In Silico Identification and Characterization of Satellite DNAs in 23 Drosophila Species from the Montium Group
Abstract
1. Introduction
2. Materials and Methods
Satellite DNA Identification
3. Results and Discussion
3.1. Identification of Satellite DNA Families in the Montium Group
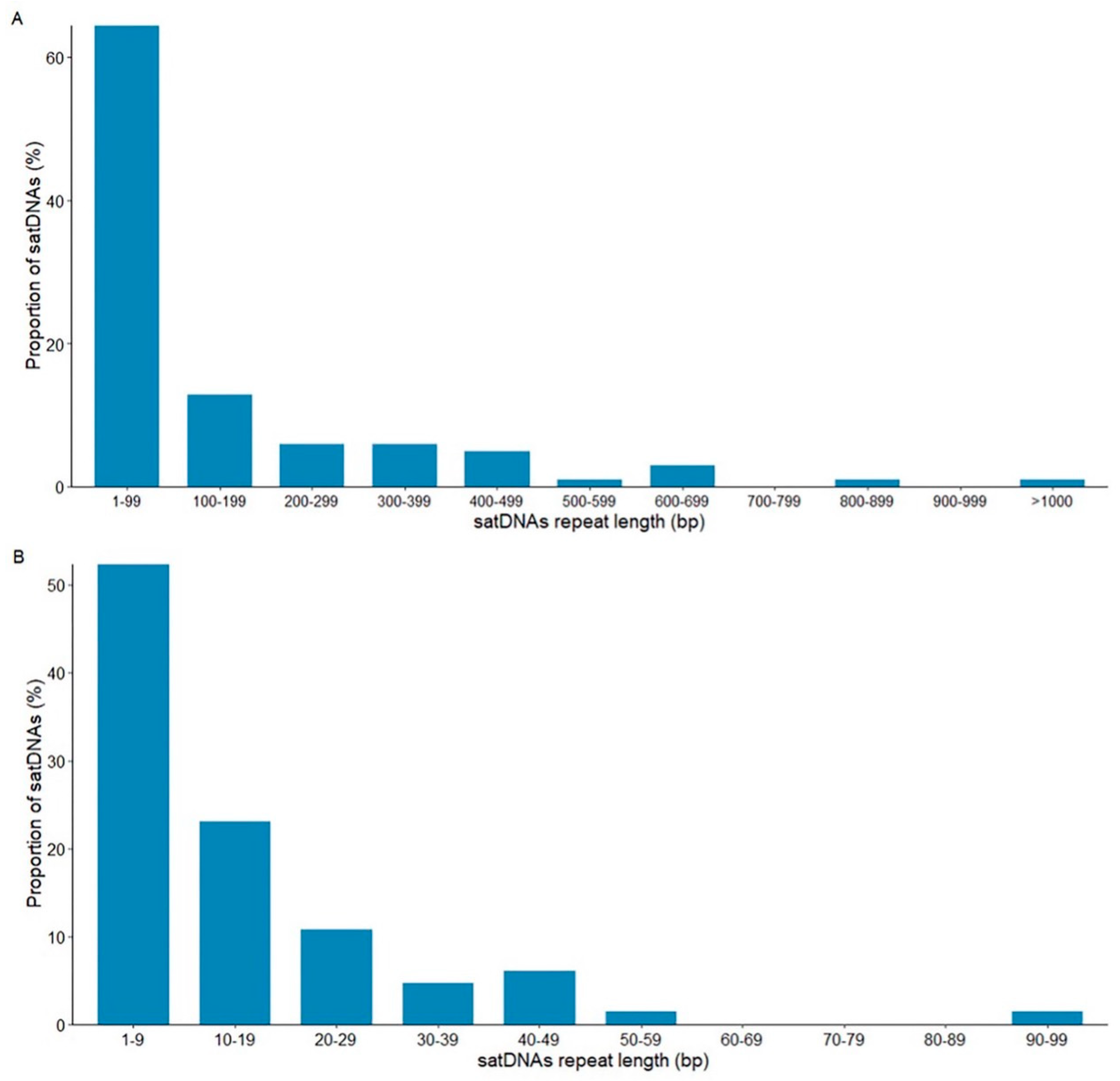
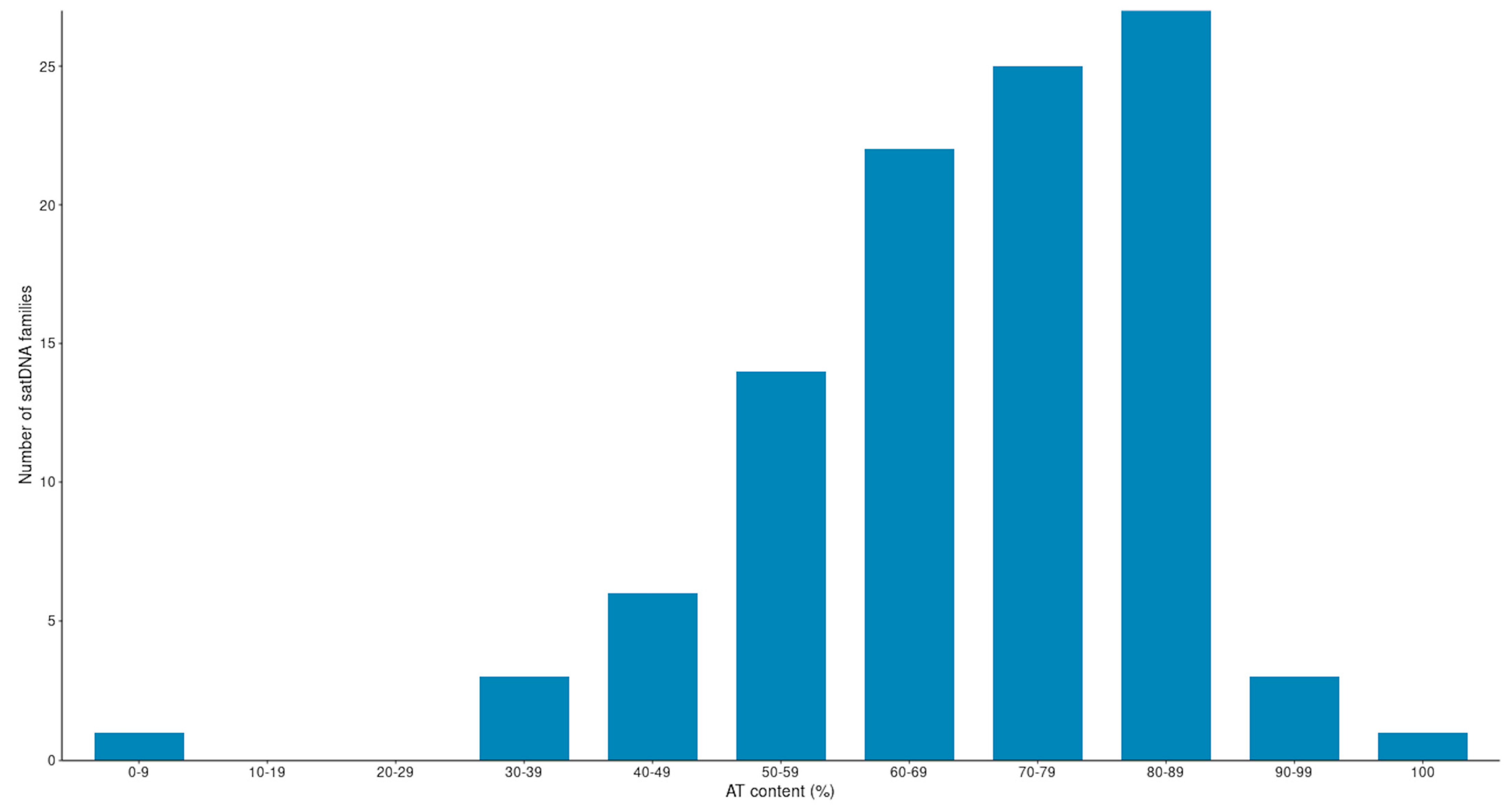
3.2. Satellite DNAs in the Montium Group: General Structural Features
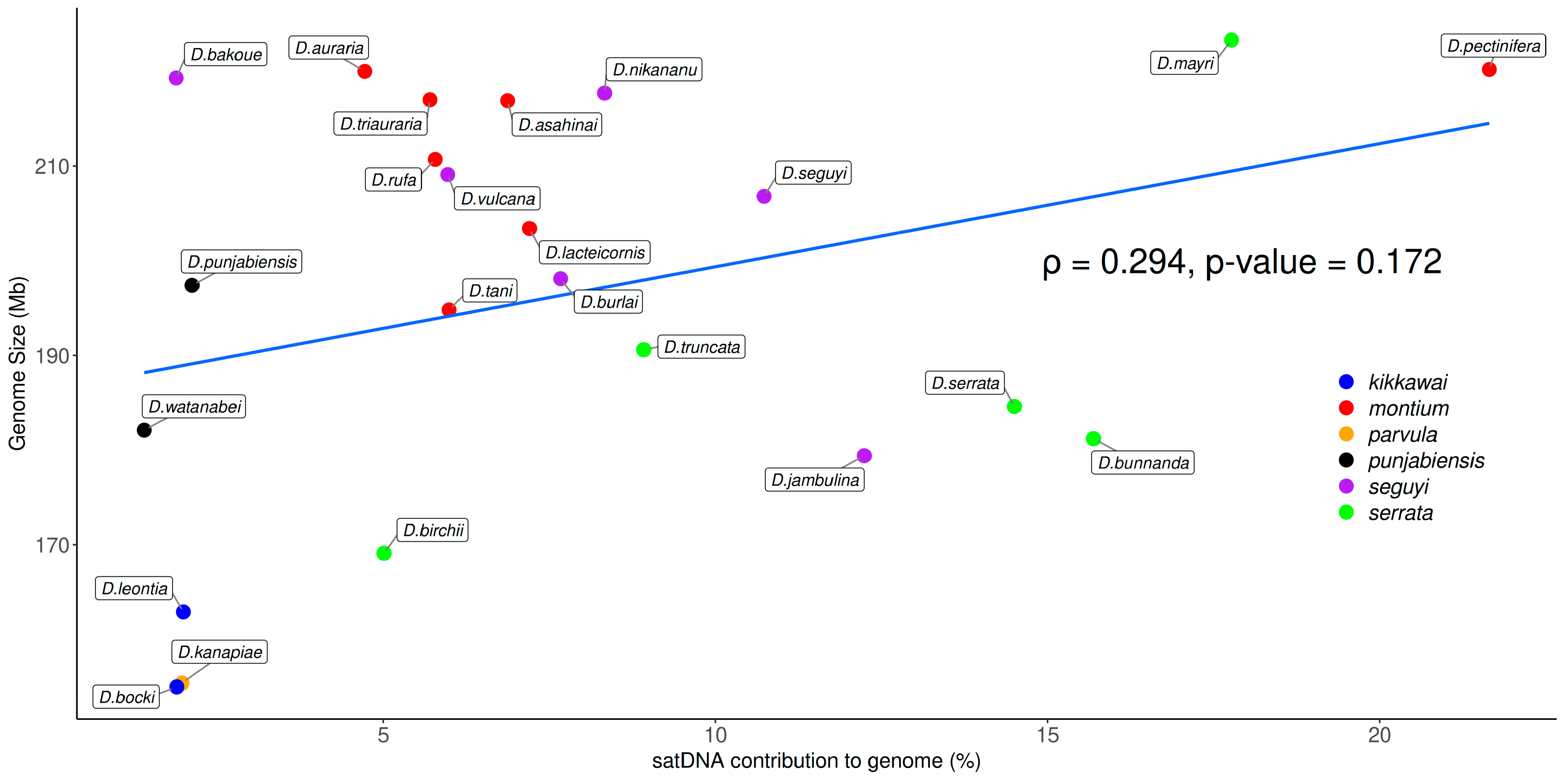
3.3. Satellite DNA Abundance and Relationship with Genome Sizes
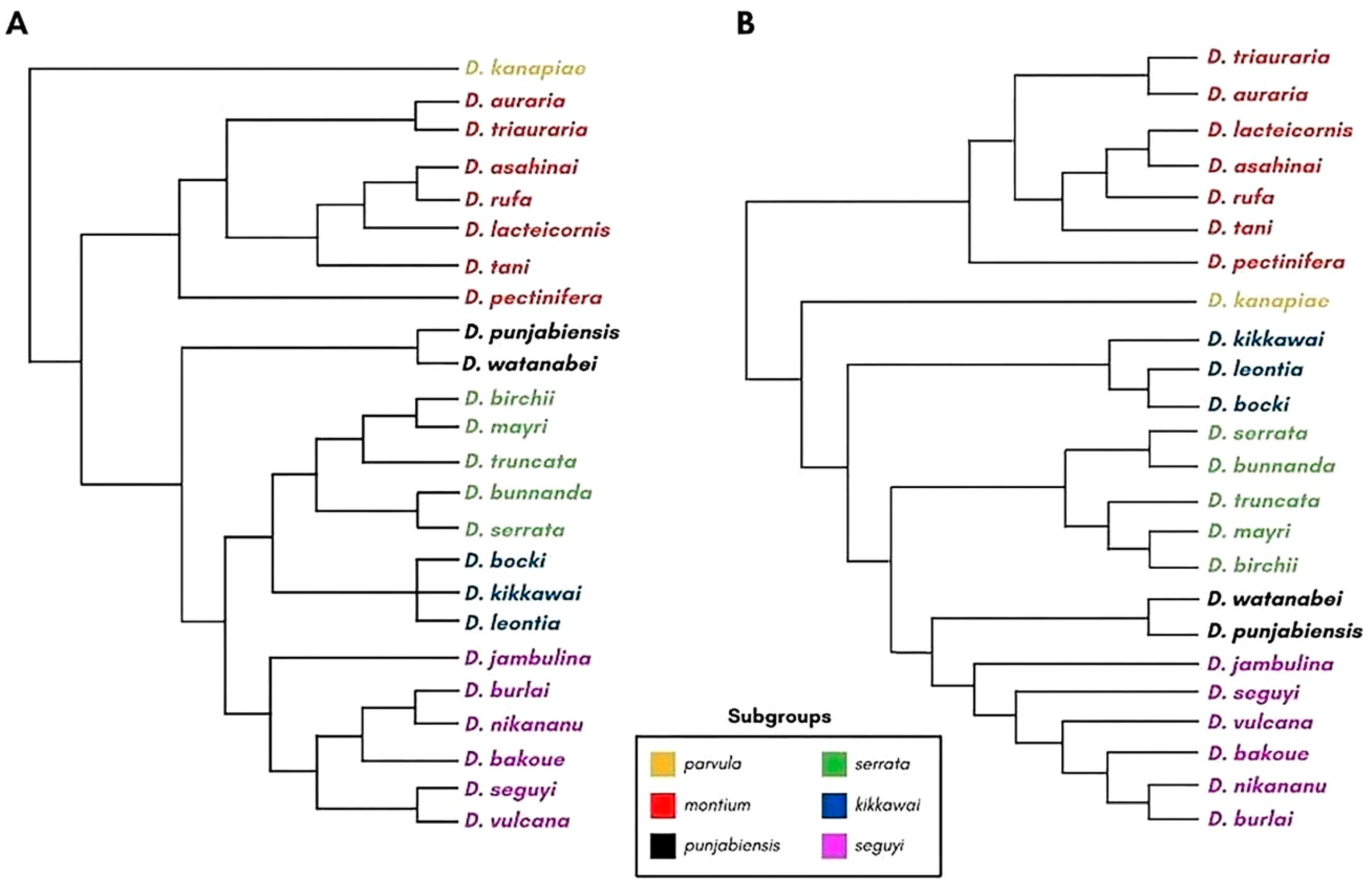
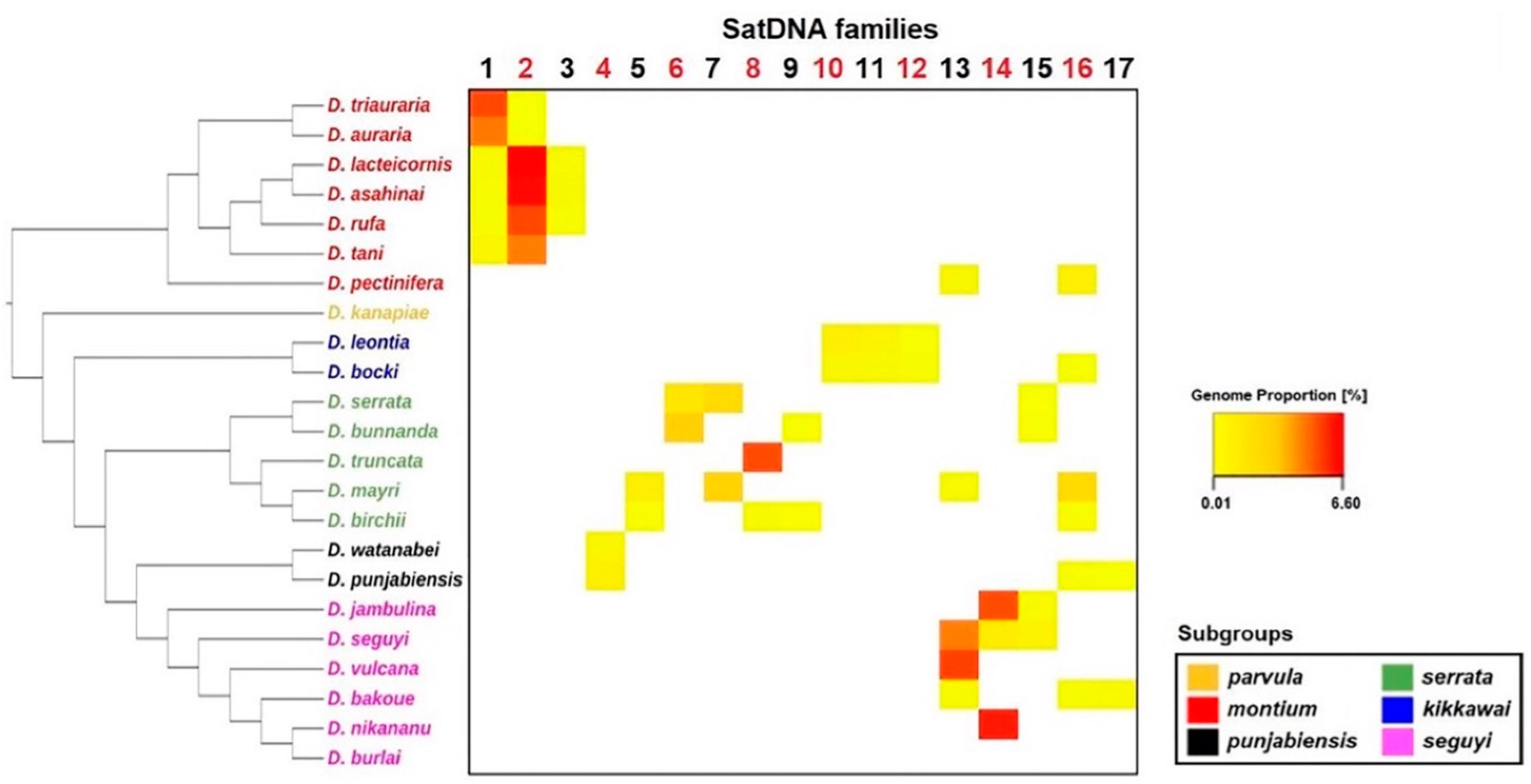
3.4. Satellite DNA Distribution across the Montium Phylogeny
3.5. Satellite DNA Emergence from DINEs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- de Koning, A.P.J.; Gu, W.; Castoe, T.A.; Batzer, M.A.; Pollock, D.D. Repetitive Elements May Comprise Over Two-Thirds of the Human Genome. PLoS Genet 2011, 7, e1002384. [Google Scholar] [CrossRef] [PubMed]
- López-Flores, I.; Garrido-Ramos, M.A. The Repetitive DNA Content of Eukaryotic Genomes. In Genome Dynamics; Garrido-Ramos, M.A., Ed.; S. Karger AG: Basel, Switzerland, 2012; Volume 7, pp. 1–28. ISBN 978-3-318-02149-3. [Google Scholar]
- Tautz, D. Notes on the Definition and Nomenclature of Tandemly Repetitive DNA Sequences. In DNA Fingerprinting: State of the Science; Pena, S.D.J., Chakraborty, R., Epplen, J.T., Jeffreys, A.J., Eds.; Birkhäuser Basel: Basel, Switzerland, 1993; pp. 21–28. ISBN 978-3-7643-2906-8. [Google Scholar]
- Charlesworth, B.; Sniegowski, P.; Stephan, W. The Evolutionary Dynamics of Repetitive DNA in Eukaryotes. Nature 1994, 371, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Plohl, M.; Meštrović, N.; Mravinac, B. Satellite DNA Evolution. In Genome Dynamics; Garrido-Ramos, M.A., Ed.; S. Karger AG: Basel, Switzerland, 2012; Volume 7, pp. 126–152. ISBN 978-3-318-02149-3. [Google Scholar]
- Garrido-Ramos, M. Satellite DNA: An Evolving Topic. Genes 2017, 8, 230. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, G.C.S.; Küttler, H.; Moreira-Filho, O.; Heslop-Harrison, J.S.; Heslop-Harrison, J.S. The 1.688 Repetitive DNA of Drosophila: Concerted Evolution at Different Genomic Scales and Association with Genes. Mol. Biol. Evol. 2011, 29, 7–11. [Google Scholar] [CrossRef]
