C8ORF88: A Novel eIF4E-Binding Protein
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequence Alignments
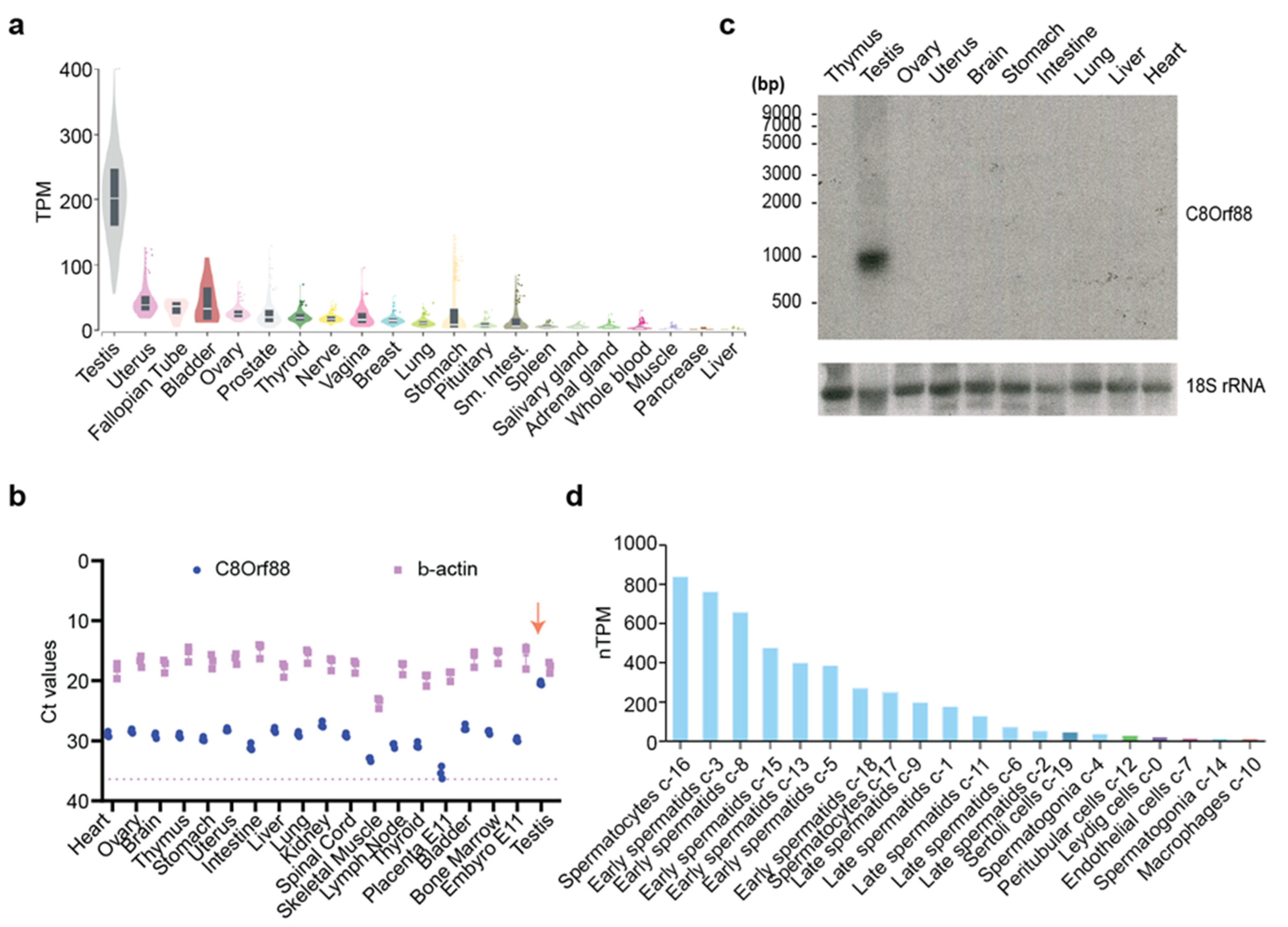
2.2. Northern Blot Analysis
2.3. RT-qPCR
- mC8Orf88_qPCR_For, 5′TGGGTCTTGAGGCGTATG3′
- mC8Orf88_qPCR_Rev, 5′ATCTGCCCACTCCACTTTGT3′
- β-actin_qPCR_For, 5′TTCCTTCTTGGGTATGGAATCC3′
- β-actin_qPCR_Rev, 5′AGGAGCAATGATCTTGATCTTC3′
2.4. Plasmids and Recombinant Proteins
2.5. Cell Culture, Transfections and Lysate Preparation
2.6. Western Blot Analysis
2.7. In Vitro Translation and m7GTP Cap-Affinity Chromatography
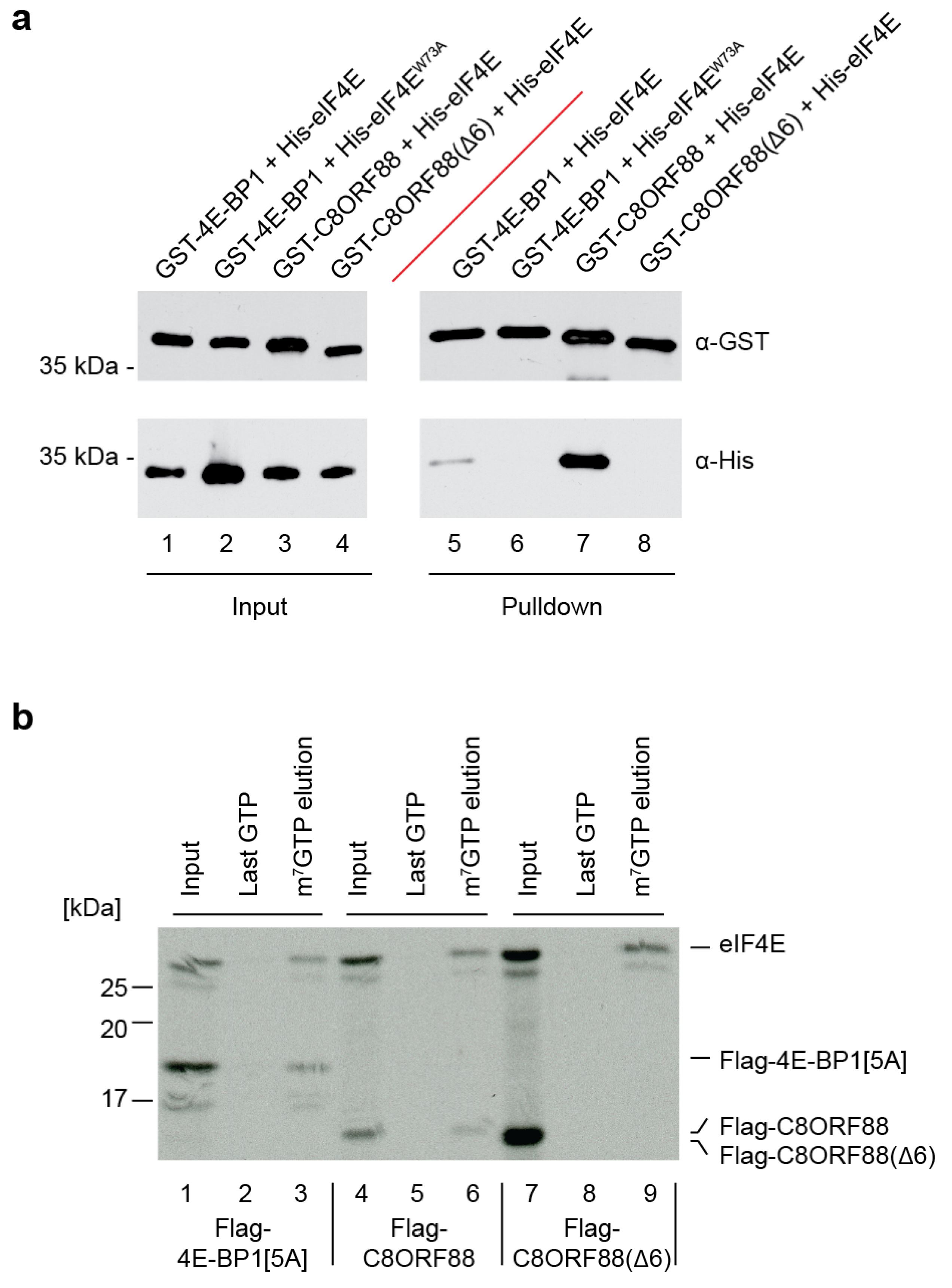
2.8. GST Pulldown
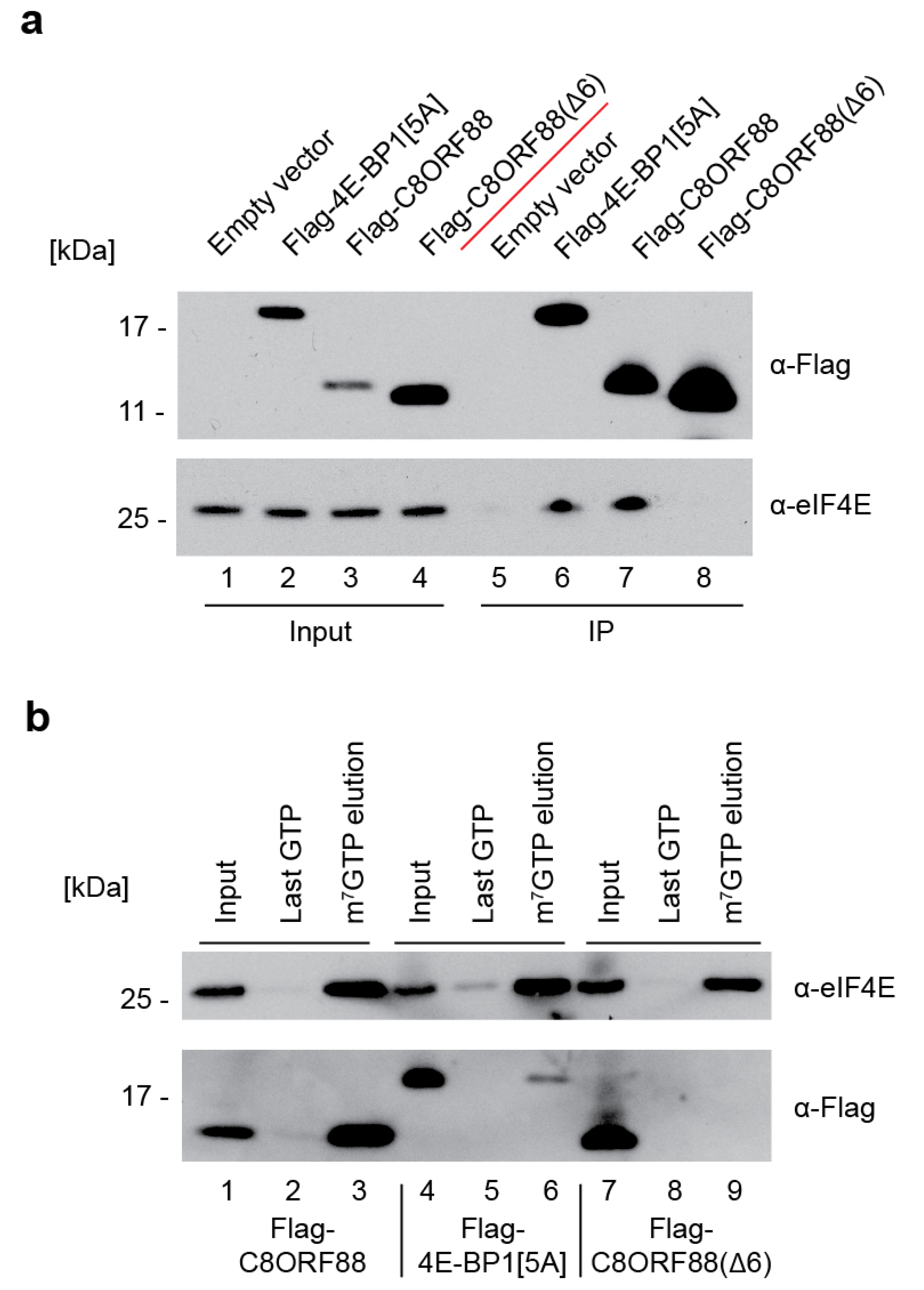
2.9. Co-Immunoprecipitation and m7GTP Cap-Affinity Chromatography
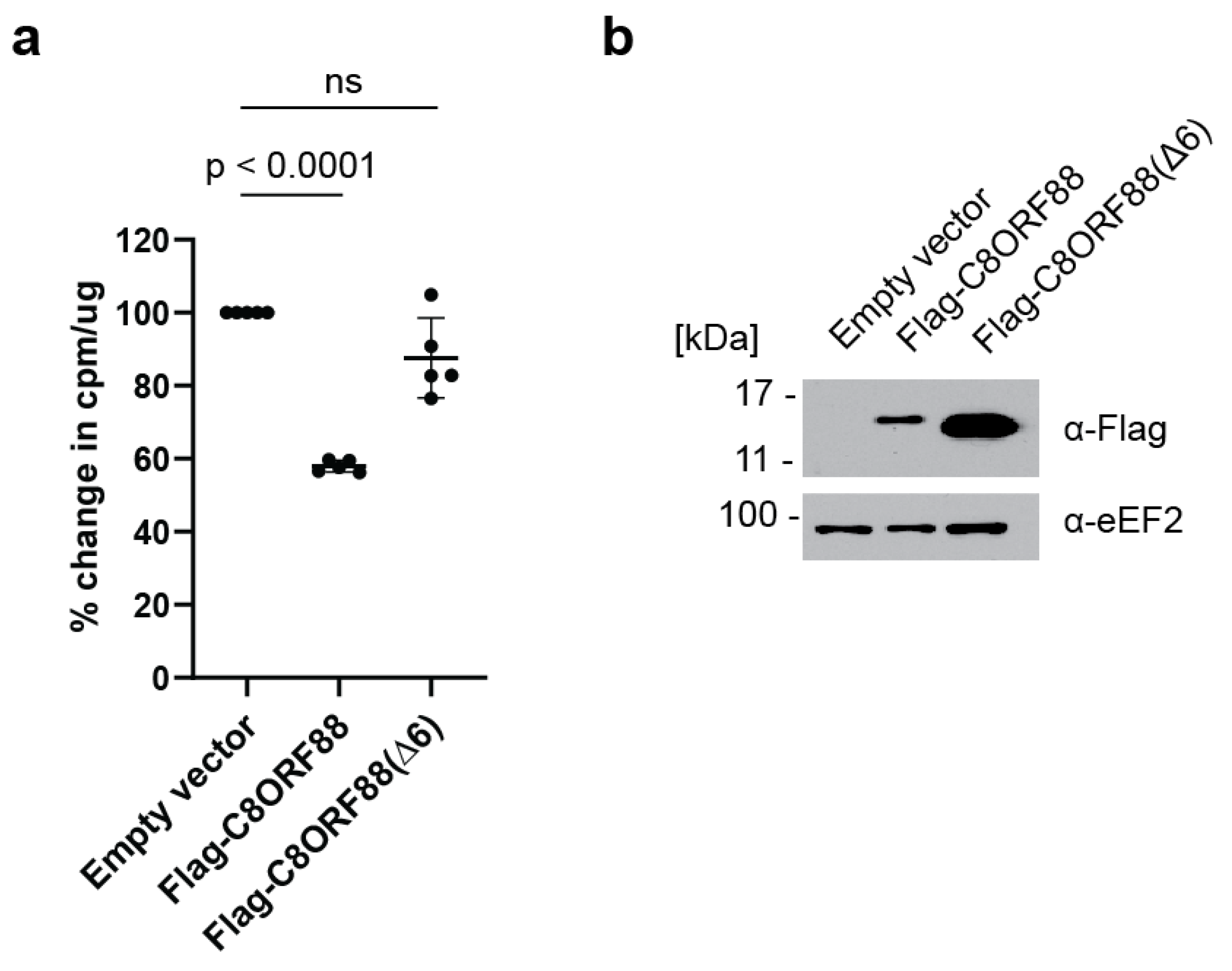
2.10. L-[35S]-Methionine/Cysteine Labelled Protein Incorporation in Cells
3. Results
3.1. Sequence Alignment Reveals Homology between C8ORF88 and the 4E-BP Homologs
3.2. Expression Profile of C8Orf88
3.3. In Vitro Association between C8ORF88 and eIF4E
3.4. C8ORF88 Interacts with Endogenous eIF4E in a Cultured Cell Line
3.5. Ectopic Expression of C8ORF88 Inhibits Translation in a Cultured Cell Line
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rhoads, R.E.; Joshi-Barve, S.; Rinker-Schaeffer, C. Mechanism of action and regulation of protein synthesis initiation factor 4E: Effects on mRNA discrimination, cellular growth rate, and oncogenesis. Prog. Nucleic Acid Res. Mol. Biol. 1993, 46, 183–219. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, J.; Schmeing, T.M.; Sonenberg, N. The multifaceted eukaryotic cap structure. Wiley Interdiscip. Rev. RNA 2021, 12, e1636. [Google Scholar] [CrossRef] [PubMed]
- Shatkin, A.J. Capping of eucaryotic mRNAs. Cell 1976, 9, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, J.; Sonenberg, N. The Organizing Principles of Eukaryotic Ribosome Recruitment. Annu. Rev. Biochem. 2019, 88, 307–335. [Google Scholar] [CrossRef] [PubMed]
