Towards a Cure for HARS Disease
Abstract
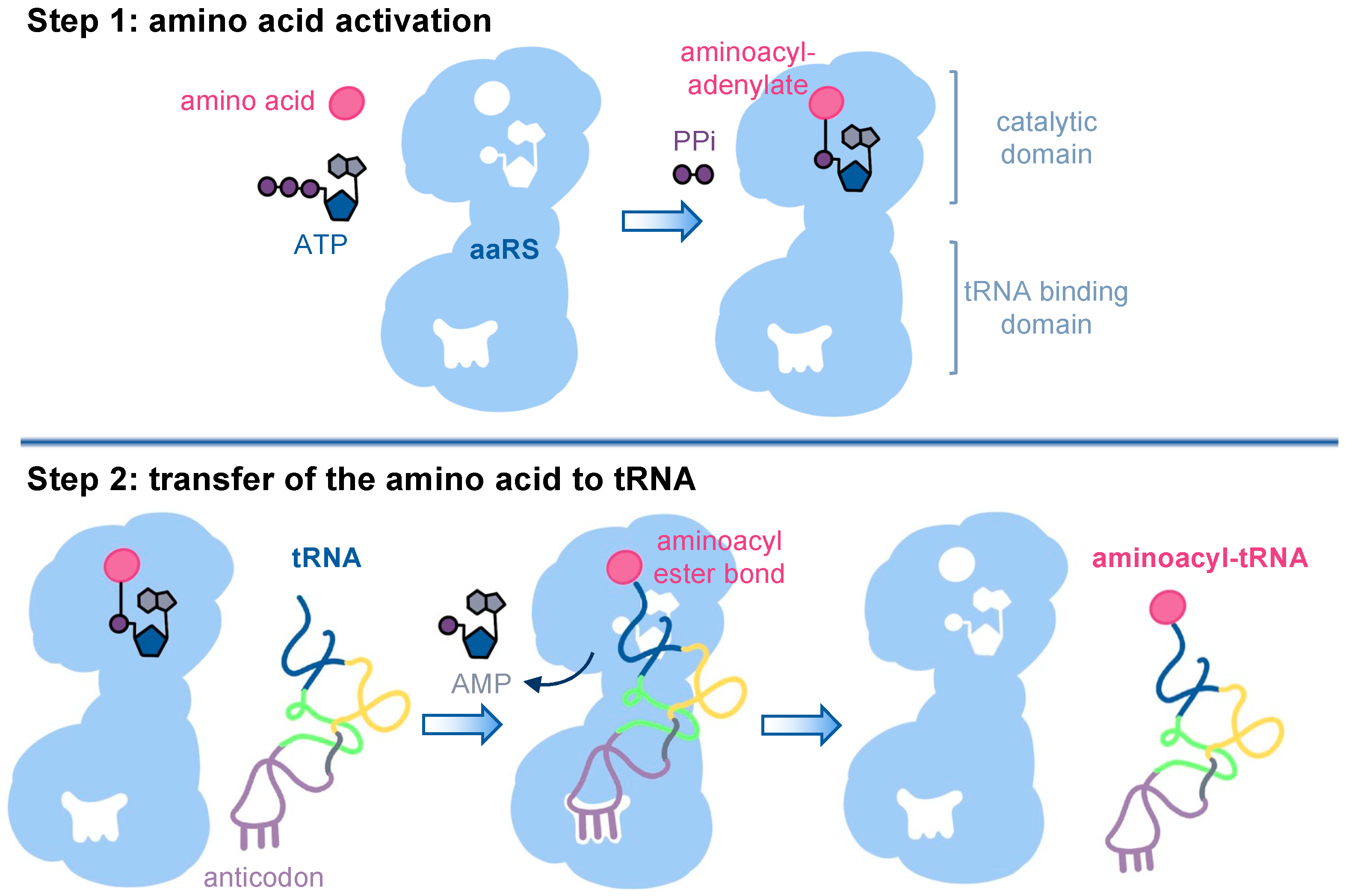
:1. Aminoacyl-tRNA Synthetases in Translation
2. Translational Fidelity Guards the Genetic Code
3. Human Histidyl-tRNA Synthetase
4. Disease-Causing HARS Mutations
4.1. aaRS Mutations in Charcot Marie Tooth Disease
4.2. Usher Syndrome Type IIIB
5. Pathways to a Cure
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schuller, A.P.; Green, R. Roadblocks and resolutions in eukaryotic translation. Nat. Rev. Mol. Cell Biol. 2018, 19, 526–541. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, G. Translation of genetic information on the ribosome. Angew. Chem. Int. Ed. 1971, 10, 638–651. [Google Scholar] [CrossRef] [PubMed]
- Moldave, K. Eukaryotic protein synthesis. Annu. Rev. Biochem. 1985, 54, 1109–1149. [Google Scholar] [CrossRef]
- O’Donoghue, P.; Luthey-Schulten, Z. On the evolution of structure in aminoacyl-tRNA synthetases. Microbiol. Mol. Biol. Rev. 2003, 67, 550–573. [Google Scholar] [CrossRef] [Green Version]
- Kwon, N.H.; Fox, P.L.; Kim, S. Aminoacyl-tRNA synthetases as therapeutic targets. Nat. Rev. Drug Discov. 2019, 18, 629–650. [Google Scholar] [CrossRef]
- Ibba, M.; Söll, D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 2000, 69, 617–650. [Google Scholar] [CrossRef]
- Chang, C.-Y.; Chien, C.-I.; Chang, C.-P.; Lin, B.-C.; Wang, C.-C. A WHEP Domain Regulates the Dynamic Structure and Activity of Caenorhabditis elegans Glycyl-tRNA Synthetase. J. Biol. Chem. 2016, 291, 16567–16575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nathanson, L.; Deutscher, M.P. Active aminoacyl-tRNA synthetases are present in nuclei as a high molecular weight multienzyme complex. J. Biol. Chem. 2000, 275, 31559–31562. [Google Scholar] [CrossRef] [Green Version]
- Gile, G.H.; Moog, D.; Slamovits, C.H.; Maier, U.-G.; Archibald, J.M. Dual Organellar Targeting of Aminoacyl-tRNA Synthetases in Diatoms and Cryptophytes. Genome Biol. Evol. 2015, 7, 1728–1742. [Google Scholar] [CrossRef] [Green Version]
- Lant, J.T.; Berg, M.D.; Heinemann, I.U.; Brandl, C.J.; O’Donoghue, P. Pathways to disease from natural variations in human cytoplasmic tRNAs. J. Biol. Chem. 2019, 294, 5294–5308. [Google Scholar] [CrossRef]
- Hasan, F.; Lant, J.T.; O’Donoghue, P. Perseverance of protein homeostasis despite mistranslation of glycine codons with alanine. Phil. Trans. R. Soc. B 2023, 378. [Google Scholar] [CrossRef] [PubMed]
- Meyer-Schuman, R.; Antonellis, A. Emerging mechanisms of aminoacyl-tRNA synthetase mutations in recessive and dominant human disease. Hum. Mol. Genet. 2017, 26, R114–R127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turvey, A.K.; Horvath, G.A.; Cavalcanti, A.R.O. Aminoacyl-tRNA synthetases in human health and disease. Front. Physiol. 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- O’Hanlon, T.P.; Miller, F. Genomic organization, transcriptional mapping, and evolutionary implications of the human bi-directional histidyl-tRNA synthetase locus (HARS/HARSL). Biochem. Biophys. Res. Commun. 2002, 294, 609–614. [Google Scholar] [CrossRef] [Green Version]