- Brajković, J.; Pezer, Ž.; Bruvo-Mađarić, B.; Sermek, A.; Feliciello, I.; Ugarković, Đ. Dispersion Profiles and Gene Associations of Repetitive DNAs in the Euchromatin of the Beetle Tribolium castaneum. G3 Genes Genomes Genet. 2018, 8, 875–886. [Google Scholar] [CrossRef]
- Sproul, J.S.; Khost, D.E.; Eickbush, D.G.; Negm, S.; Wei, X.; Wong, I.; Larracuente, A.M. Dynamic Evolution of Euchromatic Satellites on the X Chromosome in Drosophila melanogaster and the simulans Clade. Mol. Biol. Evol. 2020, 37, 2241–2256. [Google Scholar] [CrossRef]
- Feliciello, I.; Pezer, Ž.; Sermek, A.; Bruvo Mađarić, B.; Ljubić, S.; Ugarković, Đ. Satellite DNA-Mediated Gene Expression Regulation: Physiological and Evolutionary Implication. In Satellite DNAs in Physiology and Evolution; Ugarković, Ð., Ed.; Progress in Molecular and Subcellular Biology; Springer International Publishing: Cham, Switzerland, 2021; Volume 60, pp. 145–167. ISBN 978-3-030-74888-3. [Google Scholar]
- Lauria Sneideman, M.P.; Meller, V.H. Drosophila Satellite Repeats at the Intersection of Chromatin, Gene Regulation and Evolution. In Satellite DNAs in Physiology and Evolution; Ugarković, Ð., Ed.; Progress in Molecular and Subcellular Biology; Springer International Publishing: Cham, Switzerland, 2021; Volume 60, pp. 1–26. ISBN 978-3-030-74888-3. [Google Scholar]
- Pathak, R.; Mamillapalli, A.; Rangaraj, N.; Kumar, R.; Vasanthi, D.; Mishra, K.; Mishra, R.K. AAGAG Repeat RNA Is an Essential Component of Nuclear Matrix in Drosophila. RNA Biol. 2013, 10, 564–571. [Google Scholar] [CrossRef]
- Jagannathan, M.; Cummings, R.; Yamashita, Y.M. A Conserved Function for Pericentromeric Satellite DNA. eLife 2018, 7, e34122. [Google Scholar] [CrossRef]
- Jagannathan, M.; Cummings, R.; Yamashita, Y.M. The Modular Mechanism of Chromocenter Formation in Drosophila. eLife 2019, 8, e43938. [Google Scholar] [CrossRef]
- Rošić, S.; Köhler, F.; Erhardt, S. Repetitive Centromeric Satellite RNA Is Essential for Kinetochore Formation and Cell Division. J. Cell Biol. 2014, 207, 335–349. [Google Scholar] [CrossRef]
- Louzada, S.; Lopes, M.; Ferreira, D.; Adega, F.; Escudeiro, A.; Gama-Carvalho, M.; Chaves, R. Decoding the Role of Satellite DNA in Genome Architecture and Plasticity—An Evolutionary and Clinical Affair. Genes 2020, 11, 72. [Google Scholar] [CrossRef] [PubMed]