- Kulak, N.A.; Pichler, G.; Paron, I.; Nagaraj, N.; Mann, M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 2014, 11, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Hiremath, L.S.; Webb, N.R.; Rhoads, R.E. Immunological detection of the messenger RNA cap-binding protein. J. Biol. Chem. 1985, 260, 7843–7849. [Google Scholar] [CrossRef] [PubMed]
- Duncan, R.; Milburn, S.C.; Hershey, J.W. Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control. Heat shock effects on eIF-4F. J. Biol. Chem. 1987, 262, 380–388. [Google Scholar] [CrossRef]
- Goss, D.J.; Carberry, S.E.; Dever, T.E.; Merrick, W.C.; Rhoads, R.E. Fluorescence study of the binding of m7GpppG and rabbit globin mRNA to protein synthesis initiation factors 4A, 4E, and 4F. Biochemistry 1990, 29, 5008–5012. [Google Scholar] [CrossRef]
- Altmann, M.; Schmitz, N.; Berset, C.; Trachsel, H. A novel inhibitor of cap-dependent translation initiation in yeast: p20 competes with eIF4G for binding to eIF4E. EMBO J. 1997, 16, 1114–1121. [Google Scholar] [CrossRef]
- Mendelsohn, B.A.; Li, A.-M.; Vargas, C.A.; Riehman, K.; Watson, A.; Fridovich-Keil, J.L. Genetic and biochemical interactions between SCP160 and EAP1 in yeast. Nucleic Acids Res. 2003, 31, 5838–5847. [Google Scholar] [CrossRef]
- Siddiqui, N.; Sonenberg, N. Signalling to eIF4E in cancer. Biochem. Soc. Trans. 2015, 43, 763–772. [Google Scholar] [CrossRef] [PubMed]
- Laplante, M.; Sabatini, D.M. mTOR Signaling in Growth Control and Disease. Cell 2012, 149, 274–293. [Google Scholar] [CrossRef] [PubMed]
- Bar-Peled, L.; Sabatini, D.M. Regulation of mTORC1 by amino acids. Trends Cell Biol. 2014, 24, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef] [PubMed]
- Gingras, A.-C.; Gygi, S.P.; Raught, B.; Polakiewicz, R.D.; Abraham, R.T.; Hoekstra, M.F.; Aebersold, R.; Sonenberg, N. Regulation of 4E-BP1 phosphorylation: A novel two-step mechanism. Genes Dev. 1999, 13, 1422–1437. [Google Scholar] [CrossRef] [PubMed]
- Marcotrigiano, J.; Gingras, A.-C.; Sonenberg, N.; Burley, S.K. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell 1999, 3, 707–716. [Google Scholar] [CrossRef] [PubMed]
