [Google Scholar] [CrossRef]
- Selli, A.; Ventura, R.V.; Fonseca, P.A.S.; Buzanskas, M.E.; Andrietta, L.T.; Balieiro, J.C.C.; Brito, L.F. Detection and visualization of heterozygosity-rich regions and runs of homozygosity in worldwide sheep populations. Animals 2021, 11, 2696. [Google Scholar] [CrossRef] [PubMed]
- Mastrangelo, S.; Ciani, E.; Sardina, M.T.; Sottile, G.; Pilla, F.; Portolano, B.; Bi. Ov. Ita Consortium. Runs of homozygosity reveal genome-wide autozygosity in Italian sheep breeds. Anim. Genet. 2018, 49, 71–81. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Di, J.; Han, B.; Chen, L.; Liu, M.; Li, W. Genome-wide scan for runs of homozygosity identifies candidate genes related to economically important traits in Chinese Merino. Animals 2020, 10, 524. [Google Scholar] [CrossRef]
- Luigi-Sierra, M.G.; Cardoso, T.F.; Martínez, A.; Pons, A.; Bermejo, L.A.; Jordana, J.; Delgado, J.V.; Adán, S.; Ugarte, E.; Arranz, J.J.; et al. Low genome-wide homozygosity in 11 Spanish ovine breeds. Anim. Genet. 2019, 50, 501–511. [Google Scholar] [CrossRef]
- Signer-Hasler, H.; Burren, A.; Ammann, P.; Drögemüller, C.; Flury, C. Runs of homozygosity and signatures of selection: A comparison among eight local Swiss sheep breeds. Anim. Genet. 2019, 50, 512–525. [Google Scholar] [CrossRef] [PubMed]
- Dzomba, E.F.; Chimonyo, M.; Pierneef, R.; Muchadeyi, F.C. Runs of homozygosity analysis of South African sheep breeds from various production systems investigated using OvineSNP50k data. BMC Genom. 2021, 22, 7. [Google Scholar] [CrossRef]
- Machová, K.; Marina, H.; Arranz, J.J.; Pelayo, R.; Rychtářová, J.; Milerski, M.; Vostrý, L.; Suárez-Vega, A. Genetic diversity of two native sheep breeds by genome-wide analysis of single nucleotide polymorphisms. Animal 2023, 17, 100690. [Google Scholar] [CrossRef]
- Lv, J.; Ge, W.; Ding, Z.; Zeng, J.; Wang, W.; Duan, H.; Zhang, Y.; Zhao, X.; Hu, J. Regulatory role of dihydrotestosterone on BMP-6 receptors in granular cells of sheep antral follicles. Gene 2022, 810, 146066. [Google Scholar] [CrossRef]
- Zhou, N.; Li, Q.; Lin, X.; Hu, N.; Liao, J.Y.; Lin, L.B.; Zhao, C.; Hu, Z.M.; Liang, X.; Xu, W.; et al. BMP2 induces chondrogenic differentiation, osteogenic differentiation and endochondral ossification in stem cells. Cell. Tissue Res. 2016, 366, 101–111. [Google Scholar] [CrossRef]
- Lu, Z.; Liu, J.; Han, J.; Yang, B. Association between BMP2 functional polymorphisms and sheep tail type. Animals 2020, 10, 739. [Google Scholar] [CrossRef]
- Luong, H.T.; Chaplin, J.; McRae, A.F.; Medland, S.E.; Willemsen, G.; Nyholt, D.R.; Henders, A.K.; Hoekstra, C.; Duffy, D.L.; Martin, N.G.; et al. Variation in BMPR1B, TGFRB1 and BMPR2 and control of dizygotic twinning. Twin. Res. Hum. Genet. 2011, 14, 408–416. [Google Scholar] [CrossRef]
