New Insights into the Genetics and Epigenetics of Aging Plasticity
Abstract
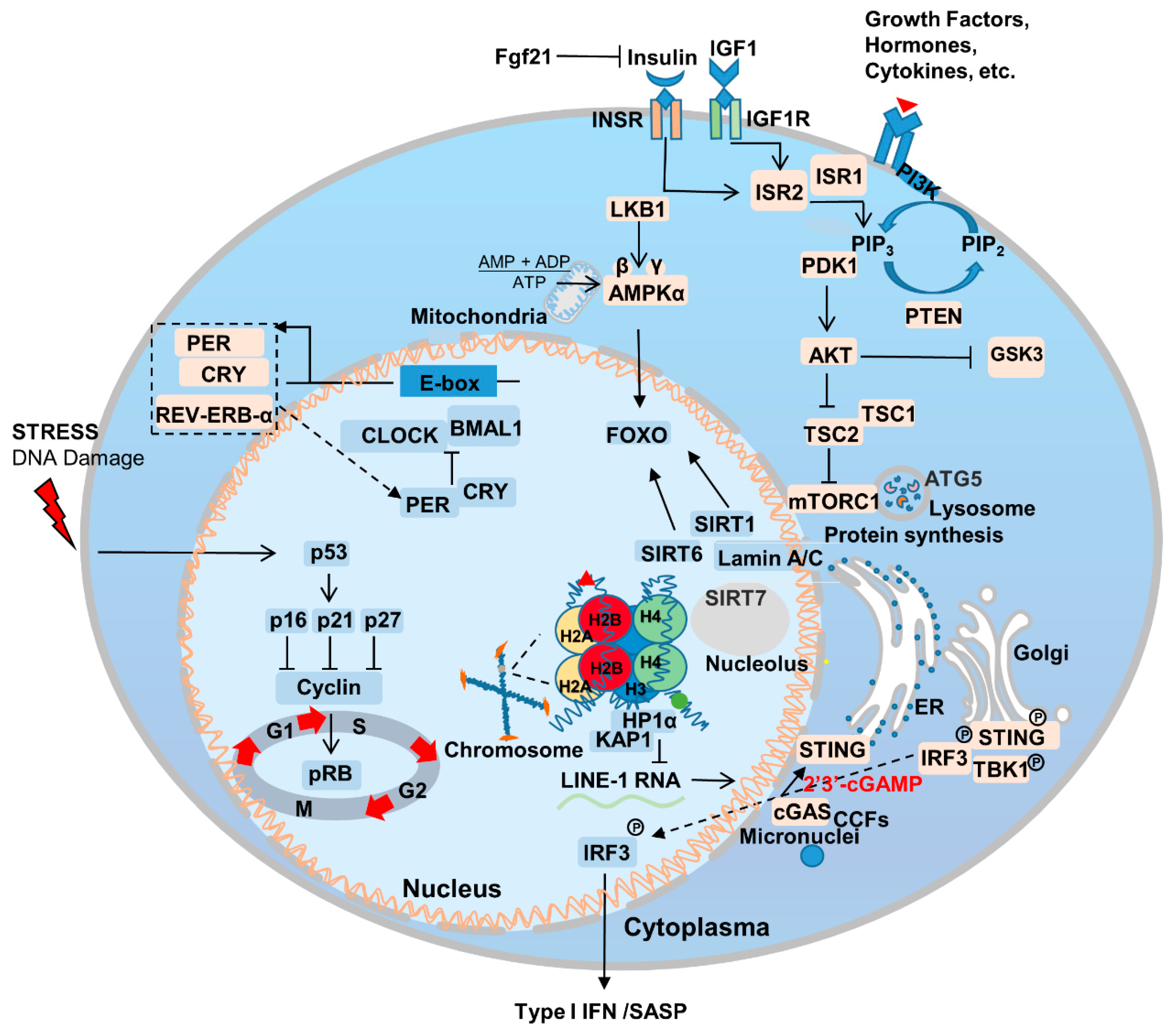
1. Introduction
2. The Genetics of Aging
| GenAge | The ageing gene database | https://genomics.senescence.info/genes/index.html | [28] |
| AnAge | Database of animal ageing and longevity | https://genomics.senescence.info/species/index.html | [28] |
| CellAge | Database of Cell Senescence Genes | https://genomics.senescence.info/cells/ | [29] |
| LongevityMap | Human longevity genetic variants | https://genomics.senescence.info/longevity/ | [30] |
| NIA Interventions Testing Program (ITP) Genetics | Conserved longevity gene prioritization | https://www.systems-genetics.org/itp-longevity | [21] |
| Aging Atlas | Transcriptomics Epigenomics Single-cell Transcriptomics Proteomics Pharmacogenomics Metabolomics | https://ngdc.cncb.ac.cn/aging/index | [31] |
2.1. Aging-Related Genes and Signaling Pathways
2.2. Nutrient Sensing
2.3. Sirtuins
2.4. Nuclear Skeleton-Associated Proteins
2.5. Immunity and Inflammation
2.6. Circadian Rhythm
3. The Epigenetics of Aging
3.1. Epigenetic Age
3.1.1. Histone Modifications
3.1.2. Chromatin Remodeling
3.2. Epigenetic Clock
3.2.1. Epigenetic Reprogramming
3.2.2. Extracellular Vesicles
3.3. Epitranscriptomics
3.3.1. m6A
3.3.2. ac4C
3.3.3. m1A and m5C
4. Interventions for Aging
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Olovnikov, A.M. Telomeres, telomerase, and aging: Origin of the theory. Exp. Gerontol. 1996, 31, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Gavrilov, L.A.; Gavrilova, N.S. Evolutionary Theories of Aging and Longevity. Sci. World J. 2002, 2, 339–356. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Otin, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef]
- Childs, B.G.; Gluscevic, M.; Baker, D.J.; Laberge, R.-M.; Marquess, D.; Dananberg, J.; van Deursen, J.M. Senescent cells: An emerging target for diseases of ageing. Nat. Rev. Drug Discov. 2017, 16, 718–735. [Google Scholar] [CrossRef]
- Hayflick, L.; Moorhead, P.S. The serial cultivation of human diploid cell strains. Exp. Cell Res. 1961, 25, 585–621. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Zamudio, R.I.; Robinson, L.; Roux, P.-F.; Bischof, O. SnapShot: Cellular Senescence Pathways. Cell 2017, 170, 816. [Google Scholar] [CrossRef]
- Hernandez-Segura, A.; Nehme, J.; Demaria, M. Hallmarks of Cellular Senescence. Trends Cell Biol. 2018, 28, 436–453. [Google Scholar] [CrossRef]
- Gasek, N.S.; Kuchel, G.A.; Kirkland, J.L.; Xu, M. Strategies for targeting senescent cells in human disease. Nat. Aging 2021, 1, 870–879. [Google Scholar] [CrossRef]
- Eriksson, M.; Brown, W.T.; Gordon, L.B.; Glynn, M.W.; Singer, J.; Scott, L.; Erdos, M.R.; Robbins, C.M.; Moses, T.Y.; Berglund, P.; et al. Recurrent de novo point mutations in lamin A cause Hutchinson–Gilford progeria syndrome. Nature 2003, 423, 293–298. [Google Scholar] [CrossRef]
- Krishnan, V.; Chow, M.Z.Y.; Wang, Z.; Zhang, L.; Liu, B.; Liu, X.; Zhou, Z. Histone H4 lysine 16 hypoacetylation is associated with defective DNA repair and premature senescence in Zmpste24-deficient mice. Proc. Natl. Acad. Sci. USA 2011, 108, 12325–12330. [Google Scholar] [CrossRef]
- Liu, B.; Wang, Z.; Zhang, L.; Ghosh, S.; Zheng, H.; Zhou, Z. Depleting the methyltransferase Suv39h1 improves DNA repair and extends lifespan in a progeria mouse model. Nat. Commun. 2013, 4, 1868. [Google Scholar] [CrossRef] [PubMed]
- Shumaker, D.K.; Dechat, T.; Kohlmaier, A.; Adam, S.A.; Bozovsky, M.R.; Erdos, M.R.; Eriksson, M.; Goldman, A.E.; Khuon, S.; Collins, F.S.; et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc. Natl. Acad. Sci. USA 2006, 103, 8703–8708. [Google Scholar] [CrossRef] [PubMed]
- Scaffidi, P.; Misteli, T. Lamin A-Dependent Nuclear Defects in Human Aging. Science 2006, 312, 1059–1063. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, J.; Suzuki, K.; Qu, J.; Wang, P.; Zhou, J.; Liu, X.; Ren, R.; Xu, X.; Ocampo, A.; et al. A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science 2015, 348, 1160–1163. [Google Scholar] [CrossRef] [PubMed]
- Ozgenc, A.; Loeb, L.A. Werner Syndrome, aging and cancer. Genome Dyn. 2006, 1, 206–217. [Google Scholar]
- Singh, P.P.; Demmitt, B.A.; Nath, R.D.; Brunet, A. The Genetics of Aging: A Vertebrate Perspective. Cell 2019, 177, 200–220. [Google Scholar] [CrossRef]
- Cagan, A.; Baez-Ortega, A.; Brzozowska, N.; Abascal, F.; Coorens, T.H.H.; Sanders, M.A.; Lawson, A.R.J.; Harvey, L.M.R.; Bhosle, S.; Jones, D.; et al. Somatic mutation rates scale with lifespan across mammals. Nature 2022, 604, 517–524. [Google Scholar] [CrossRef]
- Vincze, O.; Colchero, F.; Lemaître, J.-F.; Conde, D.A.; Pavard, S.; Bieuville, M.; Urrutia, A.O.; Ujvari, B.; Boddy, A.M.; Maley, C.C.; et al. Cancer risk across mammals. Nature 2021, 601, 263–267. [Google Scholar] [CrossRef]