- Flynn, J.M.; Hu, K.B.; Clark, A.G. Three Recent Sex Chromosome-to-Autosome Fusions in a Drosophila virilis Strain with High Satellite Content. bioRxiv 2021, 2021-06. [Google Scholar]
- Ferree, P.M.; Barbash, D.A. Species-Specific Heterochromatin Prevents Mitotic Chromosome Segregation to Cause Hybrid Lethality in Drosophila. PLoS Biol. 2009, 7, e1000234. [Google Scholar] [CrossRef]
- Ugarkovic, D. Functional Elements Residing within Satellite DNAs. EMBO Rep. 2005, 6, 1035–1039. [Google Scholar] [CrossRef]
- Ahmad, S.F.; Singchat, W.; Jehangir, M.; Suntronpong, A.; Panthum, T.; Malaivijitnond, S.; Srikulnath, K. Dark Matter of Primate Genomes: Satellite DNA Repeats and Their Evolutionary Dynamics. Cells 2020, 9, 2714. [Google Scholar] [CrossRef] [PubMed]
- Strachan, T.; Webb, D.; Dover, G.A. Transition Stages of Molecular Drive in Multiple-Copy DNA Families in Drosophila. EMBO J. 1985, 4, 1701–1708. [Google Scholar] [CrossRef] [PubMed]
- Lohe, A.R.; Hilliker, A.J.; Roberts, P.A. Mapping Simple Repeated DNA Sequences in Heterochromatin of Drosophila melanogaster. Genetics 1993, 134, 1149–1174. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, L.; Sperlich, D. Gradual evolution of a specific satellite DNA family in Drosophila ambigua, D. tristis, and D. obscura. Mol. Biol. Evol. 1993, 10, 647–659. [Google Scholar] [CrossRef]
- Kuhn, G.C.S. Satellite DNA Transcripts Have Diverse Biological Roles in Drosophila. Heredity 2015, 115, 1–2. [Google Scholar] [CrossRef]
- Khost, D.E.; Eickbush, D.G.; Larracuente, A.M. Single-Molecule Sequencing Resolves the Detailed Structure of Complex Satellite DNA Loci in Drosophila melanogaster. Genome Res. 2017, 27, 709–721. [Google Scholar] [CrossRef]
- Kuhn, G.C.S.; Heringer, P.; Dias, G.B. Structure, Organization, and Evolution of Satellite DNAs: Insights from the Drosophila repleta and D. virilis Species Groups. In Satellite DNAs in Physiology and Evolution; Ugarković, Ð., Ed.; Progress in Molecular and Subcellular Biology; Springer International Publishing: Cham, Switzerland, 2021; Volume 60, pp. 27–56. ISBN 978-3-030-74888-3. [Google Scholar]
- Novák, P.; Robledillo, L.Á.; Koblížková, A.; Vrbová, I.; Neumann, P.; Macas, J. TAREAN: A computational tool for identification and characterization of satellite DNA from unassembled short reads. Nucleic Acids Res. 2017, 45, e111. [Google Scholar] [CrossRef] [PubMed]
