Identification and Functional Characterization of Two Major Loci Associated with Resistance against Brown Planthoppers (Nilaparvata lugens (Stål)) Derived from Oryza nivara
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Mapping Population
2.2. DNA Extraction, PCR, and Electrophoresis
2.3. Marker Analysis
2.4. Targeted Mapping of BPH Locus
2.5. Sample Preparation for Quantitative Real-Time PCR (qRT-PCR)
2.6. Total RNA Isolation and cDNA Synthesis
2.7. Quantitative Real-Time PCR (qRT-PCR)
| Gene ID | Gene Name | Gene Description |
|---|---|---|
| Os04g12540.1 | OsRLK-1 | receptor-like protein kinase, putative, expressed |
| Os04g12560.1 | OsRLK-2 | receptor-like protein kinase, putative, expressed |
| Os04g12580.1 | OsRLK-3 | receptor-like protein kinase, putative, expressed |
| Os04g15580.1 | STPKR | serine/threonine-protein kinase receptor precursor, putative, expressed |
| Os04g15630.1 | Xa21 | xa21, putative, expressed |
| Os04g15650.1 | LRR | leucine-rich repeat family protein, expressed |
| Os04g15660.1 | RK | receptor kinase, putative, expressed |
| Os04g18650.1 | AP2/ERF | APETALA2/ethylene-responsive element binding protein 34 |
| Os04g19800.1 | F.BOX | F-box domain containing protein, expressed |
| Os04g20180.1 | LGDSL | lipase, GDSL-domain-containing protein |
2.8. Sequencing and Data Analysis
3. Results
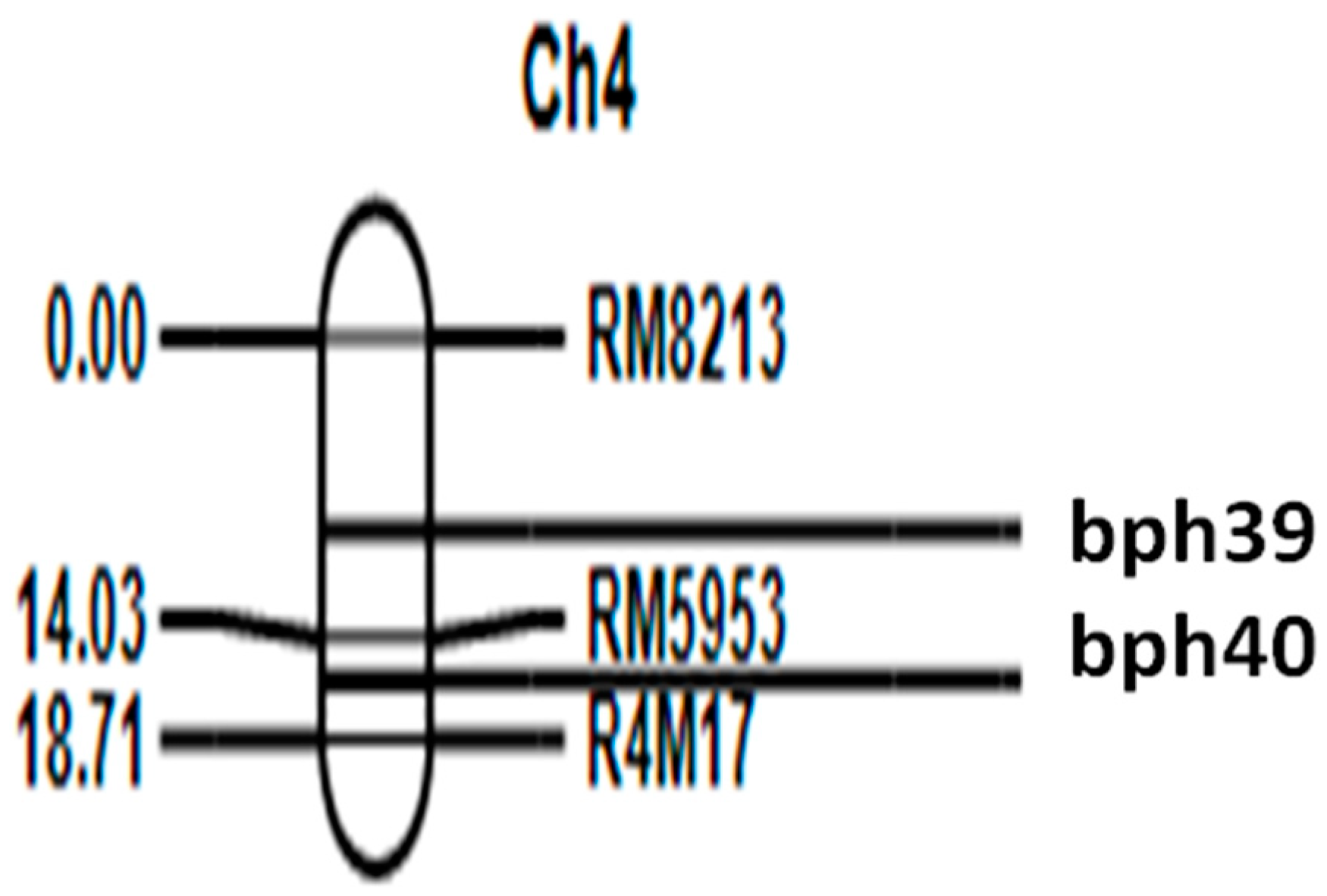
3.1. Construction of Linkage Map and Mapping of BPH-Resistant QTLs
3.2. Identification and Expression Analysis of BPH-Resistant Candidate Genes
3.3. Sequence Variation Analysis of the STPKR Gene
3.4. Translated Peptide Sequences and Amino Acid Variations
3.5. Variations in Regulatory Regions of STPKR Gene
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, C.N.; Cheng, C.C. The population levels of Nilaparvata lugens (Stål) in relation to the yield loss of rice. Plant Prot. Bull. 1978, 20, 197–209. [Google Scholar]
- Normile, D. Reinventing rice to feed the world. Science 2008, 321, 330–333. [Google Scholar] [CrossRef]
- Heong, K.L. Are planthopper problems due to breakdown in ecosystem services? In Plant Hoppers—New Threats to the Sustainability of Intensive Rice Production Systems in Asia; Heong, K.L., Hardy, B., Eds.; International Rice Research Institute: Los Banos, Philippines, 2009; pp. 221–232. [Google Scholar]
- Zhu, P.; Zheng, X.; Xu, H.; Johnson, A.C.; Heong, K.L.; Gurr, G.M.; Lu, Z. Nitrogen fertilization of rice plants improves ecological fitness of an entomophagous predator but dampens its impact on prey, the rice brown planthopper, Nilaparvata lugens. J. Pest Sci. 2020, 93, 747–755. [Google Scholar] [CrossRef]
- Wu, S.-F.; Zeng, B.; Zheng, C.; Mu, X.-C.; Zhang, Y.; Hu, J.; Zhang, S.; Gao, C.-F.; Shen, J.-L. The evolution of insecticide resistance in the brown planthopper (Nilaparvata lugens Stål) of China in the period 2012–2016. Sci. Rep. 2018, 8, 4586. [Google Scholar] [CrossRef]
- Haritha, G.; Malathi, S.; Divya, B.; Swamy, B.P.M.; Mangrauthia, S.K.; Sarla, N. Oryza nivara Sharma et Shastry. In The Wild Oryza Genomes; Mondal, T., Henry, R., Eds.; Compendium of Plant Genomes; Springer International Publishing: Cham, Switzerland, 2018; pp. 207–238. [Google Scholar] [CrossRef]