- Gomes, N.M.V.; Ryder, O.A.; Houck, M.L.; Charter, S.J.; Walker, W.; Forsyth, N.R.; Austad, S.N.; Venditti, C.; Pagel, M.; Shay, J.W.; et al. Comparative biology of mammalian telomeres: Hypotheses on ancestral states and the roles of telomeres in longevity determination. Aging Cell 2011, 10, 761–768. [Google Scholar] [CrossRef]
- Muñoz-Lorente, M.A.; Cano-Martin, A.C.; Blasco, M.A. Mice with hyper-long telomeres show less metabolic aging and longer lifespans. Nat. Commun. 2019, 10, 4723. [Google Scholar] [CrossRef]
- Sleiman, M.B.; Roy, S.; Gao, A.W.; Sadler, M.C.; von Alvensleben, G.V.G.; Li, H.; Sen, S.; Harrison, D.E.; Nelson, J.F.; Strong, R.; et al. Sex- and age-dependent genetics of longevity in a heterogeneous mouse population. Science 2022, 377, eabo3191. [Google Scholar] [CrossRef]
- Zhao, H.; Ji, Q.; Wu, Z.; Wang, S.; Ren, J.; Yan, K.; Wang, Z.; Hu, J.; Chu, Q.; Hu, H.; et al. Destabilizing heterochromatin by APOE mediates senescence. Nat. Aging 2022, 2, 303–316. [Google Scholar] [CrossRef]
- Timmers, P.R.H.J.; Tiys, E.S.; Sakaue, S.; Akiyama, M.; Kiiskinen, T.T.J.; Zhou, W.; Hwang, S.-J.; Yao, C.; Kamatani, Y.; Deelen, J.; et al. Mendelian randomization of genetically independent aging phenotypes identifies LPA and VCAM1 as biological targets for human aging. Nat. Aging 2022, 2, 19–30. [Google Scholar] [CrossRef]
- Lemaître, J.-F.; Ronget, V.; Tidière, M.; Allainé, D.; Berger, V.; Cohas, A.; Colchero, F.; Conde, D.A.; Garratt, M.; Liker, A.; et al. Sex differences in adult lifespan and aging rates of mortality across wild mammals. Proc. Natl. Acad. Sci. USA 2020, 117, 8546–8553. [Google Scholar] [CrossRef] [PubMed]
- Trifunovic, A. Mitochondrial DNA and ageing. Biochim. Biophys. Acta 2006, 1757, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Ellegren, H. Sex-chromosome evolution: Recent progress and the influence of male and female heterogamety. Nat. Rev. Genet. 2011, 12, 157–166. [Google Scholar] [CrossRef]
- Kanfi, Y.; Naiman, S.; Amir, G.; Peshti, V.; Zinman, G.; Nahum, L.; Bar-Joseph, Z.; Cohen, H.Y. The sirtuin SIRT6 regulates lifespan in male mice. Nature 2012, 483, 218–221. [Google Scholar] [CrossRef]
- Tacutu, R.; Thornton, D.; Johnson, E.; Budovsky, A.; Barardo, D.; Craig, T.; Diana, E.; Lehmann, G.; Toren, D.; Wang, J.; et al. Human Ageing Genomic Resources: New and updated databases. Nucleic Acids Res. 2017, 46, D1083–D1090. [Google Scholar] [CrossRef] [PubMed]
- Avelar, R.A.; Ortega, J.G.; Tacutu, R.; Tyler, E.J.; Bennett, D.; Binetti, P.; Budovsky, A.; Chatsirisupachai, K.; Johnson, E.; Murray, A.; et al. A multidimensional systems biology analysis of cellular senescence in aging and disease. Genome Biol. 2020, 21, 91. [Google Scholar] [CrossRef]
- Budovsky, A.; Craig, T.; Wang, J.; Tacutu, R.; Csordas, A.; Lourenço, J.; Fraifeld, V.E.; de Magalhães, J.P. LongevityMap: A database of human genetic variants associated with longevity. Trends Genet. 2013, 29, 559–560. [Google Scholar] [CrossRef]
- Aging Atlas, C. Aging Atlas: A multi-omics database for aging biology. Nucleic Acids Res. 2021, 49, D825–D830. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Wang, J.; Chan, K.M.; Tjia, W.M.; Deng, W.; Guan, X.; Huang, J.-D.; Li, K.M.; Chau, P.Y.; Chen, D.J.; et al. Genomic instability in laminopathy-based premature aging. Nat. Med. 2005, 11, 780–785. [Google Scholar] [CrossRef] [PubMed]
- Garagnani, P.; Marquis, J.; Delledonne, M.; Pirazzini, C.; Marasco, E.; Kwiatkowska, K.M.; Iannuzzi, V.; Bacalini, M.G.; Valsesia, A.; Carayol, J.; et al. Whole-genome sequencing analysis of semi-supercentenarians. Elife 2021, 10, e57849. [Google Scholar] [CrossRef]
- Deelen, J.; Evans, D.S.; Arking, D.E.; Tesi, N.; Nygaard, M.; Liu, X.; Wojczynski, M.K.; Biggs, M.L.; van der Spek, A.; Atzmon, G.; et al. A meta-analysis of genome-wide association studies identifies multiple longevity genes. Nat. Commun. 2019, 10, 3669. [Google Scholar] [CrossRef]
- Lin, J.-R.; Sin-Chan, P.; Napolioni, V.; Torres, G.G.; Mitra, J.; Zhang, Q.; Jabalameli, M.R.; Wang, Z.; Nguyen, N.; Gao, T.; et al. Rare genetic coding variants associated with human longevity and protection against age-related diseases. Nat. Aging 2021, 1, 783–794. [Google Scholar] [CrossRef]
- Franceschi, C.; Garagnani, P.; Parini, P.; Giuliani, C.; Santoro, A. Inflammaging: A new immune–metabolic viewpoint for age-related diseases. Nat. Rev. Endocrinol. 2018, 14, 576–590. [Google Scholar] [CrossRef]
- Amorim, J.A.; Coppotelli, G.; Rolo, A.P.; Palmeira, C.M.; Ross, J.M.; Sinclair, D.A. Mitochondrial and metabolic dysfunction in ageing and age-related diseases. Nat. Rev. Endocrinol. 2022, 18, 243–258. [Google Scholar] [CrossRef]
- Efeyan, A.; Comb, W.C.; Sabatini, D.M. Nutrient-sensing mechanisms and pathways. Nature 2015, 517, 302–310. [Google Scholar] [CrossRef]
- Kulkarni, A.S.; Gubbi, S.; Barzilai, N. Benefits of Metformin in Attenuating the Hallmarks of Aging. Cell Metab. 2020, 32, 15–30. [Google Scholar] [CrossRef]