- Peter, D.; Igreja, C.; Weber, R.; Wohlbold, L.; Weiler, C.; Ebertsch, L.; Weichenrieder, O.; Izaurralde, E. Molecular architecture of 4E-BP translational inhibitors bound to eIF4E. Mol. Cell 2015, 57, 1074–1087. [Google Scholar] [CrossRef]
- Igreja, C.; Peter, D.; Weiler, C.; Izaurralde, E. 4E-BPs require non-canonical 4E-binding motifs and a lateral surface of eIF4E to repress translation. Nat. Commun. 2014, 5, 4790. [Google Scholar] [CrossRef]
- Kinkelin, K.; Veith, K.; Grünwald, M.; Bono, F. Crystal structure of a minimal eIF4E–Cup complex reveals a general mechanism of eIF4E regulation in translational repression. RNA 2012, 18, 1624–1634. [Google Scholar] [CrossRef]
- Grüner, S.; Peter, D.; Weber, R.; Wohlbold, L.; Chung, M.-Y.; Weichenrieder, O.; Valkov, E.; Igreja, C.; Izaurralde, E. The Structures of eIF4E-eIF4G Complexes Reveal an Extended Interface to Regulate Translation Initiation. Mol. Cell 2016, 64, 467–479. [Google Scholar] [CrossRef]
- Dong, Y.; Srour, O.; Lukhovitskaya, N.; Makarian, J.; Baumberger, N.; Galzitskaya, O.; Elser, D.; Schepetilnikov, M.; Ryabova, L.A. Functional analogs of mammalian 4E-BPs reveal a role for TOR in global plant translation. Cell Rep. 2023, 42, 112892. [Google Scholar] [CrossRef] [PubMed]
- Musa, J.; Orth, M.F.; Dallmayer, M.; Baldauf, M.; Pardo, C.; Rotblat, B.; Kirchner, T.; Leprivier, G.; Grünewald, T.G.P. Eukaryotic initiation factor 4E-binding protein 1 (4E-BP1): A master regulator of mRNA translation involved in tumorigenesis. Oncogene 2016, 35, 4675–4688. [Google Scholar] [CrossRef] [PubMed]
- Braunstein, S.; Karpisheva, K.; Pola, C.; Goldberg, J.; Hochman, T.; Yee, H.; Cangiarella, J.; Arju, R.; Formenti, S.C.; Schneider, R.J. A hypoxia-controlled cap-dependent to cap-independent translation switch in breast cancer. Mol. Cell 2007, 28, 501–512. [Google Scholar] [CrossRef] [PubMed]
