Effects of Wnt10a and Wnt10b Double Mutations on Tooth Development
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Preparation of crRNA–tracrRNA Duplex Ribonucleoprotein (RNP) Complex Assembly
2.3. Mice, Embryo Collection, Electroporation, and Transfer
2.4. Stereomicroscope and Micro-Computed Tomography (µCT)
2.5. Mouse Genotyping
2.6. Statistical Analysis
3. Results
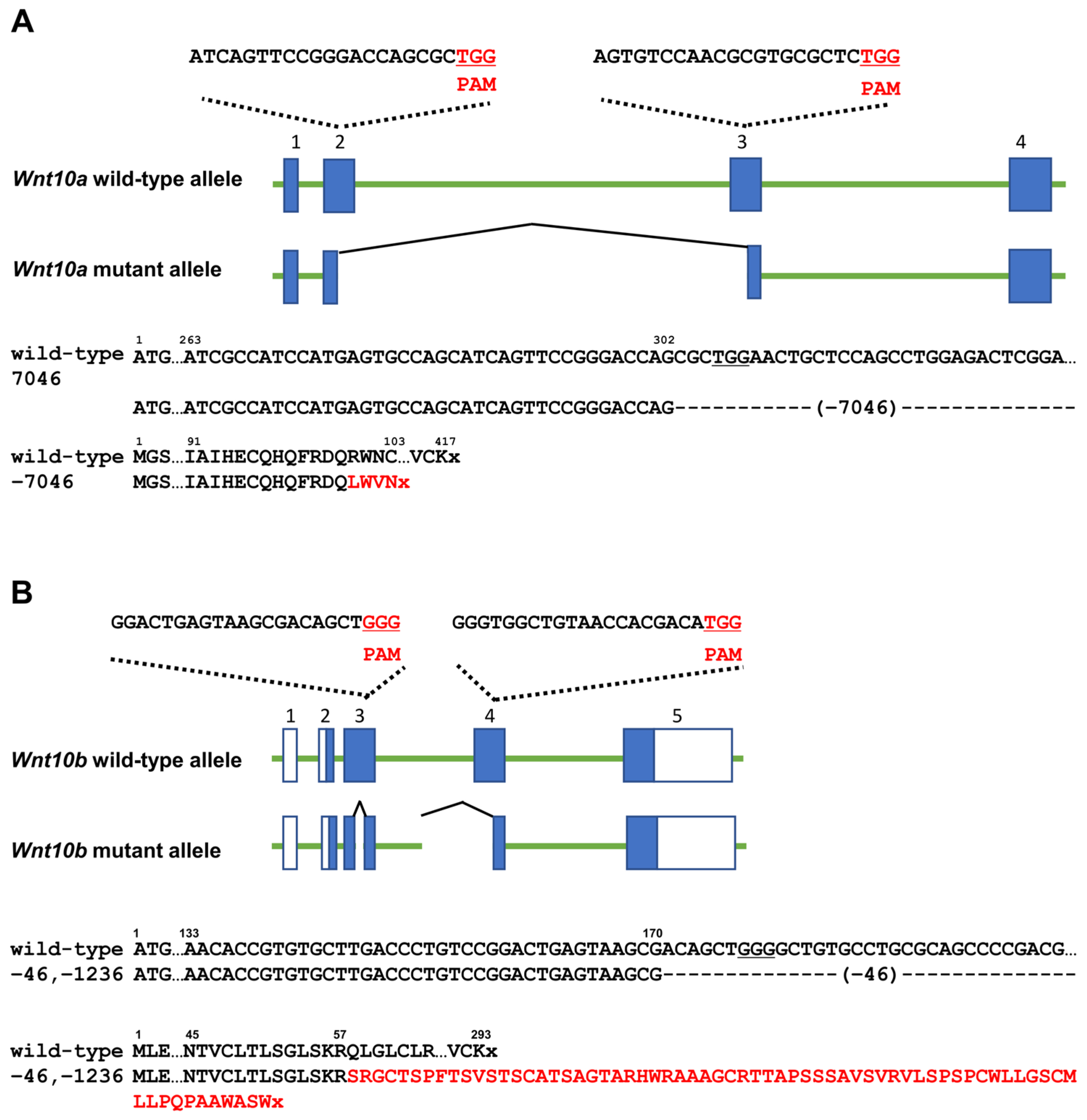
3.1. Generation of Each Wnt10a or Wnt10b Mutant