- Kenyon, C.; Chang, J.; Gensch, E.; Rudner, A.; Tabtiang, R. A C. elegans mutant that lives twice as long as wild type. Nature 1993, 366, 461–464. [Google Scholar] [CrossRef]
- Johnson, S.C.; Rabinovitch, P.S.; Kaeberlein, M. mTOR is a key modulator of ageing and age-related disease. Nature 2013, 493, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 169, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Unnikrishnan, A.; Deepa, S.S.; Herd, H.R.; Richardson, A. Chapter 19—Extension of Life Span in Laboratory Mice. In Conn’s Handbook of Models for Human Aging, 2nd ed.; Ram, J.L., Conn, P.M., Eds.; Academic Press: Cambridge, MA, USA, 2018; pp. 245–270. [Google Scholar]
- McCay, C.M.; Maynard, L.A.; Sperling, G.; Barnes, L.L. Retarded growth, life span, ultimate body size and age changes in the albino rat after feeding diets restricted in calories: Four figures. J. Nutr. 1939, 18, 1–13. [Google Scholar] [CrossRef]
- Fontana, L.; Nehme, J.; Demaria, M. Caloric restriction and cellular senescence. Mech. Ageing Dev. 2018, 176, 19–23. [Google Scholar] [CrossRef]
- Mattison, J.A.; Colman, R.J.; Beasley, T.M.; Allison, D.B.; Kemnitz, J.W.; Roth, G.S.; Ingram, D.K.; Weindruch, R.; de Cabo, R.; Anderson, R.M. Caloric restriction improves health and survival of rhesus monkeys. Nat. Commun. 2017, 8, 14063. [Google Scholar] [CrossRef]
- Anderson, R.M.; Bitterman, K.J.; Wood, J.G.; Medvedik, O.; Sinclair, D.A. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature 2003, 423, 181–185. [Google Scholar] [CrossRef]
- Panowski, S.H.; Wolff, S.; Aguilaniu, H.; Durieux, J.; Dillin, A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature 2007, 447, 550–555. [Google Scholar] [CrossRef]
- Vermeij, W.P.; Dollé, M.E.T.; Reiling, E.; Jaarsma, D.; Payan-Gomez, C.; Bombardieri, C.R.; Wu, H.; Roks, A.J.M.; Botter, S.M.; Van Der Eerden, B.C.; et al. Restricted diet delays accelerated ageing and genomic stress in DNA-repair-deficient mice. Nature 2016, 537, 427–431. [Google Scholar] [CrossRef]
- Xie, K.; Neff, F.; Markert, A.; Rozman, J.; Aguilar-Pimentel, J.A.; Amarie, O.V.; Becker, L.; Brommage, R.; Garrett, L.; Henzel, K.S.; et al. Every-other-day feeding extends lifespan but fails to delay many symptoms of aging in mice. Nat. Commun. 2017, 8, 155. [Google Scholar] [CrossRef]
- Mattison, J.A.; Roth, G.S.; Beasley, T.M.; Tilmont, E.M.; Handy, A.M.; Herbert, R.L.; Longo, D.L.; Allison, D.B.; Young, J.E.; Bryant, M.; et al. Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature 2012, 489, 318–321. [Google Scholar] [CrossRef]
- Colman, R.J.; Anderson, R.M.; Johnson, S.C.; Kastman, E.K.; Kosmatka, K.J.; Beasley, T.M.; Allison, D.B.; Cruzen, C.; Simmons, H.A.; Kemnitz, J.W.; et al. Caloric Restriction Delays Disease Onset and Mortality in Rhesus Monkeys. Science 2009, 325, 201–204. [Google Scholar] [CrossRef] [PubMed]
- Parsons, P.A. The limit to human longevity: An approach through a stress theory of ageing. Mech. Ageing Dev. 1996, 87, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Kanarek, N.; Petrova, B.; Sabatini, D.M. Dietary modifications for enhanced cancer therapy. Nature 2020, 579, 507–517. [Google Scholar] [CrossRef]
- Kalaany, N.Y.; Sabatini, D.M. Tumours with PI3K activation are resistant to dietary restriction. Nature 2009, 458, 725–731. [Google Scholar] [CrossRef]
- Richardson, N.E.; Konon, E.N.; Schuster, H.S.; Mitchell, A.T.; Boyle, C.; Rodgers, A.C.; Finke, M.; Haider, L.R.; Yu, D.; Flores, V.; et al. Lifelong restriction of dietary branched-chain amino acids has sex-specific benefits for frailty and life span in mice. Nat. Aging 2021, 1, 73–86. [Google Scholar] [CrossRef]
- Kennedy, B.K.; Austriaco, N.R., Jr.; Zhang, J.; Guarente, L. Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae. Cell 1995, 80, 485–496. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Wang, Y.; Liu, L.; Jiang, Z.; Li, X.; Tong, R.; He, J.; Shi, J. Research progress on sirtuins family members and cell senescence. Eur. J. Med. Chem. 2020, 193, 112207. [Google Scholar] [CrossRef]
- Cohen, H.Y.; Miller, C.; Bitterman, K.J.; Wall, N.R.; Hekking, B.; Kessler, B.; Howitz, K.T.; Gorospe, M.; de Cabo, R.; Sinclair, D.A. Calorie Restriction Promotes Mammalian Cell Survival by Inducing the SIRT1 Deacetylase. Science 2004, 305, 390–392. [Google Scholar] [CrossRef]
- Das, A.; Huang, G.X.; Bonkowski, M.S.; Longchamp, A.; Li, C.; Schultz, M.B.; Kim, L.-J.; Osborne, B.; Joshi, S.; Lu, Y.; et al. Impairment of an Endothelial NAD+-H2S Signaling Network Is a Reversible Cause of Vascular Aging. Cell 2019, 176, 944–945. [Google Scholar] [CrossRef]
- Osmanagic-Myers, S.; Kiss, A.; Manakanatas, C.; Hamza, O.; Sedlmayer, F.; Szabo, P.L.; Fischer, I.; Fichtinger, P.; Podesser, B.K.; Eriksson, M.; et al. Endothelial progerin expression causes cardiovascular pathology through an impaired mechanoresponse. J. Clin. Investig. 2019, 129, 531–545. [Google Scholar] [CrossRef]