- Hu, J.; Xiao, C.; He, Y. Recent progress on the genetics and molecular breeding of brown planthopper resistance in rice. Rice 2016, 9, 30. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Xue, Y.; Zhou, H.; Li, Y.; Usman, B.; Jiao, X.; Wang, X.; Liu, F.; Qin, B.; Li, R.; et al. High-resolution mapping and breeding application of a novel brown planthopper resistance gene derived from wild rice (Oryza. rufipogon Griff). Rice 2019, 12, 41. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.; Sarao, P.S.; Bhatia, D.; Neelam, K.; Kaur, A.; Mangat, G.S.; Brar, D.S.; Singh, K. High-resolution genetic mapping of a novel brown planthopper resistance locus, Bph34 in Oryza sativa L. X Oryza nivara (Sharma & Shastry) derived interspecific F2 population. Theor. Appl. Genet. 2018, 131, 1163–1171. [Google Scholar] [CrossRef] [PubMed]
- Akanksha, S.; Lakshmi, V.J.; Singh, A.K.; Deepthi, Y.; Chirutkar, P.M.; Ramdeen; Balakrishnan, D.; Sarla, N.; Mangrauthia, S.K.; Ram, T. Genetics of novel brown planthopper Nilaparvata lugens (Stål) resistance genes in derived introgression lines from the interspecific cross O. sativa var. Swarna × O. nivara. J. Genet. 2019, 98, 113. [Google Scholar] [CrossRef]
- Heinrichs, E.A.; Medrano, F.G.; Rapusas, H.R. Genetic Evaluation for Insect Resistance in Rice; International Rice Research Institute: Manila, Philippines, 1985. [Google Scholar]
- Saghai Maroof, A.M.; Biyashav, R.M.; Yang, G.P.; Zhang, Q.; Allard, R.W. Extraordinary polymorphic microsatellite DNA in barley: Species diversity, chromosomal locations and population dynamics. Proc. Natl. Acad. Sci. USA 1994, 91, 5466–5470. [Google Scholar] [CrossRef]
- Meng, L.; Li, H.; Zhang, L.; Wang, J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015, 3, 269–283. [Google Scholar] [CrossRef]
- Sailaja, B.; Anjum, N.; Prasanth, V.V.; Sarla, N.; Subrahmanyam, D.; Voleti, S.R.; Viraktamath, B.C.; Mangrauthia, S.K. Comparative Study of Susceptible and Tolerant Genotype Reveals Efficient Recovery and Root System Contributes to Heat Stress Tolerance in Rice. Plant Mol. Biol. Rep. 2014, 32, 1228–1240. [Google Scholar] [CrossRef]
- Mangrauthia, S.K.; Bhogireddy, S.; Agarwal, S.; Prasanth, V.V.; Voleti, S.R.; Neelamraju, S.; Subrahmanyam, D. Genome-wide changes in microRNA expression during short and prolonged heat stress and recovery in contrasting rice cultivars. J. Exp. Bot. 2017, 68, 2399–2412. [Google Scholar] [CrossRef]
- Wang, X.; Duan, C.-G.; Tang, K.; Wang, B.; Zhang, H.; Lei, M.; Lu, K.; Mangrauthia, S.K.; Wang, P.; Zhu, G.; et al. RNA-binding protein regulates plant DNA methylation by controlling mRNA processing at the intronic heterochromatin-containing gene IBM1. Proc. Natl. Acad. Sci. USA 2013, 110, 15467–15472. [Google Scholar] [CrossRef]
- Fujita, D.; Kohli, A.; Horgan, F.G. Rice resistance to planthoppers and leafhoppers. Crit. Rev. Plant Sci. 2013, 32, 162–191. [Google Scholar] [CrossRef]
- Cheema, K.K.; Bains, N.S.; Mangat, G.S.; Das, A.; Vikal, Y.; Brar, D.S.; Khush, G.S.; Singh, K. Development of high yielding IR64 × Oryza rufipogon (Griff.) introgression lines and identification of introgressed alien chromosome segments using SSR markers. Euphytica 2008, 160, 401–409. [Google Scholar] [CrossRef]