- Petroulakis, E.; Parsyan, A.; Dowling, R.J.; LeBacquer, O.; Martineau, Y.; Bidinosti, M.; Larsson, O.; Alain, T.; Rong, L.; Mamane, Y.; et al. p53-dependent translational control of senescence and transformation via 4E-BPs. Cancer Cell 2009, 16, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, E.; Pérez-Tenorio, G.; Amin, R.; Bostner, J.; Skoog, L.; Fornander, T.; Sgroi, D.C.; Nordenskjöld, B.; Hallbeck, A.-L.; Stål, O. The mTOR effectors 4EBP1 and S6K2 are frequently coexpressed, and associated with a poor prognosis and endocrine resistance in breast cancer: A retrospective study including patients from the randomised Stockholm tamoxifen trials. Breast Cancer Res. 2013, 15, R96. [Google Scholar] [CrossRef] [PubMed]
- Rutkovsky, A.C.; Yeh, E.S.; Guest, S.T.; Findlay, V.J.; Muise-Helmericks, R.C.; Armeson, K.; Ethier, S.P. Eukaryotic initiation factor 4E-binding protein as an oncogene in breast cancer. BMC Cancer 2019, 19, 491. [Google Scholar] [CrossRef] [PubMed]
- Maracci, C.; Motta, S.; Romagnoli, A.; Costantino, M.; Perego, P.; Di Marino, D. The mTOR/4E-BP1/eIF4E Signalling Pathway as a Source of Cancer Drug Targets. Curr. Med. Chem. 2022, 29, 3501–3529. [Google Scholar] [CrossRef]
- Gkogkas, C.G.; Khoutorsky, A.; Ran, I.; Rampakakis, E.; Nevarko, T.; Weatherill, D.B.; Vasuta, C.; Yee, S.; Truitt, M.; Dallaire, P.; et al. Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature 2013, 493, 371–377. [Google Scholar] [CrossRef]
- Tsukumo, Y.; Alain, T.; Fonseca, B.D.; Nadon, R.; Sonenberg, N. Translation control during prolonged mTORC1 inhibition mediated by 4E-BP3. Nat. Commun. 2016, 7, 11776. [Google Scholar] [CrossRef]
- Martina, J.A.; Diab, H.I.; Li, L.; Lim, J.-A.; Patange, S.; Raben, N.; Puertollano, R. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci. Signal. 2014, 7, ra9. [Google Scholar] [CrossRef]
- Roczniak-Ferguson, A.; Petit, C.S.; Froehlich, F.; Qian, S.; Ky, J.; Angarola, B.; Walther, T.C.; Ferguson, S.M. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 2012, 5, ra42. [Google Scholar] [CrossRef] [PubMed]
- Roux, K.J.; Kim, D.I.; Burke, B.; May, D.G. BioID: A Screen for Protein-Protein Interactions. Curr. Protoc. Protein Sci. 2018, 91, 19.23.1–19.23.15. [Google Scholar] [CrossRef] [PubMed]
