A Proposal of the Ur-RNAome
Abstract
:1. Introduction
2. Methods
2.1. Data Sources
2.2. Reconstruction of Sequences of Ancient RNAomes
2.3. Grouping and Assembly of RNAs
2.4. MSAs of RNAs
2.5. Sequence Logos
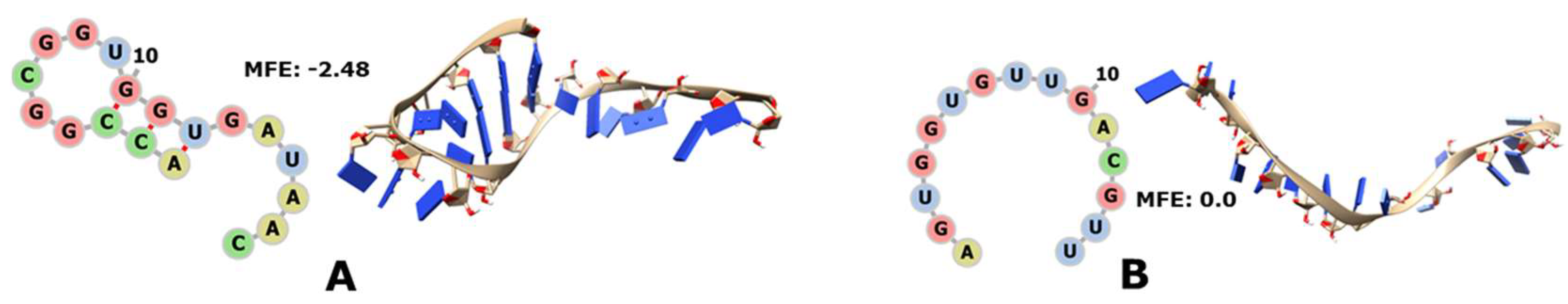
2.6. Representation in 2D of the Recovered Fragments
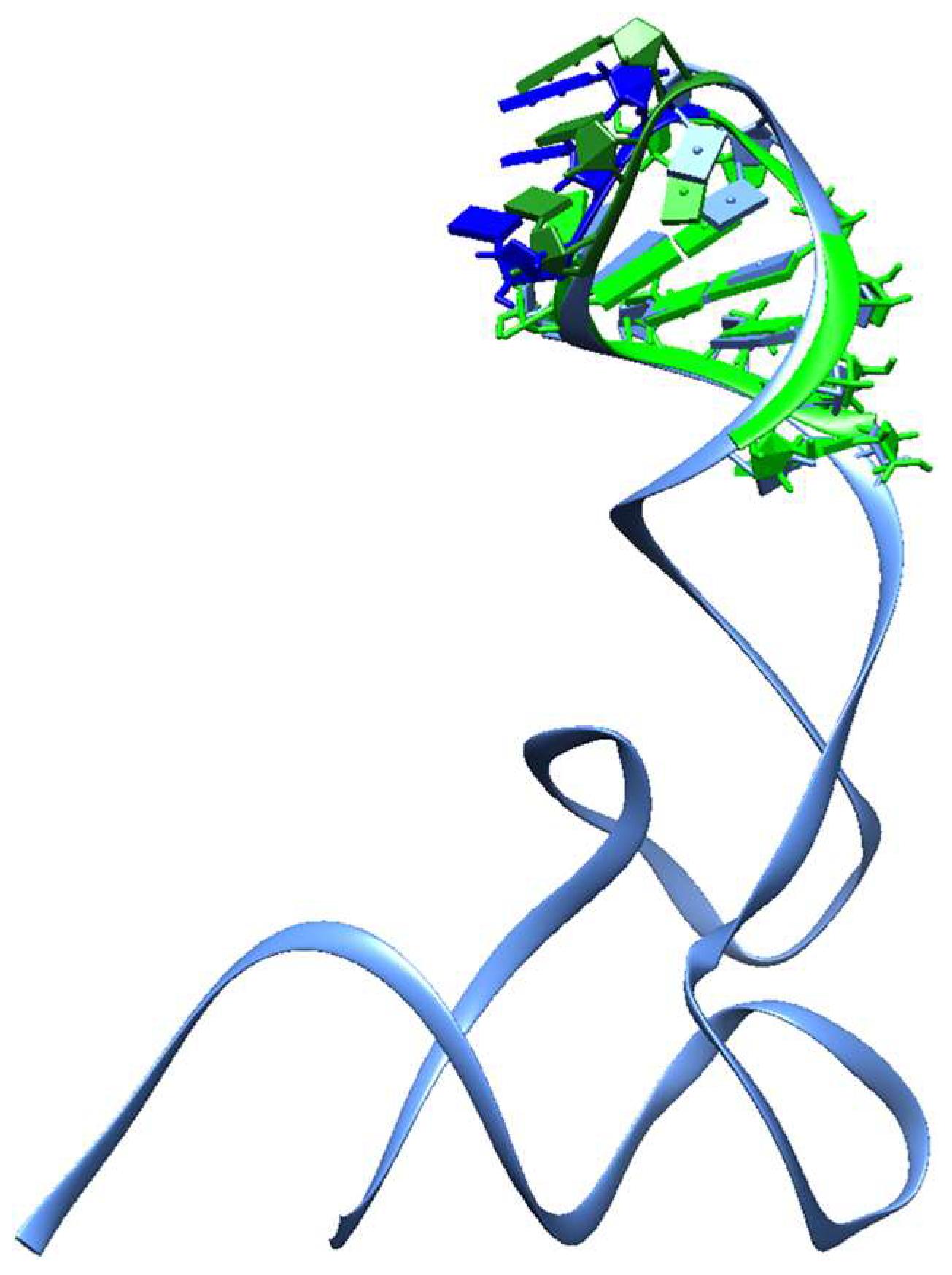
2.7. Representation in 3D of the Reconstructed Fragments
2.8. Negative Controls
3. Results and Analyses
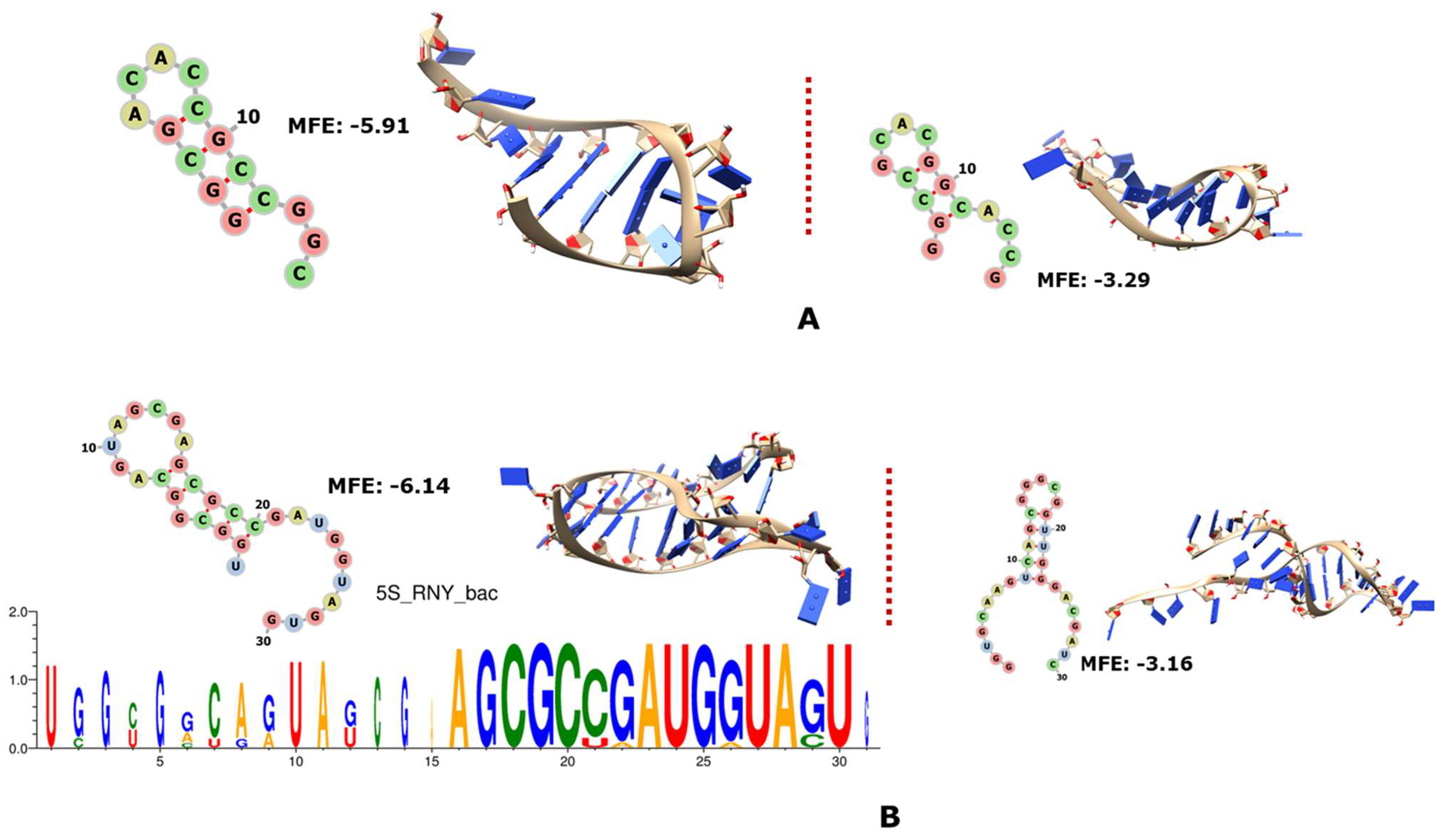
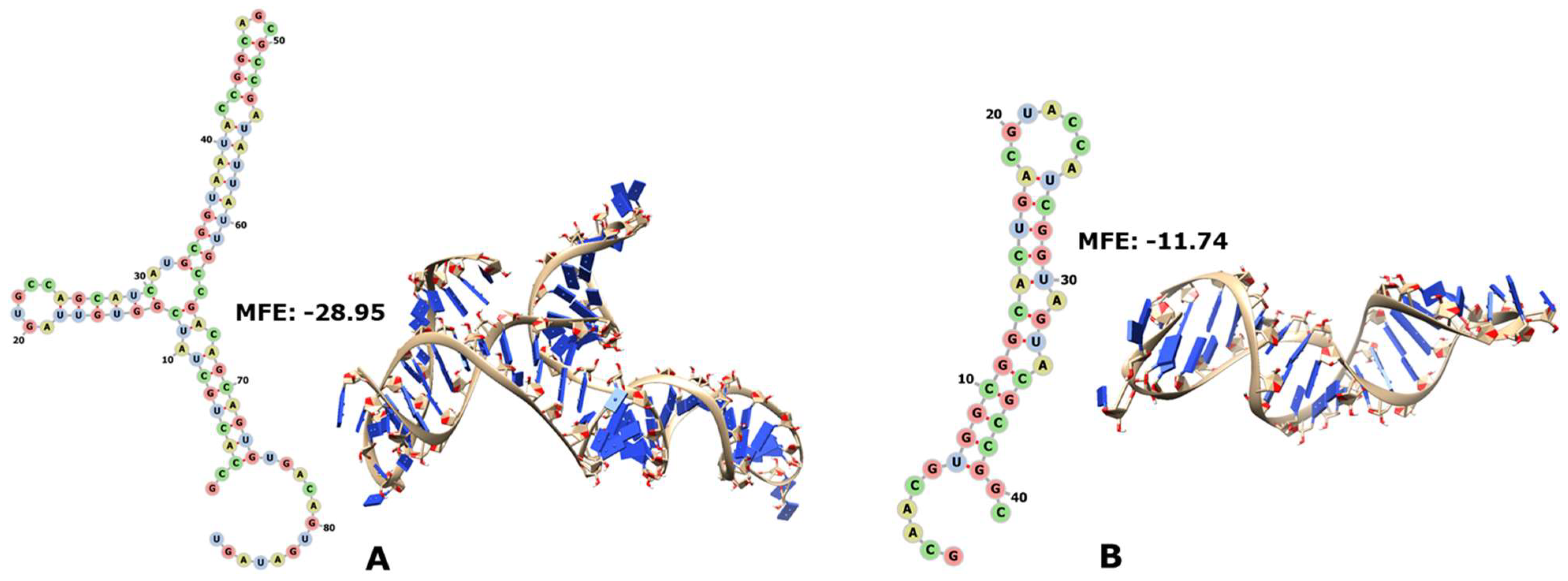
3.1. Ribosomal RNAs
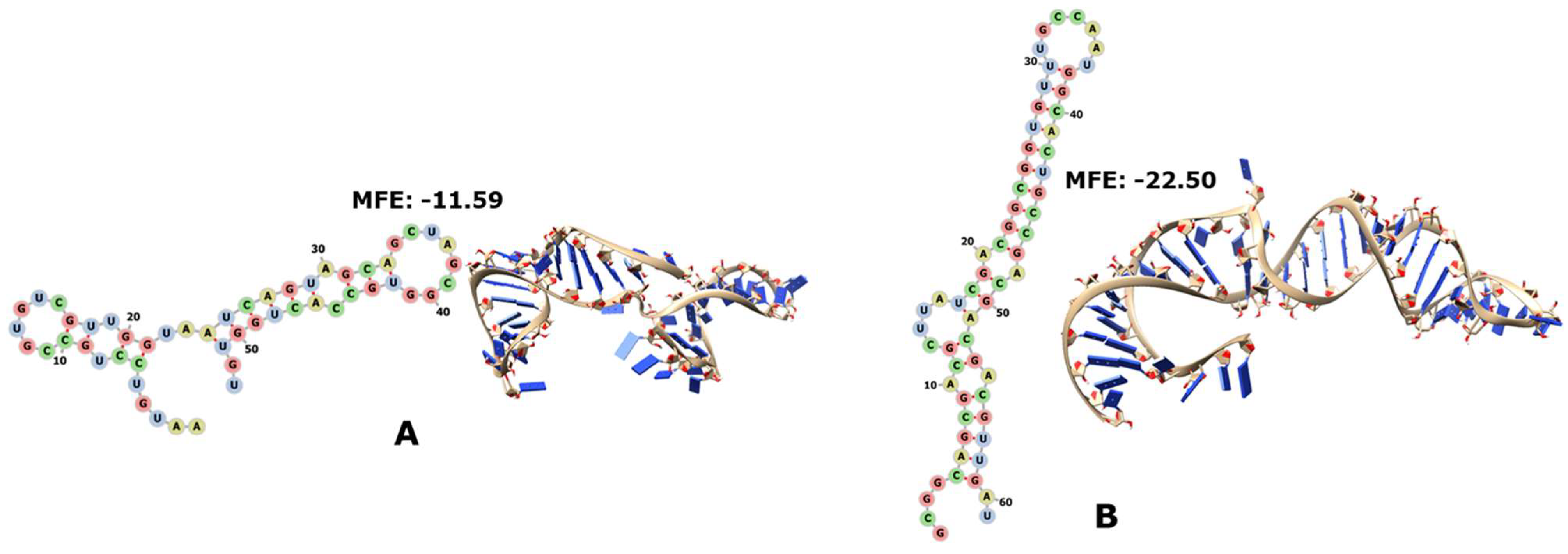
3.2. RNase P, SRP, and 6S
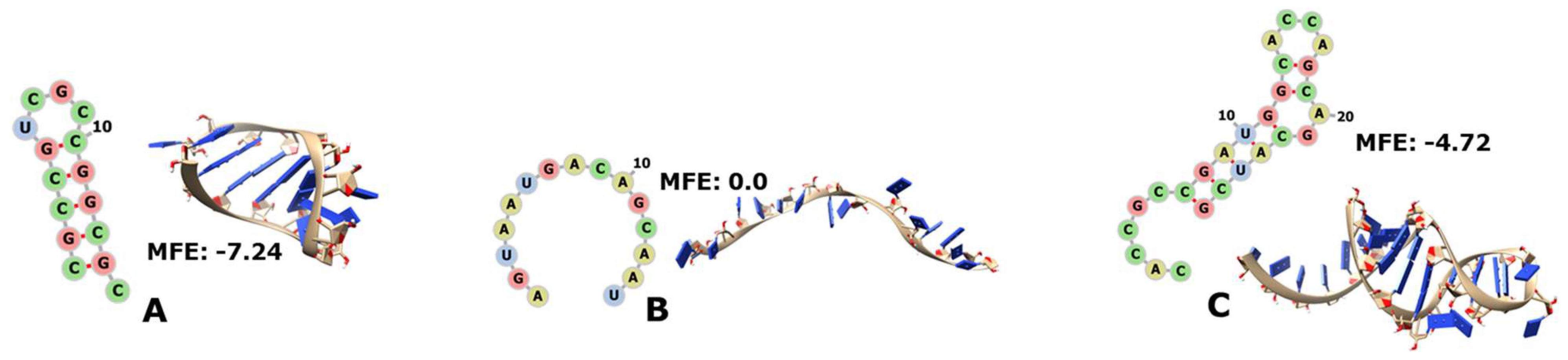
3.3. tRNAs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kruger, K.; Grabowski, P.J.; Zaug, A.J.; Sands, J.; Gottschling, D.E.; Cech, T.R. Self-Splicing RNA: Autoexcision and Autocyclization of the Ribosomal RNA Intervening Sequence of Tetrahymena. Cell 1982, 31, 147–157. [Google Scholar] [CrossRef]
- Guerrier-Takada, C.; Gardiner, K.; Marsh, T.; Pace, N.; Altman, S. The RNA Moiety of Ribonuclease P Is the Catalytic Subunit of the Enzyme. Cell 1983, 35, 849–857. [Google Scholar] [CrossRef]
- Gilbert, W. Origin of Life: The RNA World. Nature 1986, 319, 618. [Google Scholar] [CrossRef]
- Brown, T.A. Genomes, 2nd ed.; Wiley-Liss: Oxford, UK, 2002; ISBN 978-0-471-25046-3. [Google Scholar]
- Eigen, M.; Gardiner, W.; Schuster, P.; Winkler-Oswatitsch, R. The Origin of Genetic Information. Sci. Am. 1981, 244, 88–119. [Google Scholar] [CrossRef] [PubMed]
- Wächtershäuser, G. The Place of RNA in the Origin and Early Evolution of the Genetic Machinery. Life 2014, 4, 1050–1091. [Google Scholar] [CrossRef]
- Chatterjee, S.; Yadav, S. The Origin of Prebiotic Information System in the Peptide/RNA World: A Simulation Model of the Evolution of Translation and the Genetic Code. Life 2019, 9, 25. [Google Scholar] [CrossRef] [PubMed]