- Sun, S.; Qin, W.; Tang, X.; Meng, Y.; Hu, W.; Zhang, S.; Qian, M.; Liu, Z.; Cao, X.; Pang, Q.; et al. Vascular endothelium–targeted Sirt7 gene therapy rejuvenates blood vessels and extends life span in a Hutchinson-Gilford progeria model. Sci. Adv. 2020, 6, eaay5556. [Google Scholar] [CrossRef]
- Wu, X.; Liu, H.; Brooks, A.; Xu, S.; Luo, J.; Steiner, R.; Mickelsen, D.M.; Moravec, C.S.; Jeffrey, A.D.; Small, E.M.; et al. SIRT6 Mitigates Heart Failure With Preserved Ejection Fraction in Diabetes. Circ. Res. 2022, 131, 926–943. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Ghosh, S.; Yang, X.; Zheng, H.; Liu, X.; Wang, Z.; Jin, G.; Zheng, B.; Kennedy, B.K.; Suh, Y.; et al. Resveratrol rescues SIRT1-dependent adult stem cell decline and alleviates progeroid features in laminopathy-based progeria. Cell Metab. 2012, 16, 738–750. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Liu, B.; Wang, Y.; Hao, Q.; Zhou, Z. Lamin A Is an Endogenous SIRT6 Activator and Promotes SIRT6-Mediated DNA Repair. Cell Rep. 2015, 13, 1396–1406. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, B.N.; Thackray, J.K.; Simonet, N.G.; Chahar, S.; Kane-Goldsmith, N.; Newkirk, S.J.; Lee, S.; Xing, J.; Verzi, M.P.; An, W.; et al. SIRT7 mediates L1 elements transcriptional repression and their association with the nuclear lamina. Nucleic Acids Res. 2019, 47, 7870–7885. [Google Scholar] [CrossRef]
- Bi, S.; Liu, Z.; Wu, Z.; Wang, Z.; Liu, X.; Wang, S.; Ren, J.; Yao, Y.; Zhang, W.; Song, M.; et al. SIRT7 antagonizes human stem cell aging as a heterochromatin stabilizer. Protein Cell 2020, 11, 483–504. [Google Scholar] [CrossRef]
- Kulik, G.; Weber, M.J. Akt-Dependent and -Independent Survival Signaling Pathways Utilized by Insulin-Like Growth Factor I. Mol. Cell Biol. 1998, 18, 6711–6718. [Google Scholar] [CrossRef]
- Jiang, B.; Wu, X.; Meng, F.; Si, L.; Cao, S.; Dong, Y.; Sun, H.; Lv, M.; Xu, H.; Bai, N.; et al. Progerin modulates the IGF-1R/Akt signaling involved in aging. Sci. Adv. 2022, 8, eabo0322. [Google Scholar] [CrossRef]
- Ibrahim, M.X.; Sayin, V.I.; Akula, M.K.; Liu, M.; Fong, L.G.; Young, S.G.; Bergo, M.O. Targeting Isoprenylcysteine Methylation Ameliorates Disease in a Mouse Model of Progeria. Science 2013, 340, 1330–1333. [Google Scholar] [CrossRef]
- Cabral, W.A.; Tavarez, U.L.; Beeram, I.; Yeritsyan, D.; Boku, Y.D.; Eckhaus, M.A.; Nazarian, A.; Erdos, M.R.; Collins, F.S. Genetic reduction of mTOR extends lifespan in a mouse model of Hutchinson-Gilford Progeria syndrome. Aging Cell 2021, 20, e13457. [Google Scholar] [CrossRef]
- Dou, Z.; Xu, C.; Donahue, G.; Shimi, T.; Pan, J.-A.; Zhu, J.; Ivanov, A.; Capell, B.C.; Drake, A.M.; Shah, P.P.; et al. Autophagy mediates degradation of nuclear lamina. Nature 2015, 527, 105–109. [Google Scholar] [CrossRef]
- Xu, C.; Wang, L.; Fozouni, P.; Evjen, G.; Chandra, V.; Jiang, J.; Lu, C.; Nicastri, M.; Bretz, C.; Winkler, J.D.; et al. SIRT1 is downregulated by autophagy in senescence and ageing. Nature 2020, 22, 1170–1179. [Google Scholar] [CrossRef] [PubMed]
- Franceschi, C.; Bonafe, M.; Valensin, S.; Olivieri, F.; De Luca, M.; Ottaviani, E.; De Benedictis, G. Inflamm-aging: An evolutionary perspective on immunosenescence. Ann. N. Y. Acad. Sci. 2000, 908, 244–254. [Google Scholar] [CrossRef]
- Baylis, D.; Bartlett, D.B.; Patel, H.P.; Roberts, H.C. Understanding how we age: Insights into inflammaging. Longev. Health 2013, 2, 8. [Google Scholar] [CrossRef] [PubMed]
- Hopfner, K.P.; Hornung, V. Molecular mechanisms and cellular functions of cGAS-STING signalling. Nat. Rev. Mol. Cell Biol. 2020, 21, 501–521. [Google Scholar] [CrossRef]
- Kim, D.H.; Bang, E.; Jung, H.J.; Noh, S.G.; Yu, B.P.; Choi, Y.J.; Chung, H.Y. Anti-Aging Effects of Calorie Restriction (CR) and CR Mimetics Based on the Senoinflammation Concept. Nutrients 2020, 12, 422. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Ge, W.; Harns, C.; Meng, X.; Zhang, Y.; Ren, J. Ablation of toll-like receptor 4 attenuates aging-induced myocardial remodeling and contractile dysfunction through NCoRI-HDAC1-mediated regulation of autophagy. J. Mol. Cell Cardiol. 2018, 119, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Tchkonia, T.; Ding, H.; Ogrodnik, M.; Lubbers, E.R.; Pirtskhalava, T.; White, T.A.; Johnson, K.O.; Stout, M.B.; Mezera, V.; et al. JAK inhibition alleviates the cellular senescence-associated secretory phenotype and frailty in old age. Proc. Natl. Acad. Sci. USA 2015, 112, E6301–E6310. [Google Scholar] [CrossRef]
- Ablasser, A.; Goldeck, M.; Cavlar, T.; Deimling, T.; Witte, G.; Röhl, I.; Hopfner, K.-P.; Ludwig, J.; Hornung, V. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 2013, 498, 380–384. [Google Scholar] [CrossRef]