- Miah, G.; Rafii, M.Y.; Ismail, M.R.; Puteh, A.B.; Rahim, H.A.; Asfaliza, R.; Latif, M.A. Blast resistance in rice: A review of conventional breeding to molecular approaches. Mol. Biol. Rep. 2013, 40, 2369–2388. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; He, W.; Nassirou, T.Y.; Zhou, W.; Yin, Y.; Dong, X.; Rao, Q.; Shi, H.; Zhao, W.; Efisue, A.; et al. Genetic diversity and phenotypic variation in an introgression line population derived from an interspecific cross between Oryza glaberrima and Oryza sativa. PLoS ONE 2016, 11, e0161746. [Google Scholar] [CrossRef]
- Qiu, Y.; Guo, J.; Jing, S.; Zhu, L.; He, G. High-resolution mapping of the brown planthopper resistance gene Bph6 in rice and characterizing its resistance in the 9311 and Nipponbare near isogenic backgrounds. Theor. Appl. Genet. 2010, 121, 1601–1611. [Google Scholar] [CrossRef]
- Qiu, Y.; Guo, J.; Jing, S.; Zhu, L.; He, G. Development and characterization of japonica rice lines carrying the brown planthopper-resistance genes BPH12 and BPH6. Theor. Appl. Genet. 2012, 124, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; You, A.; Yang, Z.; Zhang, F.; He, R.; Zhu, L.; He, G. High-resolution genetic mapping at the Bph15 locus for brown planthopper resistance in rice (Oryza sativa L.). Theor. Appl. Genet. 2004, 110, 182–191. [Google Scholar] [CrossRef]
- Sun, L.; Su, C.; Wang, C.; Zhai, H.; Wan, J. Mapping of a major resistance gene to the brown planthopper in the rice cultivar Rathu Heenati. Breed. Sci. 2005, 55, 391–396. [Google Scholar] [CrossRef]
- Rahman, L.; Jiang, W.; Chu, S.H.; Qiao, Y.; Ham, T.-H.; Woo, M.-O.; Lee, J.; Khanam, M.S.; Chin, J.-H.; Jeung, J.-U.; et al. High-resolution mapping of two rice brown planthopper resistance genes, Bph20(t) and Bph21(t), originating from Oryza minuta. Theor. Appl. Genet. 2009, 119, 1237–1246. [Google Scholar] [CrossRef]
- Huang, D.; Qiu, Y.; Zhang, Y.; Huang, F.; Meng, J.; Wei, S.; Li, R.; Chen, B. Fine mapping and characterization of BPH27, a brown planthopper resistance gene from wild rice (Oryza rufipogon Griff.). Theor. Appl. Genet. 2013, 126, 219–229. [Google Scholar] [CrossRef]
- Hu, J.; Chang, X.; Zou, L.; Tang, W.; Wu, W. Identification and fine mapping of Bph33, a new brown planthopper resistance gene in rice (Oryza sativa L.). Rice 2018, 11, 55. [Google Scholar] [CrossRef] [PubMed]
- Naik, S.B.; Divya, D.; Sahu, N.; Sundaram, R.M.; Sarao, P.S.; Singh, K.; Lakshmi, V.J.; Bentur, J.S. A new gene Bph33(t) conferring resistance to brown planthopper (BPH), Nilaparvata lugens (Stål) in rice line RP2068-18-3-5. Euphytica 2018, 214, 53. [Google Scholar] [CrossRef]
- Akanksha, I.; Lakshmi, V.J.; Chirutkar, P.; Rao, L.S.; Sarla, N.; Ram, T. Identification of new sources for brown planthopper resistance from rice germplasm and introgression lines derived from O. nivara. Oryza-Int. J. Rice 2017, 54, 272. [Google Scholar] [CrossRef]
- Jairin, J.; Phengrat, K.; Teangdeerith, S.; Vanavichit, A.; Toojinda, T. Mapping of a broad-spectrum brown planthopper resistance gene, Bph3, on rice chromosome 6. Mol. Breed. 2007, 19, 35–44. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, H.; Chen, H.; Liu, Y.; He, J.; Kang, H.; Sun, Z.; Pan, G.; Wang, Q.; Hu, J.; et al. A gene cluster encoding lectin receptor kinases confers broad-spectrum and durable insect resistance in rice. Nat. Biotechnol. 2015, 33, 301–305. [Google Scholar] [CrossRef]