- Wasmuth, J.J.; Carlock, L.R. Chromosomal localization of human gene for histidyl-tRNA synthetase: Clustering of genes encoding aminoacyl-tRNA synthetases on human chromosome 5. Somat. Cell Mol. Genet. 1986, 12, 513–517. [Google Scholar] [CrossRef]
- Tsui, F.W.; Siminovitch, L. Isolation, structure and expression of mammalian genes for histidyl-tRNA synthetase. Nucleic Acids Res. 1987, 15, 3349–3367. [Google Scholar] [CrossRef] [Green Version]
- Waldron, A.; Wilcox, C.; Francklyn, C.; Ebert, A. Knock-Down of Histidyl-tRNA Synthetase Causes Cell Cycle Arrest and Apoptosis of Neuronal Progenitor Cells in vivo. Front. Cell Dev. Biol. 2019, 7, 67. [Google Scholar] [CrossRef]
- Xu, Z.; Wei, Z.; Zhou, J.J.; Ye, F.; Lo, W.-S.; Wang, F.; Lau, C.-F.; Wu, J.; Nangle, L.A.; Chiang, K.P.; et al. Internally deleted human tRNA synthetase suggests evolutionary pressure for repurposing. Structure 2012, 20, 1470–1477. [Google Scholar] [CrossRef] [Green Version]
- Zampieri, S.; Ghirardello, A.; Iaccarino, L.; Tarricone, E.; Gambari, P.F.; Doria, A. Anti-Jo-1 antibodies. Autoimmunity 2005, 38, 73–78. [Google Scholar] [CrossRef]
- Chen, A.W.; Jayasinghe, M.I.; Chung, C.Z.; Rao, B.S.; Kenana, R.; Heinemann, I.U.; Jackman, J.E. The Role of 3′ to 5′ Reverse RNA Polymerization in tRNA Fidelity and Repair. Genes 2019, 10, 250. [Google Scholar] [CrossRef]
- Heinemann, I.U.; Nakamura, A.; O’Donoghue, P.; Eiler, D.; Söll, D. tRNAHis-guanylyltransferase establishes tRNAHis identity. Nucleic Acids Res. 2011, 40, 333–344. [Google Scholar] [CrossRef] [Green Version]
- Jackman, J.E.; Gott, J.M.; Gray, M.W. Doing it in reverse: 3′-to-5′ polymerization by the Thg1 superfamily. RNA 2012, 18, 886–899. [Google Scholar] [CrossRef] [Green Version]
- Desai, R.; Kim, K.; Büchsenschütz, H.C.; Chen, A.W.; Bi, Y.; Mann, M.R.; Turk, M.; Chung, C.; Heinemann, I.U. Minimal requirements for reverse polymerization and tRNA repair by tRNAHis guanylyltransferase. RNA Biol. 2017, 15, 614–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heinemann, I.U.; O’Donoghue, P.; Madinger, C.; Benner, J.; Randau, L.; Noren, C.J.; Söll, D. The appearance of pyrrolysine in tRNAHis guanylyltransferase by neutral evolution. Proc. Natl. Acad. Sci. USA 2009, 106, 21103–21108. [Google Scholar] [CrossRef] [Green Version]
- Heinemann, I.U.; Randau, L.; Tomko, R.J.; Söll, D. 3′-5′ tRNAHis guanylyltransferase in bacteria. FEBS Lett. 2010, 584, 3567–3572. [Google Scholar] [CrossRef] [Green Version]
- Connolly, S.A.; Rosen, A.E.; Musier-Forsyth, K.; Francklyn, C.S. G−1:C73 recognition by an arginine cluster in the active site of Escherichia coli histidyl-tRNA synthetase. Biochemistry 2004, 43, 962–969. [Google Scholar] [CrossRef] [PubMed]
- Rao, B.S.; Jackman, J.E. Life without post-transcriptional addition of G-1: Two alternatives for tRNAHis identity in Eukarya. RNA 2014, 21, 243–253. [Google Scholar] [CrossRef] [Green Version]