- Benner, S.A.; Bell, E.A.; Biondi, E.; Brasser, R.; Carell, T.; Kim, H.-J.; Mojzsis, S.J.; Omran, A.; Pasek, M.A.; Trail, D. When Did Life Likely Emerge on Earth in an RNA-First Process? ChemSystemsChem 2020, 2, e1900035. [Google Scholar] [CrossRef]
- Lehman, N. The RNA World: 4,000,000,050 Years Old. Life 2015, 5, 1583–1586. [Google Scholar] [CrossRef]
- Eigen, M.; Schuster, P. The Hypercycle—A Principle of Natural Self-Organization Part C: The Realistic Hypercycle. Naturwissenschaften 1978, 65, 341–369. [Google Scholar] [CrossRef]
- Eigen, M.; Lindemann, B.; Winkler-Oswatitsch, R.; Clarke, C.H. Pattern Analysis of 5S rRNA. Proc. Natl. Acad. Sci. USA 1985, 82, 2437–2441. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, J. Amplification of the Sequences Displaying the Pattern RNY in the RNA World: The Translation → Translation/Replication Hypothesis. J. Theor. Biol. 2002, 219, 521–537. [Google Scholar] [CrossRef]
- Eigen, M.; Winkler-Oswatitsch, R. Transfer-RNA: The Early Adaptor. Naturwissenschaften 1981, 68, 217–228. [Google Scholar] [CrossRef]
- Eigen, M.; Winkler-Oswatitsch, R. Transfer-RNA, an Early Gene? Naturwissenschaften 1981, 68, 282–292. [Google Scholar] [CrossRef] [PubMed]
- Delaye, L.; Lazcano, A. Prebiological Evolution and the Physics of the Origin of Life. Phys. Life Rev. 2005, 2, 47–64. [Google Scholar] [CrossRef] [PubMed]
- Kun, Á.; Szilágyi, A.; Könnyű, B.; Boza, G.; Zachar, I.; Szathmáry, E. The Dynamics of the RNA World: Insights and Challenges. Ann. N. Y. Acad. Sci. 2015, 1341, 75–95. [Google Scholar] [CrossRef] [PubMed]
- Szilágyi, A.; Zachar, I.; Scheuring, I.; Kun, Á.; Könnyű, B.; Czárán, T. Ecology and Evolution in the RNA World Dynamics and Stability of Prebiotic Replicator Systems. Life 2017, 7, 48. [Google Scholar] [CrossRef] [PubMed]