- Rajkumar, G.; Weerasena, J.; Fernando, K.; Liyanage, A.; Silva, R. Genetic differentiation among Sri Lankan traditional rice (Oryza sativa) varieties and wild rice species by AFLP markers. Nord. J. Bot. 2011, 29, 238–243. [Google Scholar] [CrossRef]
- Sarao, P.S.; Sahi, G.K.; Neelam, K.; Mangat, G.S.; Patra, B.C.; Singh, K. Donors for resistance to brown planthopper Nilaparvata lugens (Stål) from wild rice species. Rice Sci. 2016, 23, 219–224. [Google Scholar] [CrossRef]
- Hu, J.; Zhou, J.; Peng, X.; Xu, H.; Liu, C.; Du, B.; Yuan, H.; Zhu, L.; He, G. The Bphi008a Gene Interacts with the Ethylene Pathway and Transcriptionally Regulates MAPK Genes in the Response of Rice to Brown Planthopper Feeding. Plant Physiol. 2011, 156, 856–872. [Google Scholar] [CrossRef]
- Lorenzo, O.; Piqueras, R.; Sánchez-Serrano, J.J.; Solano, R. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 2003, 15, 165–178. [Google Scholar] [CrossRef]
- Gao, M.; Yin, X.; Yang, W.; Lam, S.M.; Tong, X.; Liu, J.; Wang, X.; Li, Q.; Shui, G.; He, Z. GDSL lipases modulate immunity through lipid homeostasis in rice. PLoS Pathog. 2017, 13, e1006724. [Google Scholar] [CrossRef]
- Truong, D.-H.; Nguyen, H.C.; Bauwens, J.; Mazzucchelli, G.; Lognay, G.; Francis, F. Plant defense in response to chewing insects: Proteome analysis of Arabidopsis thaliana damaged by Plutella xylostella. J. Plant Interact. 2018, 13, 30–36. [Google Scholar] [CrossRef]
- Song, W.-Y.; Wang, G.-L.; Chen, L.-L.; Kim, H.-S.; Pi, L.-Y.; Holsten, T.; Gardner, J.; Wang, B.; Zhai, W.-X.; Zhu, L.-H.; et al. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 1995, 270, 1804–1806. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Loh, Y.-T.; Bressan, R.A.; Martin, G.B. The tomato gene Pti1 encodes a serine/threonine kinase that is phosphorylated by Pto and is involved in the hypersensitive response. Cell 1995, 83, 925–935. [Google Scholar] [CrossRef] [PubMed]
- Afzal, A.J.; Wood, A.J.; Lightfoot, D.A. Plant receptor-like serine threonine kinases: Roles in signaling and plant defense. Mol. Plant Microbe Interact. 2008, 21, 507–517. [Google Scholar] [CrossRef]
- Lin, Z.-J.D.; Liebrand, T.W.; Yadeta, K.A.; Coaker, G.L. PBL13 is a serine/threonine protein kinase that negatively regulates Arabidopsis immune responses. Plant Physiol. 2015, 169, 2950–2962. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, S. Dof1 and Dof2 transcription factors are associated with expression of multiple genes involved in carbon metabolism in maize. Plant J. 2000, 21, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Simpson, S.D.; Nakashima, K.; Narusaka, Y.; Seki, M.; Shinozaki, K.; Kazuko, Y.S. Two different novel cis-acting elements of erd1, a clpA homologous Arabidopsis gene function in induction by dehydration stress and dark-induced senescence. Plant J. 2003, 33, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Després, C.; Chubak, C.; Rochon, A.; Clark, R.; Bethune, T.; Desveaux, D.; Fobert, P.R. The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 2003, 15, 2181–2191. [Google Scholar] [CrossRef] [PubMed]