- Rudinger, J.; Florentz, C.; Giegé, R. Histidylation by yeast HisRS of tRNA or tRNA-like structure relies on residues–1 and 73 but is dependent on the RNA context. Nucleic Acids Res. 1994, 22, 5031–5037. [Google Scholar] [CrossRef] [PubMed]
- Blocquel, D.; Sun, L.; Matuszek, Z.; Li, S.; Weber, T.; Kuhle, B.; Kooi, G.; Wei, N.; Baets, J.; Pan, T.; et al. CMT disease severity correlates with mutation-induced open conformation of histidyl-tRNA synthetase, not aminoacylation loss, in patient cells. Proc. Natl. Acad. Sci. USA 2019, 116, 19440–19448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Q.; Wang, C.; Liu, Y.; Xie, W. Structural basis for recognition of G-1-containing tRNA by histidyl-tRNA synthetase. Nucleic Acids Res. 2015, 43, 2980–2990. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Himeno, H.; Hasegawa, T.; Ueda, T.; Watanabe, K.; Miura, K.-I.; Shimizu, M. Role of the extra G-C pair at the end of the acceptor stem of tRNAHb in aminoacylation. Nucleic Acids Res. 1989, 17, 7855–7863. [Google Scholar] [CrossRef] [PubMed]
- Hawko, S.A.; Francklyn, C.S. Covariation of a specificity-determining structural motif in an aminoacyl-tRNA synthetase and a tRNA identity element. Biochemistry 2001, 40, 1930–1936. [Google Scholar] [CrossRef] [PubMed]
- Guth, E.C.; Francklyn, C.S. Kinetic discrimination of tRNA identity by the conserved motif 2 loop of a class II aminoacyl-tRNA synthetase. Mol. Cell 2007, 25, 531–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guth, E.; Farris, M.; Bovee, M.; Francklyn, C.S. Asymmetric amino acid activation by class II histidyl-tRNA synthetase from Escherichia coli. J. Biol. Chem. 2009, 284, 20753–20762. [Google Scholar] [CrossRef] [Green Version]
- Guth, E.; Connolly, S.H.; Bovee, M.; Francklyn, C.S. A substrate-assisted concerted mechanism for aminoacylation by a class II aminoacyl-tRNA synthetase. Biochemistry 2005, 44, 3785–3794. [Google Scholar] [CrossRef]
- Lee, J.S.; Park, S.G.; Park, H.; Seol, W.; Lee, S.; Kim, S. Interaction network of human aminoacyl-tRNA synthetases and subunits of elongation factor 1 complex. Biochem. Biophys. Res. Commun. 2002, 291, 158–164. [Google Scholar] [CrossRef]
- Vester, A.; Velez-Ruiz, G.; McLaughlin, H.M.; Program, N.C.S.; Lupski, J.R.; Talbot, K.; Vance, J.M.; Züchner, S.; Roda, R.H.; Fischbeck, K.H.; et al. A loss-of-function variant in the human histidyl-tRNA synthetase (HARS) gene is neurotoxic in vivo. Hum. Mutat. 2012, 34, 191–199. [Google Scholar] [CrossRef] [Green Version]
- Brozkova, D.S.; Deconinck, T.; Griffin, L.B.; Ferbert, A.; Haberlova, J.; Mazanec, R.; Lassuthova, P.; Roth, C.; Pilunthanakul, T.; Rautenstrauss, B.; et al. Loss of function mutations in HARS cause a spectrum of inherited peripheral neuropathies. Brain 2015, 138, 2161–2172. [Google Scholar] [CrossRef] [Green Version]
- Royer-Bertrand, B.; Tsouni, P.; Mullen, P.; Campos Xavier, B.; Mittaz Crettol, L.; Lobrinus, A.J.; Ghika, J.; Baumgartner, M.R.; Rivolta, C.; Superti-Furga, A.; et al. Peripheral neuropathy and cognitive impairment associated with a novel monoallelic HARS variant. Ann. Clin. Transl. Neurol. 2019, 6, 1072–1080. [Google Scholar] [CrossRef]