- Szilágyi, A.; Könnyű, B.; Czárán, T. Dynamics and Stability in Prebiotic Information Integration: An RNA World Model from First Principles. Sci. Rep. 2020, 10, 51. [Google Scholar] [CrossRef]
- Schimmel, P.; de Pouplana, L.R. Transfer RNA: From Minihelix to Genetic Code. Cell 1995, 81, 983–986. [Google Scholar] [CrossRef]
- Ribas de Pouplana, L.; Buechter, D.; Sardesai, N.Y.; Schimmel, P. Functional Analysis of Peptide Motif for RNA Microhelix Binding Suggests New Family of RNA-Binding Domains. EMBO J. 1998, 17, 5449–5457. [Google Scholar] [CrossRef] [PubMed]
- Mizuuchi, R.; Lehman, N. Limited Sequence Diversity Within a Population Supports Prebiotic RNA Reproduction. Life 2019, 9, 20. [Google Scholar] [CrossRef]
- Giulio, M.D. On the Origin of the Transfer RNA Molecule. J. Theor. Biol. 1992, 159, 199–214. [Google Scholar] [CrossRef] [PubMed]
- Dick, T.P.; Schamel, W.W.A. Molecular Evolution of Transfer RNA from Two Precursor Hairpins: Implications for the Origin of Protein Synthesis. J. Mol. Evol. 1995, 41, 1–9. [Google Scholar] [CrossRef]
- de Farias, S.T.; José, M.V. Transfer RNA: The Molecular Demiurge in the Origin of Biological Systems. Prog. Biophys. Mol. Biol. 2020, 153, 28–34. [Google Scholar] [CrossRef]
- Farias, S.T.; Rêgo, T.G.; José, M.V. Origin and Evolution of the Peptidyl Transferase Center from Proto-tRNAs. FEBS Open Bio 2014, 4, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Torres de Farias, S.; Gaudêncio Rêgo, T.; José, M.V. Peptidyl Transferase Center and the Emergence of the Translation System. Life 2017, 7, 21. [Google Scholar] [CrossRef]
- Prosdocimi, F.; Zamudio, G.S.; Palacios-Pérez, M.; Torres de Farias, S.; José, M.V. The Ancient History of Peptidyl Transferase Center Formation as Told by Conservation and Information Analyses. Life 2020, 10, 134. [Google Scholar] [CrossRef]
- de Farias, S.T.; do Rêgo, T.G.; José, M.V. Evolution of Transfer RNA and the Origin of the Translation System. Front. Genet. 2014, 5, 303. [Google Scholar] [CrossRef]