- Redman, J.; Whitcraft, J.; Johnson, C.; Arias, J. Abiotic and biotic stress differentially stimulate as-1 element activity in Arabidopsis. Plant Cell Rep. 2002, 21, 180–185. [Google Scholar] [CrossRef]
- Eulgem, T.; Paul, J.R.; Elmon, S.; Klaus, H.; Imre, S.E. Early nuclear events in plant defence signalling: Rapid gene activation by WRKY transcription factors. EMBO J. 1999, 18, 4689–4699. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.-L.; Xie, Z.; Zou, X.L.; Casaretto, J.; Ho, T.-H.D.; Shen, Q.J. A Rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol. 2004, 134, 1500–1513. [Google Scholar] [CrossRef]
- Gowik, U.; Janet, B.; Meryem, A.; Schlue, U.; Koczor, M.; Streubel, M.; Westhoff, P. cis-Regulatory elements for mesophyll-specific gene expression in the C4 plant Flaveria trinervia, the promoter of the C4 phosphoenolpyruvate carboxylase gene. Plant Cell 2004, 16, 1077–1090. [Google Scholar] [CrossRef]
- AFilichkin, S.; Jeffrey, M.L.; Monteros, A.; Liu, P.P.; Hiroyuki, N. A novel endo-β-mannanase gene in tomato LeMAN5 is associated with anther and pollen development. Plant Physiol. 2004, 134, 1080–1087. [Google Scholar] [CrossRef]
- Hartmann, U.; Martin, S.; Frank, M.; Ralf, S.; Weisshaar, B. Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol. Biol. 2005, 57, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Takeshi, U.; Takuya, I.; Motoaki, S.; Kazuo, S.; Kazuko, Y.S. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Masaru, O.; Siddhartha, K.; Byeong-ha, L.; Xuhui, H.; Manu, A.; Zhu, J.K. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003, 17, 1043–1054. [Google Scholar] [CrossRef]
- Foster, R.; Izawa, T.; Chua, N. Plant bZIP proteins gather at ACGT elements. FASEB J. 1994, 8, 192–200. [Google Scholar] [CrossRef]
- Izawa, T.; Foster, R.; Nakajima, M.; Shimamoto, K.; Chua, N.H. The rice bZIP transcriptional activator RITA-1 is highly expressed during seed development. Plant Cell 1994, 6, 1277–1287. [Google Scholar]
- Ross, E.J.H.; Stone, J.M.; Elowsky, C.G.; Arredondo-Peter, R.; Klucas, R.V.; Sarath, G. Activation of the Oryza sativa non-symbiotic haemoglobin-2 promoter by the cytokinin-regulated transcription factor, ARR1. J. Exp. Bot. 2004, 55, 1721–1731. [Google Scholar] [CrossRef] [PubMed]
- Stougaard, J.; Jørgensen, J.-E.; Christensen, T.; Kühle, A.; Marcker, K.A. Interdependence and nodule specificity of cis-acting regulatory elements in the soybean leghemoglobin lbc 3 and N23 gene promoters. Mol. Genet. Genom. 1990, 220, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Fehlberg, V.; Vieweg, M.F.; Dohmann, E.M.N.; Hohnjec, N.; Pühler, A.; Perlick, A.M.; Küster, H. The promoter of the leghaemoglobin gene VfLb29: Functional analysis and identification of modules necessary for its activation in the infected cells of root nodules and in the arbuscule-containing cells of mycorrhizal roots. J. Exp. Bot. 2005, 56, 799–806. [Google Scholar] [CrossRef]
- Hwang, Y.-S.; Karrer, E.; Thomas, B.; Chen, L.; Rodriguez, R. Three cis-elements required for rice α-amylase Amy3D expression during sugar starvation. Plant Mol. Biol. 1998, 36, 331–341. [Google Scholar] [CrossRef]