- Bronski, M.J.; Martinez, C.C.; Weld, H.A.; Eisen, M.B. Whole Genome Sequences of 23 Species from the Drosophila montium Species Group (Diptera: Drosophilidae): A Resource for Testing Evolutionary Hypotheses. G3 Genes Genomes Genet. 2020, 10, 1443–1455. [Google Scholar] [CrossRef] [PubMed]
- Yassin, A. Phylogenetic Biogeography and Classification of the Drosophila montium Species Group (Diptera: Drosophilidae). Ann. Société Entomol. Fr. (N.S.) 2018, 54, 167–175. [Google Scholar] [CrossRef]
- Conner, W.R.; Delaney, E.K.; Bronski, M.J.; Ginsberg, P.S.; Wheeler, T.B.; Richardson, K.M.; Peckenpaugh, B.; Kim, K.J.; Watada, M.; Hoffmann, A.A.; et al. A Phylogeny for the Drosophila montium Species Group: A Model Clade for Comparative Analyses. Mol. Phylogenetics Evol. 2021, 158, 107061. [Google Scholar] [CrossRef] [PubMed]
- Han, M.V.; Zmasek, C.M. PhyloXML: XML for Evolutionary Biology and Comparative Genomics. BMC Bioinform. 2009, 10, 356. [Google Scholar] [CrossRef]
- Baimai, V. Metaphase Karyotypes of Certain Species of the Drosophila montium Subgroup. JPN J. Genet. 1980, 55, 165–175. [Google Scholar] [CrossRef]
- Venkat, S.; Ranganath, R.A. Localization and Characterization of Heterochromatin among Four Species of the montium Subgroup of Drosophila. Cytologia 2007, 72, 279–286. [Google Scholar] [CrossRef]
- Ma, J.; Jackson, S.A. Retrotransposon Accumulation and Satellite Amplification Mediated by Segmental Duplication Facilitate Centromere Expansion in Rice. Genome Res. 2006, 16, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, G.C.S.; Teo, C.H.; Schwarzacher, T.; Heslop-Harrison, J.S. Evolutionary Dynamics and Sites of Illegitimate Recombination Revealed in the Interspersion and Sequence Junctions of Two Nonhomologous Satellite DNAs in Cactophilic Drosophila Species. Heredity 2009, 102, 453–464. [Google Scholar] [CrossRef]
- Wei, K.H.-C.; Grenier, J.K.; Barbash, D.A.; Clark, A.G. Correlated Variation and Population Differentiation in Satellite DNA Abundance among Lines of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2014, 111, 18793–18798. [Google Scholar] [CrossRef]