- Joshi, B.; Cameron, A.; Jagus, R. Characterization of mammalian eIF4E-family members. Eur. J. Biochem. 2004, 271, 2189–2203. [Google Scholar] [CrossRef] [PubMed]
- Chapat, C.; Jafarnejad, S.M.; Matta-Camacho, E.; Hesketh, G.G.; Gelbart, I.A.; Attig, J.; Gkogkas, C.G.; Alain, T.; Stern-Ginossar, N.; Fabian, M.R.; et al. Cap-binding protein 4EHP effects translation silencing by microRNAs. Proc. Natl. Acad. Sci. USA 2017, 114, 5425–5430. [Google Scholar] [CrossRef] [PubMed]
- Christie, M.; Igreja, C. eIF4E-homologous protein (4EHP): A multifarious cap-binding protein. FEBS J. 2021, 290, 266–285. [Google Scholar] [CrossRef] [PubMed]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Higgins, D.G. Clustal omega. Curr. Protoc. Bioinform. 2014, 48, 3.13.1–3.13.16. [Google Scholar] [CrossRef] [PubMed]
- Rebay, I.; Fehon, R.G. Preparation of insoluble GST fusion proteins. Cold Spring Harb. Protoc. 2009, 2009, pdb.prot4997. [Google Scholar] [CrossRef]
- Boussif, O.; Lezoualc’h, F.; Zanta, M.A.; Mergny, M.D.; Scherman, D.; Demeneix, B.; Behr, J.P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine. Proc. Natl. Acad. Sci. USA 1995, 92, 7297–7301. [Google Scholar] [CrossRef]
- Kingston, R.E.; Chen, C.A.; Rose, J.K. Calcium phosphate transfection. Curr. Protoc. Mol. Biol. 2003, 63, 9.1.1–9.1.11. [Google Scholar] [CrossRef]
- Stepinski, J.; Waddell, C.; Stolarski, R.; Darzynkiewicz, E.; Rhoads, R.E. Synthesis and properties of mRNAs containing the novel “anti-reverse” cap analogs 7-methyl(3′-O-methyl)GpppG and 7-methyl (3′-deoxy)GpppG. RNA 2001, 7, 1486–1495. [Google Scholar] [PubMed]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Pyronnet, S.; Imataka, H.; Gingras, A.C.; Fukunaga, R.; Hunter, T.; Sonenberg, N. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J. 1999, 18, 270–279. [Google Scholar] [CrossRef] [PubMed]
- Rapley, J.; Oshiro, N.; Ortiz-Vega, S.; Avruch, J. The mechanism of insulin-stimulated 4E-BP protein binding to mammalian target of rapamycin (mTOR) complex 1 and its contribution to mTOR complex 1 signaling. J. Biol. Chem. 2011, 286, 38043–38053. [Google Scholar] [CrossRef] [PubMed]