- Kim, D.W.; Lee, S.H.; Choi, S.B.; Won, S.K.; Heo, Y.K.; Cho, M.; Park, Y.; Cho, H.T. Functional conservation of a root hair cell-specific cis-element in angiosperms with different root hair distribution patterns. Plant Cell 2006, 18, 2958–2970. [Google Scholar] [CrossRef]
- Chaubet, N.; Martine, F.; Bernadette, C.; Pierre, B.; Claude, G. Identification of cis-elements regulating the expression of an Arabidopsis histone H4 gene. Plant J. 1996, 10, 425–435. [Google Scholar] [CrossRef]
- Jiang, C.; Iu, B.; Singh, J. Requirement of a CCGAC cis-acting element for cold induction of the BN115 gene from winter Brassica napus. Plant Mol. Biol. 1996, 30, 679–684. [Google Scholar] [CrossRef]
- Dubouzet, J.G.; Sakuma, Y.; Ito, Y.; Kasuga, M.; Dubouzet, E.G.; Miura, S.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 2003, 33, 751–763. [Google Scholar] [CrossRef] [PubMed]
- Xue, G.-P. An AP2 domain transcription factor HvCBF1 activates expression of cold-responsive genes in barley through interaction with a (G/a)(C/t)CGAC motif. Biochim. Biophys. Acta (BBA)-Gene Struct. Expr. 2002, 1577, 63–72. [Google Scholar] [CrossRef]
- Yamauchi, D. ATGACGT motif in the 5′-upstream region of α-amylase gene from Vigna mungo is a cis-element for expression in cotyledons of germinated seeds. Plant. Cell Physiol. 2001, 42, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, H.; Sato, K.; Berberich, T.; Miyazaki, A.; Ozaki, R.; Imai, R.; Kusano, T. LIP19, a basic region leucine zipper protein, is a Fos-like molecular switch in the cold signaling of rice plants. Plant Cell Physiol. 2005, 46, 1623–1634. [Google Scholar] [CrossRef]
- Martínez, M.I.; Chrispeels, M.J. Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 2003, 15, 561–576. [Google Scholar] [CrossRef]
- Brown, R.L.; Kemal, K.; Ken, C.M.; Don, J.M.; John, M.M. A role for the GCC-box in jasmonate-mediated activation of the PDF1. 2 gene of Arabidopsis. Plant Physiol. 2003, 132, 1020–1032. [Google Scholar] [CrossRef] [PubMed]
- Chakravarthy, S.; Robert, P.T.; Mark, A.; Pierre, R.F.; Charles, D.; Gregory, B.M. The tomato transcription factor Pti4 regulates defense-related gene expression via GCC box and non-GCC box cis elements. Plant Cell 2003, 15, 3033–3050. [Google Scholar] [CrossRef] [PubMed]
- von Gromoff, E.D.; Michael, S.; Ulrike, O.; Christoph, F.B. Identification of a plastid response element that acts as an enhancer within the Chlamydomonas HSP70A promoter. Nucleic Acids Res. 2006, 34, 4767–4779. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.; Lankai, G.; Ming-Che, S. Promoter analysis of the nuclear gene encoding the chloroplast glyceraldehyde-3-phosphate dehydrogenase B subunit of Arabidopsis thaliana. Plant Mol. Biol. 2001, 46, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Xiu-Ling, C.; Zong-Yang, W.; Meng-Min, H. An interaction between a MYC protein and an EREBP protein is involved in transcriptional regulation of the rice Wx gene. J. Biol. Chem. 2003, 278, 47803–47811. [Google Scholar] [CrossRef] [PubMed]
- Boter, M.; Ruíz-Rivero, O.; Abdeen, A.; Salomé, P. Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 2004, 18, 1577–1591. [Google Scholar] [CrossRef] [PubMed]
- Rogers, H.J.; Bate, N.; Combe, J.; Sullivan, J.; Sweetman, J.; Swan, C.; Lonsdale, D.M.; Twell, D. Functional analysis of cis-regulatory elements within the promoter of the tobacco late pollen gene g10. Plant Mol. Biol. 2001, 45, 577–585. [Google Scholar] [CrossRef]