- Palacios-Pérez, M.; Andrade-Díaz, F.; José, M.V. A Proposal of the Ur-Proteome. Orig. Life Evol. Biosph. 2018, 48, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Maizels, N.; Weiner, A.M. Phylogeny from Function: Evidence from the Molecular Fossil Record That tRNA Originated in Replication, Not Translation. Proc. Natl. Acad. Sci. USA 1994, 91, 6729–6734. [Google Scholar] [CrossRef]
- José, M.V.; Zamudio, G.S.; Palacios-Pérez, M.; Bobadilla, J.R.; de Farías, S.T. Symmetrical and Thermodynamic Properties of Phenotypic Graphs of Amino Acids Encoded by the Primeval RNY Code. Orig. Life Evol. Biosph. 2015, 45, 77–83. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Kinouchi, M.; Kurokawa, K. tRNAfinder: A Software System to Find All tRNA Genes in the DNA Sequence Based on the Cloverleaf Secondary Structure. J. Comput. Aided Chem. 2006, 9, 116–124. [Google Scholar] [CrossRef]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-Line: Integrating Search and Context for Analysis of Transfer RNA Genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Brudno, M.; Steinkamp, R.; Morgenstern, B. The CHAOS/DIALIGN WWW Server for Multiple Alignment of Genomic Sequences. Nucleic Acids Res. 2004, 32, W41–W44. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A Unified Bioinformatics Toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A Sequence Logo Generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neuböck, R.; Hofacker, I.L. The Vienna RNA Websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef]
- Kerpedjiev, P.; Hammer, S.; Hofacker, I.L. Forna (Force-Directed RNA): Simple and Effective Online RNA Secondary Structure Diagrams. Bioinformatics 2015, 31, 3377–3379. [Google Scholar] [CrossRef]