- Dou, Z.; Ghosh, K.; Vizioli, M.G.; Zhu, J.; Sen, P.; Wangensteen, K.J.; Simithy, J.; Lan, Y.; Lin, Y.; Zhou, Z.; et al. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 2017, 550, 402–406. [Google Scholar] [CrossRef]
- Glück, S.; Guey, B.; Gulen, M.F.; Wolter, K.; Kang, T.-W.; Schmacke, N.A.; Bridgeman, A.; Rehwinkel, J.; Zender, L.; Ablasser, A. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nature 2017, 19, 1061–1070. [Google Scholar] [CrossRef] [PubMed]
- Sladitschek-Martens, H.L.; Guarnieri, A.; Brumana, G.; Zanconato, F.; Battilana, G.; Xiccato, R.L.; Panciera, T.; Forcato, M.; Bicciato, S.; Guzzardo, V.; et al. AP/TAZ activity in stromal cells prevents ageing by controlling cGAS-STING. Nature 2022, 607, 790–798. [Google Scholar] [CrossRef]
- Patke, A.; Young, M.W.; Axelrod, S. Molecular mechanisms and physiological importance of circadian rhythms. Nat. Rev. Mol. Cell Biol. 2020, 21, 67–84. [Google Scholar] [CrossRef] [PubMed]
- Janich, P.; Meng, Q.-J.; Benitah, S.A. Circadian control of tissue homeostasis and adult stem cells. Curr. Opin. Cell Biol. 2014, 31, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Musiek, E.S.; Holtzman, D.M. Mechanisms linking circadian clocks, sleep, and neurodegeneration. Science 2016, 354, 1004–1008. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Solanas, G.; Peixoto, F.O.; Bee, L.; Symeonidi, A.; Schmidt, M.S.; Brenner, C.; Masri, S.; Benitah, S.A.; Sassone-Corsi, P. Circadian Reprogramming in the Liver Identifies Metabolic Pathways of Aging. Cell 2017, 170, 664–677.e11. [Google Scholar] [CrossRef]
- Solanas, G.; Peixoto, F.O.; Perdiguero, E.; Jardí, M.; Ruiz-Bonilla, V.; Datta, D.; Symeonidi, A.; Castellanos, A.; Welz, P.-S.; Caballero, J.M.; et al. Aged Stem Cells Reprogram Their Daily Rhythmic Functions to Adapt to Stress. Cell 2017, 170, 678–692.e20. [Google Scholar] [CrossRef]
- Yamamoto, R.; Chung, R.; Vazquez, J.M.; Sheng, H.; Steinberg, P.L.; Ioannidis, N.M.; Sudmant, P.H. Tissue-specific impacts of aging and genetics on gene expression patterns in humans. Nat. Commun. 2022, 13, 5803. [Google Scholar] [CrossRef]
- Kaplanis, J.; Gordon, A.; Shor, T.; Weissbrod, O.; Geiger, D.; Wahl, M.; Gershovits, M.; Markus, B.; Sheikh, M.; Gymrek, M.; et al. Quantitative analysis of population-scale family trees with millions of relatives. Science 2018, 360, 171–175. [Google Scholar] [CrossRef]
- Zhang, W.; Qu, J.; Liu, G.-H.; Belmonte, J.C.I. The ageing epigenome and its rejuvenation. Nat. Rev. Mol. Cell Biol. 2020, 21, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Grewal, S.I.S.; Jia, S. Heterochromatin revisited. Nat. Rev. Genet. 2007, 8, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Tessarz, P.; Kouzarides, T. Histone core modifications regulating nucleosome structure and dynamics. Nat. Rev. Mol. Cell Biol. 2014, 15, 703–708. [Google Scholar] [CrossRef] [PubMed]
- Millán-Zambrano, G.; Burton, A.; Bannister, A.J.; Schneider, R. Histone post-translational modifications—cause and consequence of genome function. Nat. Rev. Genet. 2022, 23, 563–580. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Wei, G.; Long, F.; Nie, H.; Tian, X.; Qu, L.; Wang, S.; Li, P.; Qiu, Y.; Wang, Y.; et al. Histone methyltransferase Smyd3 is a new regulator for vascular senescence. Aging Cell 2020, 19, e13212. [Google Scholar] [CrossRef]
- Ibarrola, J.; Kim, S.K.; Lu, Q.; DuPont, J.J.; Creech, A.; Sun, Z.; A Hill, M.; Jaffe, J.D.; Jaffe, I.Z. Smooth muscle mineralocorticoid receptor as an epigenetic regulator of vascular ageing. Cardiovasc. Res. 2022, 118, 3386–3400. [Google Scholar] [CrossRef]
- Shchukina, I.; Bagaitkar, J.; Shpynov, O.; Loginicheva, E.; Porter, S.; Mogilenko, D.A.; Wolin, E.; Collins, P.; Demidov, G.; Artomov, M.; et al. Enhanced epigenetic profiling of classical human monocytes reveals a specific signature of healthy aging in the DNA methylome. Nat. Aging 2021, 1, 124–141. [Google Scholar] [CrossRef]
- Zhu, D.; Wu, X.; Zhou, J.; Li, X.; Huang, X.; Li, J.; Wu, J.; Bian, Q.; Wang, Y.; Tian, Y. NuRD mediates mitochondrial stress-induced longevity via chromatin remodeling in response to acetyl-CoA level. Sci. Adv. 2020, 6, eabb2529. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, X.; Huang, L.; Guan, Y.; Huang, X.; Tian, X.; Zhang, L.; Tao, W. ATF3 drives senescence by reconstructing accessible chromatin profiles. Aging Cell 2021, 20, e13315. [Google Scholar] [CrossRef]
- Hu, H.; Ji, Q.; Song, M.; Ren, J.; Liu, Z.; Wang, Z.; Liu, X.; Yan, K.; Hu, J.; Jing, Y.; et al. ZKSCAN3 counteracts cellular senescence by stabilizing heterochromatin. Nucleic Acids Res. 2020, 48, 6001–6018. [Google Scholar] [CrossRef]
- Zhang, X.; Jiang, Q.; Li, J.; Zhang, S.; Cao, Y.; Xia, X.; Cai, D.; Tan, J.; Chen, J.; Han, J.-D.J. KCNQ1OT1 promotes genome-wide transposon repression by guiding RNA–DNA triplexes and HP1 binding. Nature 2022, 24, 1617–1629. [Google Scholar] [CrossRef]