- Abbott, J.A.; Meyer-Schuman, R.; Lupo, V.; Feely, S.; Mademan, I.; Oprescu, S.N.; Griffin, L.B.; Alberti, M.A.; Casasnovas, C.; Aharoni, S.; et al. Substrate interaction defects in histidyl-tRNA synthetase linked to dominant axonal peripheral neuropathy. Hum. Mutat. 2017, 39, 415–432. [Google Scholar] [CrossRef] [PubMed]
- Lupo, V.; García-García, F.; Sancho, P.; Tello, C.; García-Romero, M.; Villarreal, L.; Alberti, A.; Sivera, R.; Dopazo, J.; Pascual-Pascual, S.I.; et al. Assessment of Targeted Next-Generation Sequencing as a Tool for the Diagnosis of Charcot-Marie-Tooth Disease and Hereditary Motor Neuropathy. J. Mol. Diagn. 2016, 18, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Puffenberger, E.G.; Jinks, R.N.; Sougnez, C.; Cibulskis, K.; Willert, R.A.; Achilly, N.P.; Cassidy, R.P.; Fiorentini, C.J.; Heiken, K.F.; Lawrence, J.J.; et al. Genetic mapping and exome sequencing identify variants associated with five novel diseases. PLoS ONE 2012, 7, e28936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Donoghue, P.; Ling, J.; Wang, Y.-S.; Söll, D. Upgrading protein synthesis for synthetic biology. Nat. Chem. Biol. 2013, 9, 594–598. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, I.U.; Rovner, A.J.; Aerni, H.R.; Rogulina, S.; Cheng, L.; Olds, W.; Fischer, J.T.; Söll, D.; Isaacs, F.J.; Rinehart, J. Enhanced phosphoserine insertion during Escherichia coli protein synthesis via partial UAG codon reassignment and release factor 1 deletion. FEBS Lett. 2012, 586, 3716–3722. [Google Scholar] [CrossRef] [Green Version]
- Guo, L.-T.; Wang, Y.-S.; Nakamura, A.; Eiler, D.; Kavran, J.M.; Wong, M.; Kiessling, L.L.; Steitz, T.A.; O’Donoghue, P.; Söll, D. Polyspecific pyrrolysyl-tRNA synthetases from directed evolution. Proc. Natl. Acad. Sci. USA 2014, 111, 16724–16729. [Google Scholar] [CrossRef] [Green Version]
- Lant, J.T.; Berg, M.; Sze, D.H.W.; Hoffman, K.S.; Akinpelu, I.C.; Turk, M.; Heinemann, I.U.; Duennwald, M.L.; Brandl, C.J.; O’Donoghue, P. Visualizing tRNA-dependent mistranslation in human cells. RNA Biol. 2017, 15, 567–575. [Google Scholar] [CrossRef] [Green Version]
- Lant, J.T.; Kiri, R.; Duennwald, M.L.; O’Donoghue, P. Formation and persistence of polyglutamine aggregates in mistranslating cells. Nucleic Acids Res. 2021, 49, 11883–11899. [Google Scholar] [CrossRef]
- Rozik, P.; Szabla, R.; Lant, J.T.; Kiri, R.; Wright, D.E.; Junop, M.; O’Donoghue, P. A novel fluorescent reporter sensitive to serine mis-incorporation. RNA Biol. 2022, 19, 221–233. [Google Scholar] [CrossRef]
- Kapur, M.; Monaghan, C.E.; Ackerman, S.L. Regulation of mRNA Translation in Neurons—A Matter of Life and Death. Neuron 2017, 96, 616–637. [Google Scholar] [CrossRef]
- Qiu, Y.; Kenana, R.; Beharry, A.; Wilhelm, S.D.P.; Hsu, S.Y.; Siu, V.M.; Duennwald, M.; Heinemann, I.U. Histidine supplementation can escalate or rescue HARS deficiency in a Charcot–Marie–Tooth Disease model. Hum. Mol. Genet. 2022, ddac239. [Google Scholar] [CrossRef]