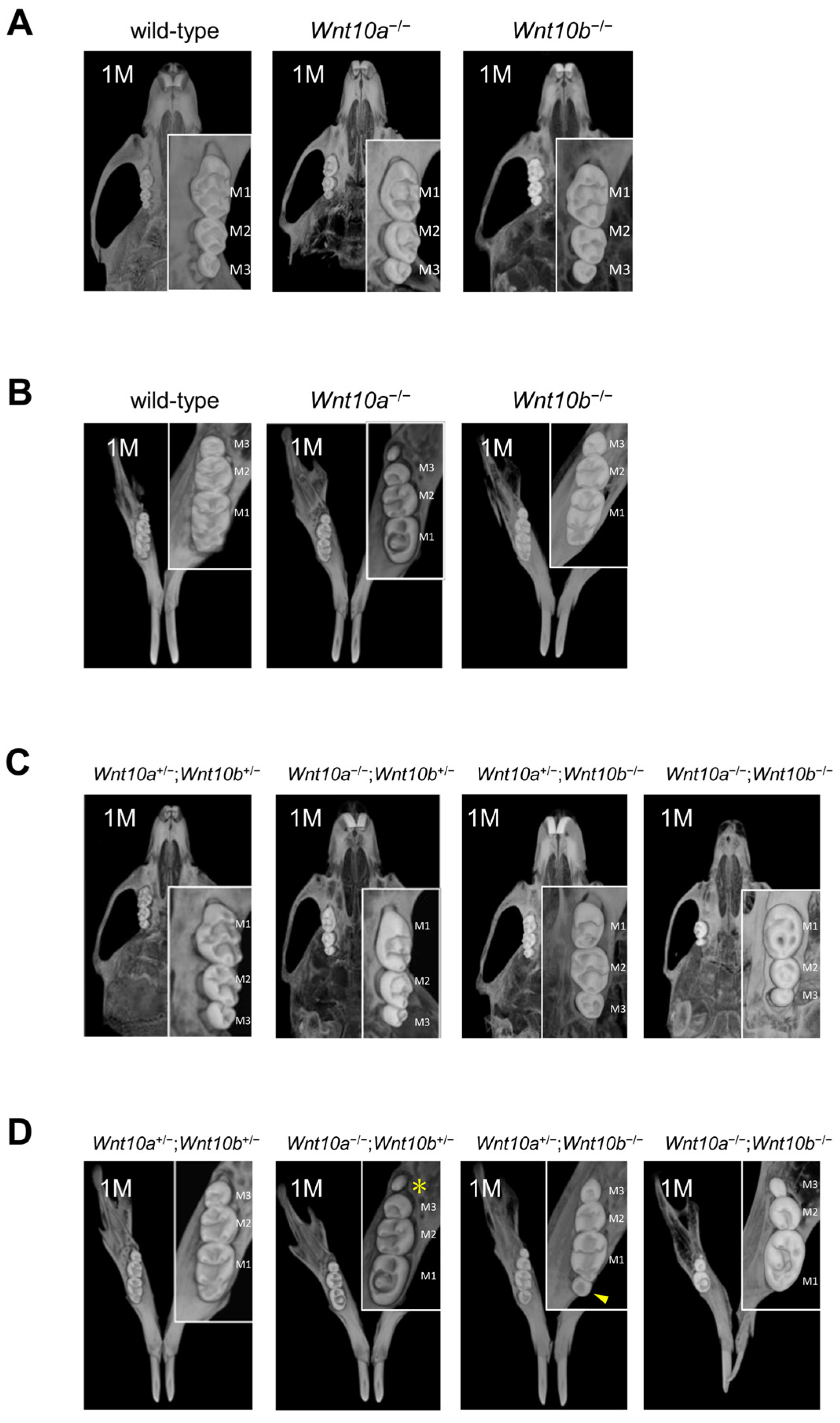
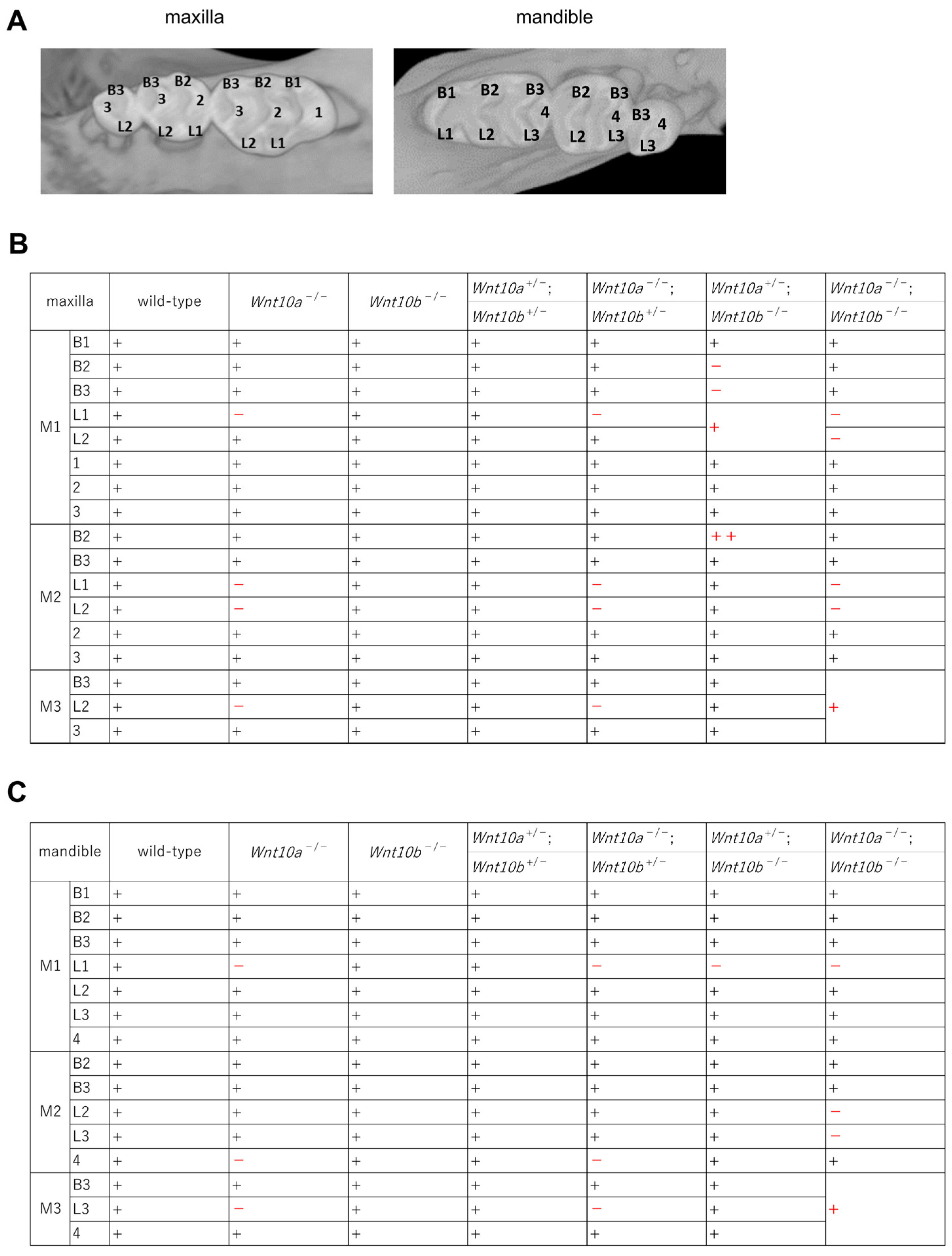
3.2. Changes in the Number and Shape of Teeth in a Single Mutant of Wnt10a and Wnt10b
3.3. Variations in the Number and Shape of Teeth in Wnt10a and Wnt10b Double Mutants