- Popenda, M.; Szachniuk, M.; Antczak, M.; Purzycka, K.J.; Lukasiak, P.; Bartol, N.; Blazewicz, J.; Adamiak, R.W. Automated 3D Structure Composition for Large RNAs. Nucleic Acids Res. 2012, 40, e112. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Williams, T.A.; Foster, P.G.; Nye, T.M.W.; Cox, C.J.; Embley, T.M. A Congruent Phylogenomic Signal Places Eukaryotes within the Archaea. Proc. R. Soc. B Biol. Sci. 2012, 279, 4870–4879. [Google Scholar] [CrossRef]
- Raymann, K.; Brochier-Armanet, C.; Gribaldo, S. The Two-Domain Tree of Life Is Linked to a New Root for the Archaea. Proc. Natl. Acad. Sci. USA 2015, 112, 6670–6675. [Google Scholar] [CrossRef]
- Spang, A.; Saw, J.H.; Jørgensen, S.L.; Zaremba-Niedzwiedzka, K.; Martijn, J.; Lind, A.E.; van Eijk, R.; Schleper, C.; Guy, L.; Ettema, T.J.G. Complex Archaea That Bridge the Gap between Prokaryotes and Eukaryotes. Nature 2015, 521, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Spang, A.; Ettema, T.J.G. Microbial Diversity: The Tree of Life Comes of Age. Nat. Microbiol. 2016, 1, 16056. [Google Scholar] [CrossRef] [PubMed]
- Cornish-Bowden, A.; Cárdenas, M.L. Life before LUCA. J. Theor. Biol. 2017, 434, 68–74. [Google Scholar] [CrossRef]
- Imachi, H.; Nobu, M.K.; Nakahara, N.; Morono, Y.; Ogawara, M.; Takaki, Y.; Takano, Y.; Uematsu, K.; Ikuta, T.; Ito, M.; et al. Isolation of an Archaeon at the Prokaryote–Eukaryote Interface. Nature 2020, 577, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Mustoe, A.M.; Brooks, C.L.; Al-Hashimi, H.M. Hierarchy of RNA Functional Dynamics. Annu. Rev. Biochem. 2014, 83, 441–466. [Google Scholar] [CrossRef]
- Moore, P.B.; Steitz, T.A. The Involvement of RNA in Ribosome Function. Nature 2002, 418, 229. [Google Scholar] [CrossRef]
- Sievers, A.; Beringer, M.; Rodnina, M.V.; Wolfenden, R. The Ribosome as an Entropy Trap. Proc. Natl. Acad. Sci. USA 2004, 101, 7897–7901. [Google Scholar] [CrossRef]