- Mullen, P.; Abbott, J.A.; Wellman, T.; Aktar, M.; Fjeld, C.; Demeler, B.; Ebert, A.M.; Francklyn, C.S. Neuropathy-associated histidyl-tRNA synthetase variants attenuate protein synthesis in vitro and disrupt axon outgrowth in developing zebrafish. FEBS J. 2020, 288, 142–159. [Google Scholar] [CrossRef] [PubMed]
- Abbott, J.A.; Guth, E.; Kim, C.; Regan, C.; Siu, V.M.; Rupar, C.A.; Demeler, B.; Francklyn, C.S.; Robey-Bond, S.M. The Usher Syndrome Type IIIB Histidyl-tRNA Synthetase Mutation Confers Temperature Sensitivity. Biochemistry 2017, 56, 3619–3631. [Google Scholar] [CrossRef] [PubMed]
- Theadom, A.; Roxburgh, R.; Macaulay, E.; O’Grady, G.; Burns, J.; Parmar, P.; Jones, K.; Rodrigues, M. Prevalence of Charcot-Marie-Tooth disease across the lifespan: A population-based epidemiological study. BMJ Open 2019, 9, e029240. [Google Scholar] [CrossRef]
- Barreto, L.C.L.S.; Oliveira, F.S.; Nunes, P.S.; de França Costa, I.M.P.; Garcez, C.A.; Goes, G.M.; Neves, E.L.A.; Quintans, J.D.S.S.; de Souza Araújo, A.A. Epidemiologic Study of Charcot-Marie-Tooth Disease: A Systematic Review. Neuroepidemiology 2016, 46, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Pareyson, D.; Marchesi, C. Diagnosis, natural history, and management of Charcot-Marie-Tooth disease. Lancet Neurol. 2009, 8, 654–667. [Google Scholar] [CrossRef]
- Szigeti, K.; Lupski, J.R. Charcot-Marie-Tooth disease. Eur. J. Hum. Genet. 2009, 17, 703–710. [Google Scholar] [CrossRef] [Green Version]
- Boerkoel, C.F.; Takashima, H.; Garcia, C.A.; Olney, R.K.; Johnson, J.; Berry, K.; Russo, P.; Kennedy, S.; Teebi, A.S.; Scavina, M.; et al. Charcot-Marie-Tooth disease and related neuropathies: Mutation distribution and genotype-phenotype correlation. Ann. Neurol. 2001, 51, 190–201. [Google Scholar] [CrossRef]
- Wei, N.; Zhang, Q.; Yang, X.-L. Neurodegenerative Charcot-Marie-Tooth disease as a case study to decipher novel functions of aminoacyl-tRNA synthetases. J. Biol. Chem. 2019, 294, 5321–5339. [Google Scholar] [CrossRef] [Green Version]
- Antonellis, A.; Ellsworth, R.E.; Sambuughin, N.; Puls, I.; Abel, A.; Lee-Lin, S.-Q.; Jordanova, A.; Kremensky, I.; Christodoulou, K.; Middleton, L.T.; et al. Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am. J. Hum. Genet. 2003, 72, 1293–1299. [Google Scholar] [CrossRef]
- Jordanova, A.; Irobi, J.; Thomas, F.P.; Van Dijck, P.; Meerschaert, K.; Dewil, M.; Dierick, I.; Jacobs, A.; De Vriendt, E.; Guergueltcheva, V.; et al. Disrupted function and axonal distribution of mutant tyrosyl-tRNA synthetase in dominant intermediate Charcot-Marie-Tooth neuropathy. Nat. Genet. 2006, 38, 197–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Latour, P.; Thauvin-Robinet, C.; Baudelet-Méry, C.; Soichot, P.; Cusin, V.; Faivre, L.; Locatelli, M.-C.; Mayençon, M.; Sarcey, A.; Broussolle, E.; et al. A major determinant for binding and aminoacylation of tRNAAla in cytoplasmic alanyl-tRNA synthetase is mutated in dominant axonal Charcot-Marie-Tooth disease. Am. J. Hum. Genet. 