- Fagerberg, L.; Hallström, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteom. 2014, 13, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.R.; Hem, V.; Katz, K.S.; Ovetsky, M.; Wallin, C.; Ermolaeva, O.; Tolstoy, I.; Tatusova, T.; Pruitt, K.D.; Maglott, D.R.; et al. Gene: A gene-centered information resource at NCBI. Nucleic Acids Res. 2015, 43, D36–D42. [Google Scholar] [CrossRef] [PubMed]
- Friday, A.J.; Keiper, B.D. Positive mRNA Translational Control in Germ Cells by Initiation Factor Selectivity. Biomed Res. Int. 2015, 2015, 327963. [Google Scholar] [CrossRef] [PubMed]
- Ciosk, R.; DePalma, M.; Priess, J.R. Translational regulators maintain totipotency in the Caenorhabditis elegans germline. Science 2006, 311, 851–853. [Google Scholar] [CrossRef]
- Henderson, M.A.; Cronland, E.; Dunkelbarger, S.; Contreras, V.; Strome, S.; Keiper, B.D. A germline-specific isoform of eIF4E (IFE-1) is required for efficient translation of stored mRNAs and maturation of both oocytes and sperm. J. Cell Sci. 2009, 122, 1529–1539. [Google Scholar] [CrossRef]
- Ghosh, S.; Lasko, P. Loss-of-function analysis reveals distinct requirements of the translation initiation factors eIF4E, eIF4E-3, eIF4G and eIF4G2 in Drosophila spermatogenesis. PLoS ONE 2015, 10, e0122519. [Google Scholar] [CrossRef]
- Buchan, J.R. mRNP granules. Assembly, function, and connections with disease. RNA Biol. 2014, 11, 1019–1030. [Google Scholar] [CrossRef] [PubMed]
- Huggins, H.P.; Keiper, B.D. Regulation of Germ Cell mRNPs by eIF4E:4EIP Complexes: Multiple Mechanisms, One Goal. Front. Cell Dev. Biol. 2020, 8, 562. [Google Scholar] [CrossRef] [PubMed]
- Voronina, A.S.; Pshennikova, E.S. mRNPs: Structure and role in development. Cell Biochem. Funct. 2021, 39, 832–843. [Google Scholar] [CrossRef] [PubMed]
- Huggins, H.P.; Subash, J.S.; Stoffel, H.; Henderson, M.A.; Hoffman, J.L.; Buckner, D.S.; Sengupta, M.S.; Boag, P.R.; Lee, M.-H.; Keiper, B.D. Distinct roles of two eIF4E isoforms in the germline of Caenorhabditis elegans. J. Cell Sci. 2020, 133, jcs237990. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef] [PubMed]