- Ken-ichi, K.; Satoshi, Y.; Fumiya, T.; Ohyama, K.; Fukuzawa, H. Cis-acting elements and DNA-binding proteins involved in CO2-responsive transcriptional activation of Cah1 encoding a periplasmic carbonic anhydrase in Chlamydomonas reinhardtii. Plant Physiol. 2003, 133, 783–793. [Google Scholar]
- Kosugi, S.; Ohashi, Y. E2F sites that can interact with E2F proteins cloned from rice are required for meristematic tissue-specific expression of rice and tobacco proliferating cell nuclear antigen promoters. Plant J. 2002, 29, 45–59. [Google Scholar] [CrossRef] [PubMed]
- Sozzani, R.; Maggio, C.; Varotto, S.; Canova, S.; Bergounioux, C.; Albani, D.; Cella, R. Interplay between Arabidopsis activating factors E2Fb and E2Fa in cell cycle progression and development. Plant Physiol. 2006, 140, 1355–1366. [Google Scholar] [CrossRef]
- Vandepoele, K.; Kobe, V.; Kobe, F.; Hennig, L.; Gerrit, T.S.B.; Gruissem, W.; de Peer, Y.V.; Inzé, D.; De Veylder, L. Genome-wide identification of potential plant E2F target genes. Plant Physiol. 2005, 139, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Lacombe, E.; Doorsselaere, J.V.; Wout, B.; Boudet, A.M.; Pettenati, J.G. Characterization of cis-elements required for vascular expression of the Cinnamoyl CoA Reductase gene and for protein–DNA complex formation. Plant J. 2000, 23, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Planchais, S.; Perennes, C.; Glab, N.; Mironov, V.; Inzé, D.; Bergounioux, C. Characterization of cis-acting element involved in cell cycle phase-independent activation of Arath; CycB1; 1 transcription and identification of putative regulatory proteins. Plant Mol. Biol. 2002, 50, 109–125. [Google Scholar] [CrossRef]
- Yang, T.; Poovaiah, B.W. A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. J. Biol. Chem. 2002, 277, 45049–45058. [Google Scholar] [CrossRef] [PubMed]
- Ellerström, M.; Stålberg, K.; Inés, E.; Rask, L. Functional dissection of a napin gene promoter: Identification of promoter elements required for embryo and endosperm-specific transcription. Plant Mol. Biol. 1996, 32, 1019–1027. [Google Scholar] [CrossRef]
- Svensson, J.; Ismail, A.M.; Palva, E.T.; Close, T.J. Dehydrins. In Sensing Signaling and Cell Adaptation; Storey, K.B., Storey, J.M., Eds.; Elsevier: Amsterdam, The Netherlands, 2002; pp. 155–171. [Google Scholar]
- Baker, S.S.; Kathy, S.W.; Thomashow, F.M. The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought-and ABA-regulated gene expression. Plant Mol. Biol. 1994, 24, 701–713. [Google Scholar] [CrossRef]
- Kropat, J.; Tottey, S.; Birkenbihl, R.P.; Nathalie, D.; Huijser, P.; Merchant, S. A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc. Natl. Acad. Sci. USA 2005, 102, 18730–18735. [Google Scholar] [CrossRef] [PubMed]
- Urao, T.; Yamaguchi-Shinozaki, K.; Urao, S.; Shinozaki, K. An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 1993, 5, 1529–1539. [Google Scholar] [PubMed]
- Solano, R.; Nieto, C.; Avila, J.; Canas, L.; Diaz, I.; Paz-Ares, J. Dual DNA binding specificity of a petal epidermis-specific MYB transcription factor (MYB. Ph3) from Petunia hybrida. EMBO J. 1995, 14, 1773–1784. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.K.; Agarwal, P.; Reddy, M.K.; Sopory, S.K. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 2006, 25, 1263–1274. [Google Scholar] [CrossRef] [PubMed]