2010, 86, 77–82. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, M.; McLaughlin, H.; Houlden, H.; Guo, M.; Yo-Tsen, L.; Hadjivassilious, M.; Speziani, F.; Yang, X.-L.; Antonellis, A.; Reilly, M.M.; et al. Exome sequencing identifies a significant variant in methionyl-tRNA synthetase (MARS) in a family with late-onset CMT2. J. Neurol. Neurosurg. Psychiatry 2013, 84, 1247–1249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, K.; Long, B.; Gogonea, V.; Deshpande, G.M.; Vasu, K.; Fox, P.L. Multimodal cotranslational interactions direct assembly of the human multi-tRNA synthetase complex. Proc. Natl. Acad. Sci. USA 2022, 119, e2205669119. [Google Scholar] [CrossRef] [PubMed]
- Millán, J.M.; Aller, E.; Jaijo, T.; Blanco-Kelly, F.; Gimenez-Pardo, A.; Ayuso, C. An Update on the Genetics of Usher Syndrome. J. Ophthalmol. 2010, 2011, 417217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aparisi, M.J.; Aller, E.; Fuster-García, C.; García-García, G.; Rodrigo, R.; Vazquez-Manrique, R.P.; Blanco-Kelly, F.; Ayuso, C.; Roux, A.-F.; Jaijo, T.; et al. Targeted next generation sequencing for molecular diagnosis of Usher syndrome. Orphanet J. Rare Dis. 2014, 9, 168. [Google Scholar] [CrossRef] [Green Version]
- Whatley, M.; Francis, A.; Ng, Z.Y.; Khoh, X.E.; Atlas, M.D.; Dilley, R.J.; Wong, E.Y.M. Usher Syndrome: Genetics and Molecular Links of Hearing Loss and Directions for Therapy. Front. Genet. 2020, 11, 565216. [Google Scholar] [CrossRef]
- Li, J.; He, Y.; Lu, Q.; Zhang, M. Mechanistic Basis of Organization of the Harmonin/USH1C-Mediated Brush Border Microvilli Tip-Link Complex. Dev. Cell 2016, 36, 179–189. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Li, J.; Yao, X.; Li, W.; Du, H.; Tang, M.; Xiong, W.; Chai, R.; Xu, Z. Loss of CIB2 Causes Profound Hearing Loss and Abolishes Mechanoelectrical Transduction in Mice. Front. Mol. Neurosci. 2017, 10, 401. [Google Scholar] [CrossRef] [Green Version]
- García-García, G.; Besnard, T.; Baux, D.; Vaché, C.; Aller, E.; Malcolm, S.; Claustres, M.; Millan, J.M.; Roux, A.-F. The contribution of GPR98 and DFNB31 genes to a Spanish Usher syndrome type 2 cohort. Mol. Vis. 2013, 19, 367–373. [Google Scholar]
- Eudy, J.D.; Weston, M.D.; Yao, S.; Hoover, D.M.; Rehm, H.L.; Ma-Edmonds, M.; Yan, D.; Ahmad, I.; Cheng, J.J.; Ayuso, C.; et al. Mutation of a gene encoding a protein with extracellular matrix motifs in usher syndrome type IIa. Science 1998, 280, 1753–1757. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Chen, Q.; Almishaal, A.; Mathur, P.D.; Zheng, T.; Tian, C.; Zheng, Q.Y.; Yang, J. The roles of USH1 proteins and PDZ domain-containing USH proteins in USH2 complex integrity in cochlear hair cells. Hum. Mol. Genet. 2016, 26, 624–636. [Google Scholar] [CrossRef] [PubMed]