- Safran, M.; Rosen, N.; Twik, M.; BarShir, R.; Stein, T.I.; Dahary, D.; Fishilevich, S.; Lancet, D. The GeneCards Suite. In Practical Guide to Life Science Databases; Abugessaisa, I., Kasukawa, T., Eds.; Springer Nature: Singapore, 2021; pp. 27–56. [Google Scholar]
- Cunningham, F.; Allen, J.E.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Bennett, R.; et al. Ensembl 2022. Nucleic Acids Res. 2022, 50, D988–D995. [Google Scholar] [CrossRef] [PubMed]
- Dowell, R.D. The similarity of gene expression between human and mouse tissues. Genome Biol. 2011, 12, 101. [Google Scholar] [CrossRef]
- Gu, X.; Su, Z. Tissue-driven hypothesis of genomic evolution and sequence-expression correlations. Proc. Natl. Acad. Sci. USA 2007, 104, 2779–2784. [Google Scholar] [CrossRef]
- Cheng, L.; Albers, P.; Berney, D.M.; Feldman, D.R.; Daugaard, G.; Gilligan, T.; Looijenga, L.H.J. Testicular cancer. Nat. Rev. Dis. Primers 2018, 4, 29. [Google Scholar] [CrossRef]
- Daugaard, G.; Gundgaard, M.G.; Mortensen, M.S.; Agerbæk, M.; Holm, N.V.; Rørth, M.; von der Maase, H.; Christensen, I.J.; Lauritsen, J. Surveillance for stage I nonseminoma testicular cancer: Outcomes and long-term follow-up in a population-based cohort. J. Clin. Oncol. 2014, 32, 3817–3823. [Google Scholar] [CrossRef]
- De Benedetti, A.; Graff, J.R. eIF-4E expression and its role in malignancies and metastases. Oncogene 2004, 23, 3189–3199. [Google Scholar] [CrossRef]
- Fingar, D.C.; Richardson, C.J.; Tee, A.R.; Cheatham, L.; Tsou, C.; Blenis, J. mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mol. Cell. Biol. 2004, 24, 200–216. [Google Scholar] [CrossRef]
- Fingar, D.C.; Salama, S.; Tsou, C.; Harlow, E.; Blenis, J. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 2002, 16, 1472–1487. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Ruggero, D. The Role of Translation Control in Tumorigenesis and Its Therapeutic Implications. Annu. Rev. Cancer Biol. 2020, 4, 437–457. [Google Scholar] [CrossRef]