- Ávila Robledillo, L.; Koblížková, A.; Novák, P.; Böttinger, K.; Vrbová, I.; Neumann, P.; Schubert, I.; Macas, J. Satellite DNA in Vicia faba Is Characterized by Remarkable Diversity in Its Sequence Composition, Association with Centromeres, and Replication Timing. Sci. Rep. 2018, 8, 5838. [Google Scholar] [CrossRef] [PubMed]
- Palacios-Gimenez, O.M.; Koelman, J.; Palmada-Flores, M.; Bradford, T.M.; Jones, K.K.; Cooper, S.J.B.; Kawakami, T.; Suh, A. Comparative Analysis of Morabine Grasshopper Genomes Reveals Highly Abundant Transposable Elements and Rapidly Proliferating Satellite DNA Repeats. BMC Biol. 2020, 18, 199. [Google Scholar] [CrossRef] [PubMed]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy Platform for Accessible, Reproducible and Collaborative Biomedical Analyses: 2018 Update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef] [PubMed]
- Novák, P.; Neumann, P.; Macas, J. Global Analysis of Repetitive DNA from Unassembled Sequence Reads Using RepeatExplorer2. Nat. Protoc. 2020, 15, 3745–3776. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- de Lima, L.G.; Svartman, M.; Kuhn, G.C.S. Dissecting the Satellite DNA Landscape in Three Cactophilic Drosophila Sequenced Genomes. G3 Genes Genomes Genet. 2017, 7, 2831–2843. [Google Scholar] [CrossRef]
- Silva, B.S.M.L.; Heringer, P.; Dias, G.B.; Svartman, M.; Kuhn, G.C.S. De Novo Identification of Satellite DNAs in the Sequenced Genomes of Drosophila virilis and D. americana Using the RepeatExplorer and TAREAN Pipelines. PLoS ONE 2019, 14, e0223466. [Google Scholar] [CrossRef]
- da Silva, M.J.; Fogarin Destro, R.; Gazoni, T.; Narimatsu, H.; Pereira dos Santos, P.S.; Haddad, C.F.B.; Parise-Maltempi, P.P. Great Abundance of Satellite DNA in Proceratophrys (Anura, Odontophrynidae) Revealed by Genome Sequencing. Cytogenet. Genome Res. 2020, 160, 141–147. [Google Scholar] [CrossRef]
- Sena, R.S.; Heringer, P.; Valeri, M.P.; Pereira, V.S.; Kuhn, G.C.S.; Svartman, M. Identification and Characterization of Satellite DNAs in Two-Toed Sloths of the Genus Choloepus (Megalonychidae, Xenarthra). Sci. Rep. 2020, 10, 19202. [Google Scholar] [CrossRef]
- DiBartolomeis, S.M.; Tartof, K.D.; Jackson, F.R. A superfamily of Drosophila satellite related (SR) DNA repeats restricted to the X chromosome euchromatin. Nucleic Acids Res. 1992, 20, 1113–1116. [Google Scholar] [CrossRef]