- Wallin, G.; Åqvist, J. The Transition State for Peptide Bond Formation Reveals the Ribosome as a Water Trap. Proc. Natl. Acad. Sci. USA 2010, 107, 1888–1893. [Google Scholar] [CrossRef]
- Jüttner, M.; Ferreira-Cerca, S. A Comparative Perspective on Ribosome Biogenesis: Unity and Diversity Across the Tree of Life. In Ribosome Biogenesis: Methods and Protocols; Entian, K.-D., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2022; pp. 3–22. ISBN 978-1-07-162501-9. [Google Scholar]
- Wang, Q.; Su, H. A Tale of Water Molecules in the Ribosomal Peptidyl Transferase Reaction. Biochemistry 2022, 61, 2241–2247. [Google Scholar] [CrossRef] [PubMed]
- Jarrous, N. Roles of RNase P and Its Subunits. Trends Genet. 2017, 33, 594–603. [Google Scholar] [CrossRef] [PubMed]
- Daniels, C.J.; Lai, L.B.; Chen, T.-H.; Gopalan, V. Both Kinds of RNase P in All Domains of Life: Surprises Galore. RNA 2019, 25, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Di Giulio, M. The RNase P, LUCA, the Ancestors of the Life Domains, the Progenote, and the Tree of Life. Biosystems 2022, 212, 104604. [Google Scholar] [CrossRef]
- Larsen, N.; Zwieb, C. SRP-RNA Sequence Alignment and Secondary Structure. Nucleic Acids Res. 1991, 19, 209–215. [Google Scholar] [CrossRef]
- Rosenblad, M.A.; Larsen, N.; Samuelsson, T.; Zwieb, C. Kinship in the SRP RNA Family. RNA Biol. 2009, 6, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Barrick, J.E.; Sudarsan, N.; Weinberg, Z.; Ruzzo, W.L.; Breaker, R.R. 6S RNA Is a Widespread Regulator of Eubacterial RNA Polymerase That Resembles an Open Promoter. RNA 2005, 11, 774–784. [Google Scholar] [CrossRef]
- Wassarman, K.M. 6S RNA: A Small RNA Regulator of Transcription. Curr. Opin. Microbiol. 2007, 10, 164–168. [Google Scholar] [CrossRef]