- Sankila, E.-M.; Pakarinen, L.; Kääriäinen, H.; Aittomäki, K.; Karjalainen, S.; Sistonen, P.; de la Chapelle, A. Assignment of an Usher syndrome type III (USH3) gene to chromosome 3q. Hum. Mol. Genet. 1995, 4, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Joensuu, T.; Hämäläinen, R.; Yuan, B.; Johnson, C.; Tegelberg, S.; Gasparini, P.; Zelante, L.; Pirvola, U.; Pakarinen, L.; Lehesjoki, A.-E.; et al. Mutations in a novel gene with transmembrane domains underlie Usher syndrome type 3. Am. J. Hum. Genet. 2001, 69, 673–684. [Google Scholar] [CrossRef] [Green Version]
- Isosomppi, J.; Västinsalo, H.; Geller, S.F.; Heon, E.; Flannery, J.G.; Sankila, E.-M. Disease-causing mutations in the CLRN1 gene alter normal CLRN1 protein trafficking to the plasma membrane. Mol. Vis. 2009, 15, 1806–1818. [Google Scholar] [PubMed]
- Dulon, D.; Papal, S.; Patni, P.; Cortese, M.; Vincent, P.F.; Tertrais, M.; Emptoz, A.; Tlili, A.; Bouleau, Y.; Michel, V.; et al. Clarin-1 gene transfer rescues auditory synaptopathy in model of Usher syndrome. J. Clin. Investig. 2018, 128, 3382–3401. [Google Scholar] [CrossRef]
- Khan, A.O.; Becirovic, E.; Betz, C.; Neuhaus, C.; Altmüller, J.; Riedmayr, L.M.; Motameny, S.; Nürnberg, G.; Nürnberg, P.; Bolz, H.J. A deep intronic CLRN1 (USH3A) founder mutation generates an aberrant exon and underlies severe Usher syndrome on the Arabian Peninsula. Sci. Rep. 2017, 7, 1411. [Google Scholar] [CrossRef] [Green Version]
- Koh, C.Y.; Wetzel, A.B.; de van der Schueren, W.J.; Hol, W.G. Comparison of histidine recognition in human and trypanosomatid histidyl-tRNA synthetases. Biochimie 2014, 106, 111–120. [Google Scholar] [CrossRef] [Green Version]
- Boczonadi, V.; Jennings, M.J.; Horvath, R. The role of tRNA synthetases in neurological and neuromuscular disorders. FEBS Lett. 2018, 592, 703–717. [Google Scholar] [CrossRef] [Green Version]
- Bauhammer, J.; Fiehn, C. Antisynthetase syndrome. Z. Rheumatol. 2019, 78, 645–655. [Google Scholar] [CrossRef]
- Soto-Mota, A.; Norwitz, N.G.; Evans, R.D.; Clarke, K. Exogenous d-β-hydroxybutyrate lowers blood glucose in part by decreasing the availability of L-alanine for gluconeogenesis. Endocrinol. Diabetes Metab. 2021, 5, e00300. [Google Scholar] [CrossRef]
- Thalacker-Mercer, A.E.; Gheller, M.E. Benefits and Adverse Effects of Histidine Supplementation. J. Nutr. 2020, 150, 2588S–2592S. [Google Scholar] [CrossRef] [PubMed]
- Shibui, Y.; Miwa, T.; Yamashita, M.; Chin, K.; Kodama, T. A 4-week Repeated Dose Toxicity Study of Glycine in Rats by Gavage Administration. J. Toxicol. Pathol. 2013, 26, 405–412. [Google Scholar] [CrossRef] [Green Version]
- Zuko, A.; Mallik, M.; Thompson, R.; Spaulding, E.L.; Wienand, A.R.; Been, M.; Tadenev, A.L.D.; van Bakel, N.; Sijlmans, C.; Santos, L.A.; et al. tRNA overexpression rescues peripheral neuropathy caused by mutations in tRNA synthetase. Science 2021, 373, 1161–1166. [Google Scholar] [CrossRef] [PubMed]
- Albers, S.; Beckert, B.; Matthies, M.C.; Mandava, C.S.; Schuster, R.; Seuring, C.; Riedner, M.; Sanyal, S.; Torda, A.E.; Wilson, D.N.; et al. Repurposing tRNAs for nonsense suppression. Nat. Commun. 2021, 12, 3850. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Y.; Mendonca, C.A.; Yukselen, O.; Muneeruddin, K.; Ren, L.; Liang, J.; Zhou, C.; Xie, J.; Li, J.; et al. AAV-delivered suppressor tRNA overcomes a nonsense mutation in mice. Nature 2022, 604, 343–348. [Google Scholar] [CrossRef]