| Gene Name | Forward Primer and Reverse Primers | Size |
|---|---|---|
| OsRLK-1 | GACCCAAGCATGCGACCTAA | |
| AGAAGCGTATGGAAGTGAGC | 114 bp | |
| OsRLK-2 | GCTCCTCTTTGACACCCCAG | |
| CAGCGAGCAGGTATGAGGAG | 111 bp | |
| OsRLK-3 | GGCTGTGTGTGACTGATTGT | |
| TGTCCTCGGTACATTGACATCC | 119 bp | |
| STPKR | TGGCTCACGAGCTAATAACAA | |
| TGATTGCCTCTTGATTTGCGTA | 120 bp | |
| Xa21 | CGCGCGTCTAGGTGATTTTG | |
| CATTCCATACTCTGGTGCAATGT | 115 bp | |
| LRR | GCGCTTCTTGGTGACTTTGG | |
| CCCATTCCATACTCTGGTGCAAT | 116 bp | |
| RK | GTACCAAGGCAGCCCAAAGA | |
| CACAGTGGATCAGAGGAGGT | 119 bp | |
| AP2/ERF | GAGGACTACAGCTACCGCAA | |
| TAGCAATCCGGGTCTCGTTT | 115 bp | |
| F.BOX | GGAAACGTCTCGGGACAGAA | |
| GGTCGTAAGGCGGGTAGAAC | 115 bp | |
| LGDSL | CGGGATCACTGCCAGAAAAA | |
| GCACGGATTTGGAAGGGCT | 119 bp |
| S. No | Nucleotide Change in RP bio4918-230S | Position of the Nucleotide Change | Type of Change | Exon Number |
|---|---|---|---|---|
| 1 | G→A | 994 bp | Transition | 1 |
| 2 | A→C | 1069 bp | Transversion | 1 |
| 3 | A→C | 1092 bp | Transversion | 1 |
| 4 | C→T | 1103 bp | Transition | 1 |
| 5 | G→C | 1359 bp | Transversion | 2 |
| 6 | G→A | 1384 bp | Transition | 2 |
| 7 | T→C | 1407 bp | Transition | 2 |
| 8 | A→T | 1429 bp | Transversion | 2 |
| 9 | G→A | 1561 bp | Transition | 3 |
| 10 | A→G | 2350 bp | Transition | 5 |
| 11 | A→T | 2353 bp | Transversion | 5 |
| 12 | T→C | 2480 bp | Transition | 6 |
| 13 | C→G | 2512 bp | Transversion | 6 |
| 14 | T→C | 2535 bp | Transition | 6 |
| 15 | T→A | 2539 bp | Transversion | 6 |
| 16 | T→G | 2558 bp | Transversion | 6 |
| 17 | G→C | 2575 bp | Transversion | 6 |
| 18 | C→A | 2617 bp | Transversion | 6 |
| 19 | T→G | 2762 bp | Transversion | 7 |
| 20 | T→A | 2763 bp | Transversion | 7 |
| 21 | A→C | 2835 bp | Transversion | 7 |
| 22 | T→C | 2880 bp | Transition | 7 |
| 23 | G→C | 2900 bp | Transversion | 7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Srivastava, A.; Pusuluri, M.; Balakrishnan, D.; Vattikuti, J.L.; Neelamraju, S.; Sundaram, R.M.; Mangrauthia, S.K.; Ram, T. Identification and Functional Characterization of Two Major Loci Associated with Resistance against Brown Planthoppers (Nilaparvata lugens (Stål)) Derived from Oryza nivara. Genes 2023, 14, 2066. https://doi.org/10.3390/genes14112066
Srivastava A, Pusuluri M, Balakrishnan D, Vattikuti JL, Neelamraju S, Sundaram RM, Mangrauthia SK, Ram T. Identification and Functional Characterization of Two Major Loci Associated with Resistance against Brown Planthoppers (Nilaparvata lugens (Stål)) Derived from Oryza nivara. Genes. 2023; 14(11):2066. https://doi.org/10.3390/genes14112066
Chicago/Turabian StyleSrivastava, Akanksha, Madhu Pusuluri, Divya Balakrishnan, Jhansi Lakshmi Vattikuti, Sarla Neelamraju, Raman Meenakshi Sundaram, Satendra Kumar Mangrauthia, and Tilathoo Ram. 2023. "Identification and Functional Characterization of Two Major Loci Associated with Resistance against Brown Planthoppers (Nilaparvata lugens (Stål)) Derived from Oryza nivara" Genes 14, no. 11: 2066. https://doi.org/10.3390/genes14112066