- Cavanagh, A.T.; Wassarman, K.M. 6S RNA, a Global Regulator of Transcription in Escherichia Coli, Bacillus Subtilis, and Beyond. Annu. Rev. Microbiol. 2014, 68, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Burenina, O.Y.; Elkina, D.A.; Hartmann, R.K.; Oretskaya, T.S.; Kubareva, E.A. Small Noncoding 6S RNAs of Bacteria. Biochemistry 2015, 80, 1429–1446. [Google Scholar] [CrossRef]
- Chen, J.; Wassarman, K.M.; Feng, S.; Leon, K.; Feklistov, A.; Winkelman, J.T.; Li, Z.; Walz, T.; Campbell, E.A.; Darst, S.A. 6S RNA Mimics B-Form DNA to Regulate Escherichia Coli RNA Polymerase. Mol. Cell 2017, 68, 388–397.e6. [Google Scholar] [CrossRef] [PubMed]
- Wassarman, K.M. 6S RNA, A Global Regulator of Transcription. Microbiol. Spectr. 2018, 6, 1–13. [Google Scholar] [CrossRef]
- Berg, J.M.; Tymoczko, J.L.; Stryer, L. Biochemistry, 5th ed.; W H Freeman: New York, NY, USA, 2002; ISBN 978-0-7167-3051-4. [Google Scholar]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002; ISBN 978-0-8153-3218-3. [Google Scholar]
- Berg, M.D.; Brandl, C.J. Transfer RNAs: Diversity in Form and Function. RNA Biol. 2021, 18, 316–339. [Google Scholar] [CrossRef] [PubMed]
- Hyeon, C.; Thirumalai, D. Chain Length Determines the Folding Rates of RNA. Biophys. J. 2012, 102, L11–L13. [Google Scholar] [CrossRef]
- Root-Bernstein, R.; Kim, Y.; Sanjay, A.; Burton, Z.F. tRNA Evolution from the Proto-tRNA Minihelix World. Transcription 2016, 7, 153–163. [Google Scholar] [CrossRef]
- Rodin, S.; Rodin, A.; Ohno, S. The Presence of Codon-Anticodon Pairs in the Acceptor Stem of tRNAs. Proc. Natl. Acad. Sci. USA 1996, 93, 4537–4542. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Schimmel, P. Chiral-Selective Aminoacylation of an RNA Minihelix. Science 2004, 305, 1253. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Su, M. The Origin of Translation: Bridging the Nucleotides and Peptides. Int. J. Mol. Sci. 2023, 24, 197. [Google Scholar] [CrossRef]