- Horvath, S.; Raj, K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 2018, 19, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Minteer, C.; Morselli, M.; Meer, M.; Cao, J.; Higgins-Chen, A.; Lang, S.M.; Pellegrini, M.; Yan, Q.; Levine, M.E. Tick tock, tick tock: Mouse culture and tissue aging captured by an epigenetic clock. Aging Cell 2022, 21, e13553. [Google Scholar] [CrossRef]
- Kabacik, S.; Lowe, D.; Fransen, L.; Leonard, M.; Ang, S.-L.; Whiteman, C.; Corsi, S.; Cohen, H.; Felton, S.; Bali, R.; et al. The relationship between epigenetic age and the hallmarks of aging in human cells. Nat. Aging 2022, 2, 484–493. [Google Scholar] [CrossRef]
- Horvath, S.; Oshima, J.; Martin, G.M.; Lu, A.T.; Quach, A.; Cohen, H.; Felton, S.; Matsuyama, M.; Lowe, D.; Kabacik, S.; et al. Epigenetic clock for skin and blood cells applied to Hutchinson Gilford Progeria Syndrome and ex vivo studies. Aging 2018, 10, 1758–1775. [Google Scholar] [CrossRef]
- Higgins-Chen, A.T.; Thrush, K.L.; Wang, Y.; Minteer, C.J.; Kuo, P.-L.; Wang, M.; Niimi, P.; Sturm, G.; Lin, J.; Moore, A.Z.; et al. A computational solution for bolstering reliability of epigenetic clocks: Implications for clinical trials and longitudinal tracking. Nat. Aging 2022, 2, 644–661. [Google Scholar] [CrossRef] [PubMed]
- Benayoun, B.; Pollina, E.A.; Brunet, A. Epigenetic regulation of ageing: Linking environmental inputs to genomic stability. Nat. Rev. Mol. Cell Biol. 2015, 16, 593–610. [Google Scholar] [CrossRef] [PubMed]
- Ocampo, A.; Reddy, P.; Martinez-Redondo, P.; Platero-Luengo, A.; Hatanaka, F.; Hishida, T.; Li, M.; Lam, D.; Kurita, M.; Beyret, E.; et al. In Vivo Amelioration of Age-Associated Hallmarks by Partial Reprogramming. Cell 2016, 167, 1719–1733.e12. [Google Scholar] [CrossRef]
- Mosteiro, L.; Pantoja, C.; Alcazar, N.; Marión, R.M.; Chondronasiou, D.; Rovira, M.; Fernandez-Marcos, P.J.; Muñoz-Martin, M.; Blanco-Aparicio, C.; Pastor, J.; et al. Tissue damage and senescence provide critical signals for cellular reprogramming in vivo. Science 2016, 354, aaf4445. [Google Scholar] [CrossRef]
- Lu, Y.; Brommer, B.; Tian, X.; Krishnan, A.; Meer, M.; Wang, C.; Vera, D.L.; Zeng, Q.; Yu, D.; Bonkowski, M.S.; et al. Reprogramming to recover youthful epigenetic information and restore vision. Nature 2020, 588, 124–129. [Google Scholar] [CrossRef]
- Browder, K.C.; Reddy, P.; Yamamoto, M.; Haghani, A.; Guillen, I.G.; Sahu, S.; Wang, C.; Luque, Y.; Prieto, J.; Shi, L.; et al. In vivo partial reprogramming alters age-associated molecular changes during physiological aging in mice. Nat. Aging 2022, 2, 243–253. [Google Scholar] [CrossRef]
- Kaur, P.; Otgonbaatar, A.; Ramamoorthy, A.; Chua, E.H.Z.; Harmston, N.; Gruber, J.; Tolwinski, N.S. Combining stem cell rejuvenation and senescence targeting to synergistically extend lifespan. Aging 2022, 14, 8270–8291. [Google Scholar] [CrossRef] [PubMed]
- Hou, P.; Li, Y.; Zhang, X.; Liu, C.; Guan, J.; Li, H.; Zhao, T.; Ye, J.; Yang, W.; Liu, K.; et al. Pluripotent Stem Cells Induced from Mouse Somatic Cells by Small-Molecule Compounds. Science 2013, 341, 651–654. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.; Wang, G.; Wang, J.; Zhang, Z.; Fu, Y.; Cheng, L.; Meng, G.; Lyu, Y.; Zhu, J.; Li, Y.; et al. Chemical reprogramming of human somatic cells to pluripotent stem cells. Nature 2022, 605, 325–331. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, M.; Satoh, A.; Lin, J.B.; Mills, K.F.; Sasaki, Y.; Rensing, N.; Wong, M.; Apte, R.S.; Imai, S.-I. Extracellular Vesicle-Contained eNAMPT Delays Aging and Extends Lifespan in Mice. Cell Metab. 2019, 30, 329–342.e5. [Google Scholar] [CrossRef]
- Sanz-Ros, J.; Romero-García, N.; Mas-Bargues, C.; Monleón, D.; Gordevicius, J.; Brooke, R.T.; Dromant, M.; Díaz, A.; Derevyanko, A.; Guío-Carrión, A.; et al. Small extracellular vesicles from young adipose-derived stem cells prevent frailty, improve health span, and decrease epigenetic age in old mice. Sci. Adv. 2022, 8, eabq2226. [Google Scholar] [CrossRef]
- Wiener, D.; Schwartz, S. The epitranscriptome beyond m6A. Nat. Rev. Genet. 2021, 22, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ao, Y.; Zhang, Z.; Mo, Y.; Peng, L.; Jiang, Y.; Wang, Z.; Liu, B. Lamin A safeguards the m6A methylase METTL14 nuclear speckle reservoir to prevent cellular senescence. Aging Cell 2020, 19, e13215. [Google Scholar] [CrossRef] [PubMed]
- Min, K.-W.; Zealy, R.W.; Davila, S.; Fomin, M.; Cummings, J.C.; Makowsky, D.; Mcdowell, C.H.; Thigpen, H.; Hafner, M.; Kwon, S.-H.; et al. Profiling of m6A RNA modifications identified an age-associated regulation of AGO2 mRNA stability. Aging Cell 2018, 17, e12753. [Google Scholar] [CrossRef]