- Lohe, A.R.; Brutlag, D.L. Adjacent Satellite DNA Segments in Drosophila. J. Mol. Biol. 1987, 194, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Jagannathan, M.; Warsinger-Pepe, N.; Watase, G.J.; Yamashita, Y.M. Comparative Analysis of Satellite DNA in the Drosophila melanogaster Species Complex. G3 Genes Genomes Genet. 2017, 7, 693–704. [Google Scholar] [CrossRef]
- de Lima, L.G.; Ruiz-Ruano, F.J. In-Depth Satellitome Analyses of 37 Drosophila Species Illuminate Repetitive DNA Evolution in the Drosophila Genus. Genome Biol. Evol. 2022, 14, evac064. [Google Scholar] [CrossRef]
- Palomeque, T.; Lorite, P. Satellite DNA in Insects: A Review. Heredity 2008, 100, 564–573. [Google Scholar] [CrossRef] [PubMed]
- Melters, D.P.; Bradnam, K.R.; Young, H.A.; Telis, N.; May, M.R.; Ruby, J.; Sebra, R.; Peluso, P.; Eid, J.; Rank, D.; et al. Comparative Analysis of Tandem Repeats from Hundreds of Species Reveals Unique Insights into Centromere Evolution. Genome Biol. 2013, 14, R10. [Google Scholar] [CrossRef]
- Gall, J.; Cohen, E.; Polan, M. Repetitive DNA Sequences in Drosophila. Chromosoma 1971, 33, 319–344. [Google Scholar] [CrossRef] [PubMed]
- Gall, J.G.; Atherton, D.D. Satellite DNA Sequences in Drosophila virilis. J. Mol. Biol. 1974, 85, 633–664. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, E.R. Two AT-Rich Satellite DNAs in the Chironomid Glyptotendipes barbipes (Staeger): Isolation and Localization in Polytene Chromosomes of G. barbipes and Chironomus thummi. Chromosoma 1980, 79, 315–328. [Google Scholar] [CrossRef]
- Ganal, M.; Hemleben, V. Different AT-Rich Satellite DNAs in Cucurbita pepo and Cucurbita maxima. Theoret. Appl. Genetics 1986, 73, 129–135. [Google Scholar] [CrossRef]
- Bosco, G.; Campbell, P.; Leiva-Neto, J.T.; Markow, T.A. Analysis of Drosophila Species Genome Size and Satellite DNA Content Reveals Significant Differences Among Strains as Well as Between Species. Genetics 2007, 177, 1277–1290. [Google Scholar] [CrossRef] [PubMed]
- Craddock, E.M.; Gall, J.G.; Jonas, M. Hawaiian Drosophila Genomes: Size Variation and Evolutionary Expansions. Genetica 2016, 144, 107–124. [Google Scholar] [CrossRef]