- Widmann, J.; Di Giulio, M.; Yarus, M.; Knight, R. tRNA Creation by Hairpin Duplication. J. Mol. Evol. 2005, 61, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Prosdocimi, F.; de Farias, S.T. Entering the Labyrinth: A Hypothesis about the Emergence of Metabolism from Protobiotic Routes. Biosystems 2022, 220, 104751. [Google Scholar] [CrossRef]
- Prosdocimi, F.; de Farias, S.T. Origin of Life: Drawing the Big Picture. Prog. Biophys. Mol. Biol. 2023, 180–181, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Abbott, J.A.; Francklyn, C.S.; Robey-Bond, S.M. Transfer RNA and Human Disease. Front. Genet. 2014, 5, 158. [Google Scholar] [CrossRef]
- Brandon, M.C.; Lott, M.T.; Nguyen, K.C.; Spolim, S.; Navathe, S.B.; Baldi, P.; Wallace, D.C. MITOMAP: A Human Mitochondrial Genome Database—2004 Update. Nucleic Acids Res. 2005, 33, D611–D613. [Google Scholar] [CrossRef] [PubMed]
- Yarham, J.W.; Elson, J.L.; Blakely, E.L.; McFarland, R.; Taylor, R.W. Mitochondrial tRNA Mutations and Disease. WIREs RNA 2010, 1, 304–324. [Google Scholar] [CrossRef]
- Pavon-Eternod, M.; Gomes, S.; Geslain, R.; Dai, Q.; Rosner, M.R.; Pan, T. tRNA Over-Expression in Breast Cancer and Functional Consequences. Nucleic Acids Res. 2009, 37, 7268–7280. [Google Scholar] [CrossRef] [PubMed]
- Glatz, C.; D’Aco, K.; Smith, S.; Sondheimer, N. Mutation in the Mitochondrial tRNAVal Causes Mitochondrial Encephalopathy, Lactic Acidosis and Stroke-like Episodes. Mitochondrion 2011, 11, 615–619. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palacios-Pérez, M.; José, M.V. A Proposal of the Ur-RNAome. Genes 2023, 14, 2158. https://doi.org/10.3390/genes14122158
Palacios-Pérez M, José MV. A Proposal of the Ur-RNAome. Genes. 2023; 14(12):2158. https://doi.org/10.3390/genes14122158
Chicago/Turabian StylePalacios-Pérez, Miryam, and Marco V. José. 2023. "A Proposal of the Ur-RNAome" Genes 14, no. 12: 2158. https://doi.org/10.3390/genes14122158