- Wu, Z.; Shi, Y.; Lu, M.; Song, M.; Yu, Z.; Wang, J.; Wang, S.; Ren, J.; Yang, Y.-G.; Liu, G.-H.; et al. METTL3 counteracts premature aging via m6A-dependent stabilization of MIS12 mRNA. Nucleic Acids Res. 2020, 48, 11083–11096. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhang, L.; Lai, C.; Peng, X.; Yu, S.; Zhou, C.; Zhang, B.; Zhang, W. Dynamic Alteration Profile and New Role of RNA m6A Methylation in Replicative and H2O2-Induced Premature Senescence of Human Embryonic Lung Fibroblasts. Int. J. Mol. Sci. 2022, 23, 9271. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Yu, Y.; Yang, M.; Huang, H.; Ma, S.; Hu, J.; Xi, Z.; Guo, H.; Yao, G.; Yang, L.; et al. YTHDF1 promotes breast cancer progression by facilitating FOXM1 translation in an m6A-dependent manner. Cell Biosci. 2022, 12, 19. [Google Scholar] [CrossRef]
- Ribeiro, R.; Macedo, J.C.; Costa, M.; Ustiyan, V.; Shindyapina, A.V.; Tyshkovskiy, A.; Gomes, R.N.; Castro, J.P.; Kalin, T.V.; Vasques-Nóvoa, F.; et al. In vivo cyclic induction of the FOXM1 transcription factor delays natural and progeroid aging phenotypes and extends healthspan. Nat. Aging 2022, 2, 397–411. [Google Scholar] [CrossRef]
- Lu, Y.; He, X.; Zhong, S. Cross-species microarray analysis with the OSCAR system suggests an INSR->Pax6->NQO1 neuro-protective pathway in aging and Alzheimer’s disease. Nucleic Acids Res. 2007, 35, W105–W114. [Google Scholar] [CrossRef]
- Zheng, Q.; Gan, H.; Yang, F.; Yao, Y.; Hao, F.; Hong, L.; Jin, L. Cytoplasmic m1A reader YTHDF3 inhibits trophoblast invasion by downregulation of m1A-methylated IGF1R. Cell Discov. 2020, 6, 12. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X.; Zhang, X.; Wang, J.; Ma, Y.; Zhang, L.; Cao, X. RNA-binding protein YTHDF3 suppresses interferon-dependent antiviral responses by promoting FOXO3 translation. Proc. Natl. Acad. Sci. USA 2019, 116, 976–981. [Google Scholar] [CrossRef]
- Hao, W.; Dian, M.; Zhou, Y.; Zhong, Q.; Pang, W.; Li, Z.; Zhao, Y.; Ma, J.; Lin, X.; Luo, R.; et al. Autophagy induction promoted by m6A reader YTHDF3 through translation upregulation of FOXO3 mRNA. Nat. Commun. 2022, 13, 5845. [Google Scholar] [CrossRef]
- Arango, D.; Sturgill, D.; Alhusaini, N.; Dillman, A.A.; Sweet, T.J.; Hanson, G.; Hosogane, M.; Sinclair, W.R.; Nanan, K.K.; Mandler, M.D.; et al. Acetylation of Cytidine in mRNA Promotes Translation Efficiency. Cell 2018, 175, 1872–1886.e24. [Google Scholar] [CrossRef]
- Larrieu, D.; Britton, S.; Demir, M.; Rodriguez, R.; Jackson, S.P. Chemical Inhibition of NAT10 Corrects Defects of Laminopathic Cells. Science 2014, 344, 527–532. [Google Scholar] [CrossRef]
- Balmus, G.; Larrieu, D.; Barros, A.C.; Collins, C.; Abrudan, M.; Demir, M.; Geisler, N.J.; Lelliott, C.J.; White, J.K.; Karp, N.A.; et al. Targeting of NAT10 enhances healthspan in a mouse model of human accelerated aging syndrome. Nat. Commun. 2018, 9, 1700. [Google Scholar] [CrossRef]
- Xu, C.; Zhang, J.; Zhang, J.; Liu, B. SIRT7 is a deacetylase of N4-acetylcytidine on ribosomal RNA. Genome Instab. Dis. 2021, 2, 253–260. [Google Scholar] [CrossRef]
- He, H.; Wang, Y.; Wang, J. ALKBH3 is dispensable in maintaining hematopoietic stem cells but forced ALKBH3 rectified the differentiation skewing of aged hematopoietic stem cells. Blood Sci. 2020, 2, 137–143. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, Z.; Yi, J.; Tang, H.; Xing, J.; Yu, M.; Tong, T.; Shang, Y.; Gorospe, M.; Wang, W. The tRNA methyltransferase NSun2 stabilizes p16INK4 mRNA by methylating the 3′-untranslated region of p16. Nat. Commun. 2012, 3, 712. [Google Scholar] [CrossRef]
- Morris, B.J.; Willcox, B.J.; Donlon, T.A. Genetic And epigenetic regulation of human aging and longevity. Biochim. Biophys. Acta. Mol. Basis Dis. 2019, 1865, 1718–1744. [Google Scholar] [CrossRef] [PubMed]
- Campisi, J.; Kapahi, P.; Lithgow, G.J.; Melov, S.; Newman, J.C.; Verdin, E. From discoveries in ageing research to therapeutics for healthy ageing. Nature 2019, 571, 183–192. [Google Scholar] [CrossRef]
- Covarrubias, A.J.; Perrone, R.; Grozio, A.; Verdin, E. NAD+ metabolism and its roles in cellular processes during ageing. Nat. Rev. Mol. Cell Biol. 2021, 22, 119–141. [Google Scholar] [CrossRef] [PubMed]
- Lagoumtzi, S.M.; Chondrogianni, N. Senolytics and senomorphics: Natural and synthetic therapeutics in the treatment of aging and chronic diseases. Free. Radic. Biol. Med. 2021, 171, 169–190. [Google Scholar] [CrossRef]
- Davidsohn, N.; Pezzone, M.; Vernet, A.; Graveline, A.; Oliver, D.; Slomovic, S.; Punthambaker, S.; Sun, X.; Liao, R.; Bonventre, J.V.; et al. A single combination gene therapy treats multiple age-related diseases. Proc. Natl. Acad. Sci. USA 2019, 116, 23505–23511. [Google Scholar] [CrossRef] [PubMed]
- Suda, M.; Shimizu, I.; Katsuumi, G.; Yoshida, Y.; Hayashi, Y.; Ikegami, R.; Matsumoto, N.; Yoshida, Y.; Mikawa, R.; Katayama, A.; et al. Senolytic vaccination improves normal and pathological age-related phenotypes and increases lifespan in progeroid mice. Nat. Aging 2021, 1, 1117–1126. [Google Scholar] [CrossRef]