- Foroughinia, G.; Fazileh, A.; Eghbalsaied, S. Expression of genes involved in BMP and estrogen signaling and AMPK production can be important factors affecting total number of antral follicles in ewes. Theriogenology 2017, 91, 36–43. [Google Scholar] [CrossRef]
- Akhatayeva, Z.; Bi, Y.; He, Y.; Khan, R.; Li, J.; Li, H.; Pan, C.; Lan, X. Survey of the relationship between polymorphisms within the BMPR1B gene and sheep reproductive traits. Anim. Biotechnol. 2023, 34, 718–727. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; He, X.; Di, R.; Chu, M. Comparison of expression patterns of six canonical clock genes of follicular phase and luteal phase in Small-tailed Han sheep. Arch. Anim. Breed. 2021, 64, 457–466. [Google Scholar] [CrossRef]
- Xiong, X.; Huang, X.; Zhu, Y.; Hai, Z.; Fei, X.; Pan, B.; Yang, Q.; Xiong, Y.; Fu, W.; Lan, D.; et al. Testis-specific knockout of Kdm2a reveals nonessential roles in male fertility but partially compromises spermatogenesis. Theriogenology 2023, 209, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Flori, L.; Gonzatti, M.I.; Thevenon, S.; Chantal, I.; Pinto, J.; Berthier, D.; Aso, P.M.; Gautier, M. A quasi-exclusive European ancestry in the Senepol tropical cattle breed highlights the importance of the slick locus in tropical adaptation. PLoS ONE 2012, 7, e36133. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Zhang, D.; Wang, Z.; Zhao, Y.; Blecker, C.; Li, S.; Zheng, X.; Chen, L. Validation of protein biological markers of lamb meat quality characteristics based on the different muscle types. Food Chem. 2023, 427, 136739. [Google Scholar] [CrossRef]
- Yuan, Z.R.; Xu, S.Z. Novel SNPs of the bovine CACNA2D1 gene and their association with carcass and meat quality traits. Mol. Biol. Rep. 2011, 38, 365–370. [Google Scholar] [CrossRef]
- Hou, G.Y.; Yuan, Z.R.; Gao, X. Genetic polymorphisms of the CACNA2D1 gene and their association with carcass and meat quality traits in cattle. Biochem. Genet. 2010, 48, 751–759. [Google Scholar] [CrossRef]
- Sbardella, A.P.; Watanabe, R.N.; da Costa, R.M.; Bernardes, P.A.; Braga, L.G.; Baldi Rey, F.S.; Lôbo, R.B.; Munari, D.P. Genome-wide association study provides insights into important genes for reproductive traits in Nelore cattle. Animals 2021, 11, 1386. [Google Scholar] [CrossRef]
- Liu, D.; Fan, W.; Xu, Y.; Yu, S.; Liu, W.; Guo, Z.; Huang, W.; Zhou, Z.; Hou, S. Genome-wide association studies demonstrate that TASP1 contributes to increased muscle fiber diameter. Heredity 2021, 126, 991–999. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Jin, H.G.; Ma, H.H.; Zhao, Z.H. Comparative analysis on genome-wide DNA methylation in longissimus dorsi muscle between Small Tailed Han and Dorper×Small Tailed Han crossbred sheep. Asian-Australas. J. Anim. Sci. 2017, 30, 1529–1539. [Google Scholar] [CrossRef] [PubMed]

| Populations | Statistics | ROH Length Category (Mb) | |||||
|---|---|---|---|---|---|---|---|
| 1+ | 2+ | 4+ | 8+ | 16+ | |||
| POP1 n = 95 | Number of ROHs per animal | Mean | 40.3 | 6.9 | 3.2 | 2.4 | 2.1 |