- Taylor-Weiner, A.; Zack, T.; O’Donnell, E.; Guerriero, J.L.; Bernard, B.; Reddy, A.; Han, G.C.; AlDubayan, S.; Amin-Mansour, A.; Schumacher, S.E.; et al. Genomic evolution and chemoresistance in germ-cell tumours. Nature 2016, 540, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Olivier, M.; Hollstein, M.; Hainaut, P. TP53 mutations in human cancers: Origins, consequences, and clinical use. Cold Spring Harb. Perspect. Biol. 2010, 2, a001008. [Google Scholar] [CrossRef] [PubMed]
- Lutzker, S.G. P53 tumour suppressor gene and germ cell neoplasia. APMIS 1998, 106, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Romano, F.J.; Rossetti, S.; Conteduca, V.; Schepisi, G.; Cavaliere, C.; Di Franco, R.; La Mantia, E.; Castaldo, L.; Nocerino, F.; Ametrano, G.; et al. Role of DNA repair machinery and p53 in the testicular germ cell cancer: A review. Oncotarget 2016, 7, 85641–85649. [Google Scholar] [CrossRef]
- Shimada, T.; Yabuki, Y.; Noguchi, T.; Tsuchida, M.; Komatsu, R.; Hamano, S.; Yamada, M.; Ezaki, Y.; Hirata, Y.; Matsuzawa, A. The Distinct Roles of LKB1 and AMPK in p53-Dependent Apoptosis Induced by Cisplatin. Int. J. Mol. Sci. 2022, 23, 10064. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pugsley, L.; Naineni, S.K.; Amiri, M.; Yanagiya, A.; Cencic, R.; Sonenberg, N.; Pelletier, J. C8ORF88: A Novel eIF4E-Binding Protein. Genes 2023, 14, 2076. https://doi.org/10.3390/genes14112076
Pugsley L, Naineni SK, Amiri M, Yanagiya A, Cencic R, Sonenberg N, Pelletier J. C8ORF88: A Novel eIF4E-Binding Protein. Genes. 2023; 14(11):2076. https://doi.org/10.3390/genes14112076
Chicago/Turabian StylePugsley, Lauren, Sai Kiran Naineni, Mehdi Amiri, Akiko Yanagiya, Regina Cencic, Nahum Sonenberg, and Jerry Pelletier. 2023. "C8ORF88: A Novel eIF4E-Binding Protein" Genes 14, no. 11: 2076. https://doi.org/10.3390/genes14112076
APA StylePugsley, L., Naineni, S. K., Amiri, M., Yanagiya, A., Cencic, R., Sonenberg, N., & Pelletier, J. (2023). C8ORF88: A Novel eIF4E-Binding Protein. Genes, 14(11), 2076. https://doi.org/10.3390/genes14112076