- Misteli, T. Farnesyltransferase inhibition in HGPS. Cell 2021, 184, 293. [Google Scholar] [CrossRef]
- Partridge, L.; Fuentealba, M.; Kennedy, B.K. The quest to slow ageing through drug discovery. Nat. Rev. Drug Discov. 2020, 19, 513–532. [Google Scholar] [CrossRef] [PubMed]
- Heckenbach, I.; Mkrtchyan, G.V.; Ezra, M.B.; Bakula, D.; Madsen, J.S.; Nielsen, M.H.; Oró, D.; Osborne, B.; Covarrubias, A.J.; Idda, M.L.; et al. Nuclear morphology is a deep learning biomarker of cellular senescence. Nat. Aging 2022, 2, 742–755. [Google Scholar] [CrossRef]
- Ryu, D.; Zhang, H.; Ropelle, E.R.; Sorrentino, V.; Mazala, D.A.; Mouchiroud, L.; Marshall, P.L.; Campbell, M.D.; Ali, A.S.; Knowels, G.M.; et al. NAD+ repletion improves muscle function in muscular dystrophy and counters global PARylation. Sci. Transl. Med. 2016, 8, 361ra139. [Google Scholar] [CrossRef] [PubMed]
- Jaskelioff, M.; Muller, F.L.; Paik, J.H.; Thomas, E.; Jiang, S.; Adams, A.C.; Sahin, E.; Kost-Alimova, M.; Protopopov, A.; Cadinanos, J.; et al. Telomerase reactivation reverses tissue degeneration in aged telomerase-deficient mice. Nature 2011, 469, 102–106. [Google Scholar] [CrossRef]
- Kuro-o, M.; Matsumura, Y.; Aizawa, H.; Kawaguchi, H.; Suga, T.; Utsugi, T.; Ohyama, Y.; Kurabayashi, M.; Kaname, T.; Kume, E.; et al. Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature 1997, 390, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Asadi Shahmirzadi, A.; Edgar, D.; Liao, C.Y.; Hsu, Y.M.; Lucanic, M.; Asadi Shahmirzadi, A.; Wiley, C.D.; Gan, G.; Kim, D.E.; Kasler, H.G.; et al. Alpha-Ketoglutarate, an Endogenous Metabolite, Extends Lifespan and Compresses Morbidity in Aging Mice. Cell Metab. 2020, 32, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Andreux, P.A.; Blanco-Bose, W.; Ryu, D.; Burdet, F.; Ibberson, M.; Aebischer, P.; Auwerx, J.; Singh, A.; Rinsch, C. The mitophagy activator urolithin A is safe and induces a molecular signature of improved mitochondrial and cellular health in humans. Nat. Metab. 2019, 1, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Baar, M.P.; Brandt, R.M.C.; Putavet, D.A.; Klein, J.D.D.; Derks, K.W.J.; Bourgeois, B.R.M.; Stryeck, S.; Rijksen, Y.; van Willigenburg, H.; Feijtel, D.A.; et al. Targeted Apoptosis of Senescent Cells Restores Tissue Homeostasis in Response to Chemotoxicity and Aging. Cell 2017, 169, 132–147. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Pirtskhalava, T.; Farr, J.N.; Weigand, B.M.; Palmer, A.K.; Weivoda, M.M.; Inman, C.L.; Ogrodnik, M.B.; Hachfeld, C.M.; Fraser, D.G.; et al. Senolytics improve physical function and increase lifespan in old age. Nat. Med. 2018, 24, 1246–1256. [Google Scholar] [CrossRef]
- Yousefzadeh, M.J.; Zhu, Y.; McGowan, S.J.; Angelini, L.; Fuhrmann-Stroissnigg, H.; Xu, M.; Ling, Y.Y.; Melos, K.I.; Pirtskhalava, T.; Inman, C.L.; et al. Fisetin is a senotherapeutic that extends health and lifespan. EBioMedicine 2018, 36, 18–28. [Google Scholar] [CrossRef]
- Oubaha, M.; Miloudi, K.; Dejda, A.; Guber, V.; Mawambo, G.; Germain, M.A.; Bourdel, G.; Popovic, N.; Rezende, F.A.; Kaufman, R.J.; et al. Senescence-associated secretory phenotype contributes to pathological angiogenesis in retinopathy. Sci. Transl. Med. 2016, 8, 362ra144. [Google Scholar] [CrossRef] [PubMed]
- Harrison, D.E.; Strong, R.; Sharp, Z.D.; Nelson, J.F.; Astle, C.M.; Flurkey, K.; Nadon, N.L.; Wilkinson, J.E.; Frenkel, K.; Carter, C.S.; et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 2009, 460, 392–395. [Google Scholar] [CrossRef] [PubMed]
- van Rhee, F.; Wong, R.S.; Munshi, N.; Rossi, J.F.; Ke, X.Y.; Fossa, A.; Simpson, D.; Capra, M.; Liu, T.; Hsieh, R.K.; et al. Siltuximab for multicentric Castleman’s disease: A randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2014, 15, 966–974. [Google Scholar] [CrossRef]
- Tompkins, B.A.; DiFede, D.L.; Khan, A.; Landin, A.M.; Schulman, I.H.; Pujol, M.V.; Heldman, A.W.; Miki, R.; Goldschmidt-Clermont, P.J.; Goldstein, B.J.; et al. Allogeneic Mesenchymal Stem Cells Ameliorate Aging Frailty: A Phase II Randomized, Double-Blind, Placebo-Controlled Clinical Trial. J. Gerontol. A Biol. Sci. Med. Sci. 2017, 72, 1513–1522. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Wang, S.; Liu, B. New Insights into the Genetics and Epigenetics of Aging Plasticity. Genes 2023, 14, 329. https://doi.org/10.3390/genes14020329
Zhang J, Wang S, Liu B. New Insights into the Genetics and Epigenetics of Aging Plasticity. Genes. 2023; 14(2):329. https://doi.org/10.3390/genes14020329
Chicago/Turabian StyleZhang, Jie, Shixiao Wang, and Baohua Liu. 2023. "New Insights into the Genetics and Epigenetics of Aging Plasticity" Genes 14, no. 2: 329. https://doi.org/10.3390/genes14020329
APA StyleZhang, J., Wang, S., & Liu, B. (2023). New Insights into the Genetics and Epigenetics of Aging Plasticity. Genes, 14(2), 329. https://doi.org/10.3390/genes14020329