- Paunovska, K.; Loughrey, D.; Dahlman, J.E. Drug delivery systems for RNA therapeutics. Nat. Rev. Genet. 2022, 23, 265–280. [Google Scholar] [CrossRef]
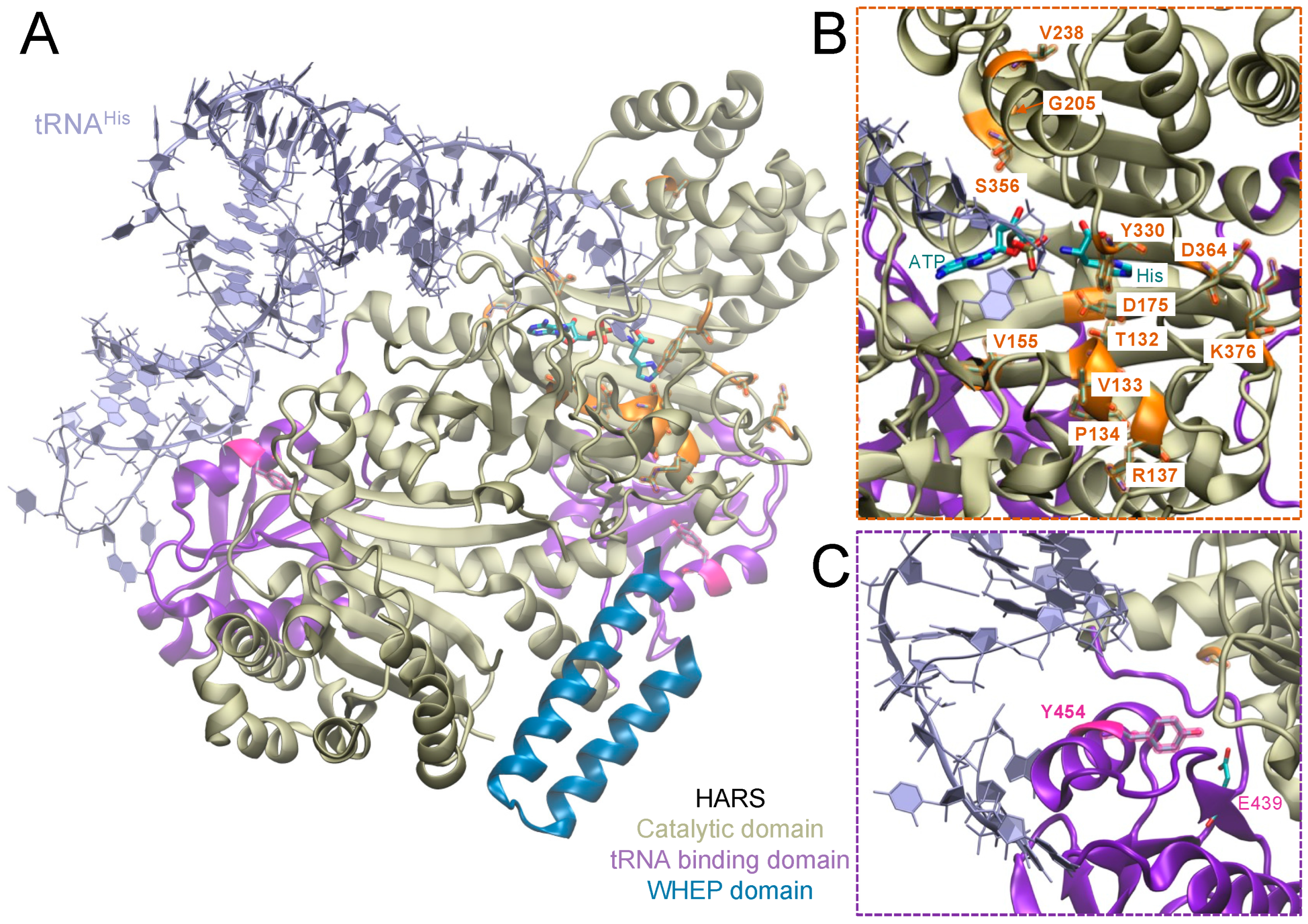

| HARS Mutation | Publication |
|---|---|
| T132I | Safka Brozkova et al., 2015 [39], Blocquel et al., 2019 [29] |
| T132S | Safka Brozkova et al., 2015 [39] |
| V133F | Royer-Bertrand et al., 2019 [40], Qiu et al., 2022 [51] |
| P134H | Safka Brozkova et al., 2015 [39], Blocquel et al., 2019 [29] |
| R137Q | Vester et al., 2013 [38], Mullen et al., 2021 [52] |
| V155G | Abbott et al., 2018 [41], Mullen et al., 2021 [52], Qiu et al., 2022 [51] |
| D175E | Safka Brozkova et al., 2015 [39], Blocquel et al., 2019 [29] |
| G205D | Vester et al., 2013 [38] |
| V238A | Vester et al., 2013 [38] |
| Y330C | Abbott et al., 2018 [41], Mullen et al., 2021 [52], Qiu et al., 2022 [51] |
| S356N | Abbott et al., 2018 [41], Qiu et al., 2022 [51] |
| D364Y | Safka Brozkova et al., 2015 [39], Blocquel et al., 2019 [29] |
| K376R | Vester et al., 2013 [38] |
| Y454S | Puffenberger et al., 2012 [43], Abbott et al., 2017 [53], Qiu et al., 2022 [51] |
| P505S | Vester et al., 2013 [38] |
| Active Site | tRNA Binding Domain | Other Domains | |
|---|---|---|---|
| AARS | N71Y, G102R, R326W, R329H, E337K | S627L, E688G, E778A, D893N | |
| GARS | E71G, L129P, D167Y, D146N, C157R, H162R, C201R, S211F, H216R, L218Q, P224L P234K/Y, M238R, G240R, P244L, S265Y, S273R, E279D, I280F, G327R, P336H/R, H418R, H472R, D500N, K510Q, G526R | M555V, S581L, G625R | K27R, K27P, A57V |
| HARS | T132I, P134H, R137Q, V155G, D175E, Y330C, S356N, D364Y | Y454S (USH3B) | |
| MARS | A397T | R618C, R737W, P800T | |
| YARS | G41R, D81I, D(153–156), E196K, E196Q | ||
| WARS | H257R, D314G |
| Associated genes/loci | USH-Type I | USH-Type II | USH-Type III |
| MYO7A/USH1B | USH2A | CLRN1 | |
| CDH23/USH1D | GPR98/USH2C | HARS | |
| PCDH15 | DFNB31/USH2D | ||
| USH1C | |||
| USH1E | |||
| USH1G | |||
| USH1H | |||
| USH1K | |||
| CIB2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wilhelm, S.D.P.; Kenana, R.; Qiu, Y.; O’Donoghue, P.; Heinemann, I.U. Towards a Cure for HARS Disease. Genes 2023, 14, 254. https://doi.org/10.3390/genes14020254
Wilhelm SDP, Kenana R, Qiu Y, O’Donoghue P, Heinemann IU. Towards a Cure for HARS Disease. Genes. 2023; 14(2):254. https://doi.org/10.3390/genes14020254
Chicago/Turabian StyleWilhelm, Sarah D. P., Rosan Kenana, Yi Qiu, Patrick O’Donoghue, and Ilka U. Heinemann. 2023. "Towards a Cure for HARS Disease" Genes 14, no. 2: 254. https://doi.org/10.3390/genes14020254
APA StyleWilhelm, S. D. P., Kenana, R., Qiu, Y., O’Donoghue, P., & Heinemann, I. U. (2023). Towards a Cure for HARS Disease. Genes, 14(2), 254. https://doi.org/10.3390/genes14020254