- Bachmann, L.; Venanzetti, F.; Sbordoni, V. Characterization of a Species-Specific Satellite DNA Family of Dolichopoda schiavazzii (Orthoptera, Rhaphidophoridae) Cave Crickets. J. Mol. Evol. 1994, 39, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.-P.; Barbash, D.A. Abundant and Species-Specific DINE-1 Transposable Elements in 12 Drosophila Genomes. Genome Biol. 2008, 9, R39. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.; Pritham, E.J. Helitrons, the Eukaryotic Rolling-Circle Transposable Elements. Microbiol. Spectr. 2015, 3, 3–4. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A Multiple Sequence Alignment Method with Reduced Time and Space Complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef]
- Dias, G.B.; Heringer, P.; Svartman, M.; Kuhn, G.C.S. Helitrons Shaping the Genomic Architecture of Drosophila: Enrichment of DINE-TR1 in α- and β-Heterochromatin, Satellite DNA Emergence, and PiRNA Expression. Chromosome. Res. 2015, 23, 597–613. [Google Scholar] [CrossRef] [PubMed]
- Kohany, O.; Gentles, A.J.; Hankus, L.; Jurka, J. Annotation, Submission and Screening of Repetitive Elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinform. 2006, 7, 474. [Google Scholar] [CrossRef]
- Allen, S.L.; Delaney, E.K.; Kopp, A.; Chenoweth, S.F. Single-Molecule Sequencing of the Drosophila serrata Genome. G3 Genes Genomes Genet. 2017, 7, 781–788. [Google Scholar] [CrossRef]
- Dias, G.B.; Svartman, M.; Delprat, A.; Ruiz, A.; Kuhn, G.C.S. Tetris Is a Foldback Transposon That Provided the Building Blocks for an Emerging Satellite DNA of Drosophila virilis. Genome Biol. Evol. 2014, 6, 1302–1313. [Google Scholar] [CrossRef]
- Dias, C.A.R.; Kuhn, G.C.S.; Svartman, M.; dos Santos Júnior, J.E.; Santos, F.R.; Pinto, C.M.; Perini, F.A. Identification and Characterization of Repetitive DNA in the Genus Didelphis Linnaeus, 1758 (Didelphimorphia, Didelphidae) and the Use of Satellite DNAs as Phylogenetic Markers. Genet. Mol. Biol. 2021, 44, e20200384. [Google Scholar] [CrossRef] [PubMed]
- Valeri, M.P.; Dias, G.B.; do Espírito Santo, A.A.; Moreira, C.N.; Yonenaga-Yassuda, Y.; Sommer, I.B.; Kuhn, G.C.S.; Svartman, M. First Description of a Satellite DNA in Manatees’ Centromeric Regions. Front. Genet. 2021, 12, 694866. [Google Scholar] [CrossRef] [PubMed]