| SD | 6.8 | 3.0 | 2.7 | 1.9 | 1.2 | ||
| Min | 26 | 1 | 0 | 0 | 0 | ||
| Max | 58 | 15 | 12 | 8 | 5 | ||
| Length of ROHs per animal (Mb) | Mean | 51.4 | 18.7 | 17.2 | 26.2 | 41.4 | |
| SD | 8.9 | 8.3 | 15.1 | 21.4 | 25.0 | ||
| Min | 31.2 | 2.2 | 0 | 0 | 0 | ||
| Max | 74.4 | 41.8 | 68.8 | 96.5 | 101.5 | ||
| POP2 n = 115 | Number of ROHs per animal | Mean | 42.2 | 8.0 | 3.9 | 3.2 | 1.6 |
| SD | 11.0 | 5.0 | 3.4 | 3.0 | 1.1 | ||
| Min | 17 | 1 | 0 | 0 | 0 | ||
| Max | 102 | 30 | 16 | 14 | 5 | ||
| Length of ROHs per animal (Mb) | Mean | 54.1 | 21.5 | 21.4 | 35.3 | 31.8 | |
| SD | 14.6 | 14.1 | 19.3 | 33.1 | 22.5 | ||
| Min | 22.5 | 2.1 | 0 | 0 | 0 | ||
| Max | 133.5 | 85.2 | 85.6 | 143.7 | 100.2 | ||
| POP3 n = 71 | Number of ROHs per animal | Mean | 40.7 | 6.4 | 2.3 | 1.8 | 1.1 |
| SD | 8.5 | 3.5 | 1.7 | 1.1 | 0.4 | ||
| Min | 24 | 1 | 0 | 0 | 0 | ||
| Max | 83 | 23 | 7 | 4 | 2 | ||
| Length of ROHs per animal (Mb) | Mean | 52.5 | 17.3 | 12.5 | 19.6 | 19.9 | |
| SD | 11.5 | 9.2 | 9.3 | 14.7 | 5.7 | ||
| Min | 30.3 | 2.3 | 0 | 0 | 0 | ||
| Max | 112 | 58.6 | 38.5 | 53.6 | 33.8 | ||
| Population | FROH > 1 Mb | FROH > 2 Mb | FROH > 4 Mb | FROH > 8 Mb | FROH > 16 Mb |
|---|---|---|---|---|---|
| Pop1 | 0.042 | 0.021 | 0.016 | 0.017 | 0.017 |
| Pop2 | 0.047 | 0.025 | 0.019 | 0.020 | 0.013 |
| Pop3 | 0.036 | 0.014 | 0.009 | 0.010 | 0.008 |
| CHR | nSNP | Start, Mb | End, Mb | Candidate Genes |
|---|---|---|---|---|
| 1 | 78 | 130.47 | 135.69 | TIAM1 |
| 2 | 36 | 218.8 | 219.93 | BMPR2 |
| 3 | 20 | 182.28 | 183.49 | MYBPC1 |
| 4 | 24 | 42.18 | 43.2 | CACNA2D1 |
| 6 | 21 | 78.04 | 79.31 | CLOCK |
| 6 | 33 | 33.61 | 35 | BMPR1B |
| 13 | 19 | 50.59 | 51.89 | BMP2 |
| 13 | 204 | 0.12 | 8.87 | TASP1 |
| 17 | 87 | 60.65 | 64.98 | KDM2B |
| 23 | 28 | 41.93 | 43.98 | MYOM1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amandykova, M.; Akhatayeva, Z.; Kozhakhmet, A.; Kapassuly, T.; Orazymbetova, Z.; Yergali, K.; Khamzin, K.; Iskakov, K.; Dossybayev, K. Distribution of Runs of Homozygosity and Their Relationship with Candidate Genes for Productivity in Kazakh Meat–Wool Sheep Breed. Genes 2023, 14, 1988. https://doi.org/10.3390/genes14111988
Amandykova M, Akhatayeva Z, Kozhakhmet A, Kapassuly T, Orazymbetova Z, Yergali K, Khamzin K, Iskakov K, Dossybayev K. Distribution of Runs of Homozygosity and Their Relationship with Candidate Genes for Productivity in Kazakh Meat–Wool Sheep Breed. Genes. 2023; 14(11):1988. https://doi.org/10.3390/genes14111988
Chicago/Turabian StyleAmandykova, Makpal, Zhanerke Akhatayeva, Altynay Kozhakhmet, Tilek Kapassuly, Zarina Orazymbetova, Kanagat Yergali, Kadyrzhan Khamzin, Kairat Iskakov, and Kairat Dossybayev. 2023. "Distribution of Runs of Homozygosity and Their Relationship with Candidate Genes for Productivity in Kazakh Meat–Wool Sheep Breed" Genes 14, no. 11: 1988. https://doi.org/10.3390/genes14111988