- Gasparotto, A.E.; Milani, D.; Martí, E.; Ferretti, A.B.S.M.; Bardella, V.B.; Hickmann, F.; Zrzavá, M.; Marec, F.; Cabral-de-Mello, D.C. A Step Forward in the Genome Characterization of the Sugarcane Borer, Diatraea saccharalis: Karyotype Analysis, Sex Chromosome System and Repetitive DNAs through a Cytogenomic Approach. Chromosoma 2022, 131, 253–267. [Google Scholar] [CrossRef] [PubMed]



| Species | Subgroup | HC satDNAs (Before Filtering) | LC satDNAs (Before Filtering) | Final Number of satDNA-like Families (After Filtering) |
|---|---|---|---|---|
| D. kanapiae | parvula | 13 | 9 | 4 |
| D. auraria | montium | 3 | 10 | 2 |
| D. triauraria | montium | 6 | 5 | 3 |
| D. asahinai | montium | 7 | 8 | 3 |
| D. rufa | montium | 4 | 7 | 3 |
| D. lacteicornis | montium | 6 | 5 | 3 |
| D. tani | montium | 7 | 9 | 4 |
| D. pectinifera | montium | 14 | 5 | 10 |
| D. punjabiensis | punjabiensis | 14 | 5 | 6 |
| D. watanabei | punjabiensis | 7 | 7 | 3 |
| D. birchii | serrata | 15 | 5 | 8 |
| D. mayri | serrata | 15 | 8 | 13 |
| D. truncata | serrata | 11 | 6 | 5 |
| D. bunnanda | serrata | 24 | 6 | 14 |
| D. serrata | serrata | 12 | 9 | 6 |
| D. bocki | kikkawai | 10 | 4 | 7 |
| D. leontia | kikkawai | 5 | 7 | 4 |
| D. jambulina | seguyi | 8 | 2 | 6 |
| D. burlai | seguyi | 11 | 7 | 7 |
| D. nikananu | seguyi | 6 | 6 | 3 |
| D. bakoue | seguyi | 25 | 8 | 9 |
| D. seguyi | seguyi | 17 | 6 | 12 |
| D. vulcana | seguyi | 5 | 8 | 3 |
| Total | 245 | 152 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, B.S.M.L.; Picorelli, A.C.R.; Kuhn, G.C.S. In Silico Identification and Characterization of Satellite DNAs in 23 Drosophila Species from the Montium Group. Genes 2023, 14, 300. https://doi.org/10.3390/genes14020300
Silva BSML, Picorelli ACR, Kuhn GCS. In Silico Identification and Characterization of Satellite DNAs in 23 Drosophila Species from the Montium Group. Genes. 2023; 14(2):300. https://doi.org/10.3390/genes14020300
Chicago/Turabian StyleSilva, Bráulio S. M. L., Agnello C. R. Picorelli, and Gustavo C. S. Kuhn. 2023. "In Silico Identification and Characterization of Satellite DNAs in 23 Drosophila Species from the Montium Group" Genes 14, no. 2: 300. https://doi.org/10.3390/genes14020300
APA StyleSilva, B. S. M. L., Picorelli, A. C. R., & Kuhn, G. C. S. (2023). In Silico Identification and Characterization of Satellite DNAs in 23 Drosophila Species from the Montium Group. Genes, 14(2), 300. https://doi.org/10.3390/genes14020300