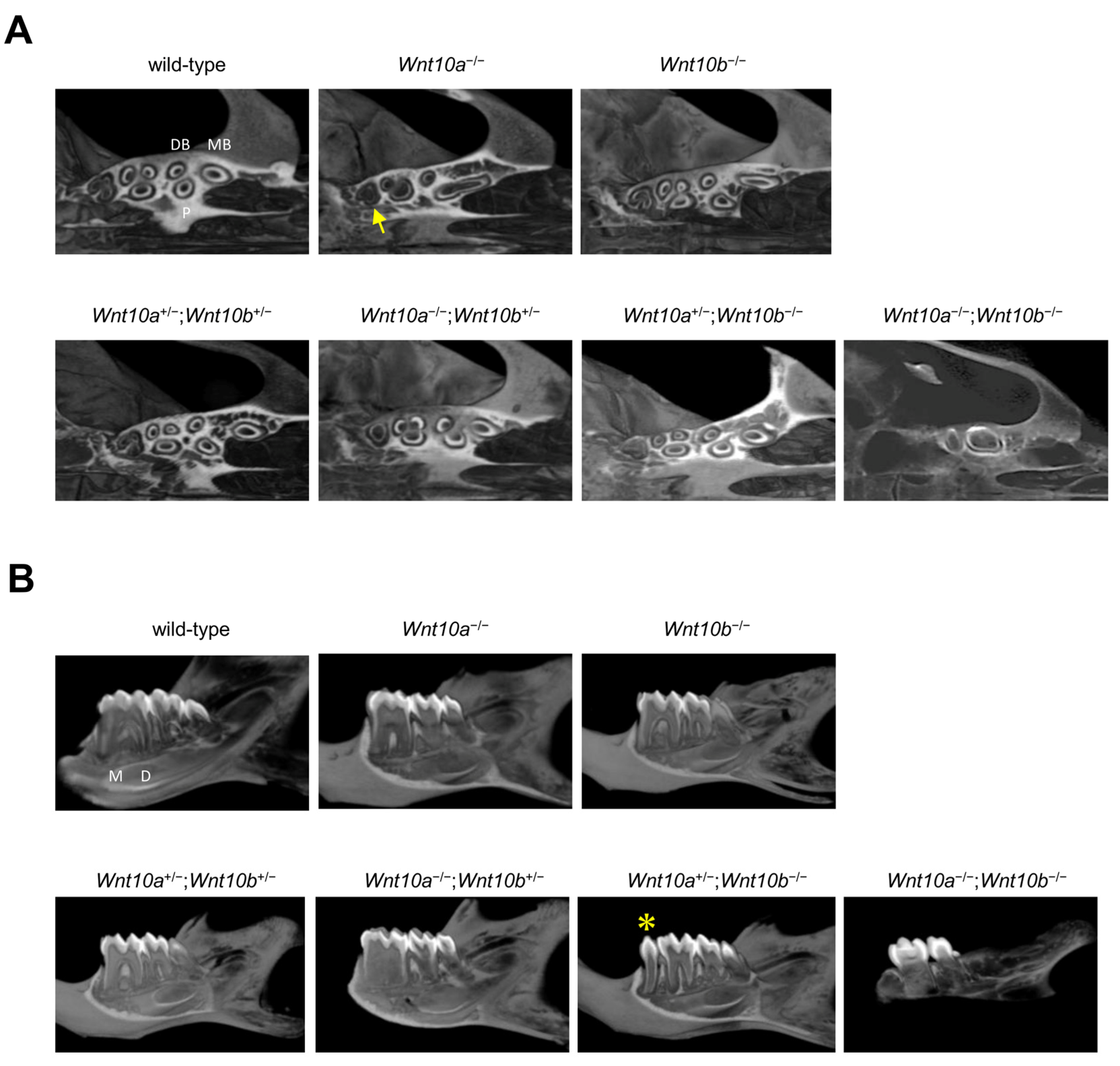
3.4. Only Double Homozygous Alleles of Wnt10a and Wnt10b Show Developmental Retardation
3.5. Root Morphology of Wnt10a and Wnt10b Mutant Mice
4. Discussion
4.1. WNT Signaling Regulates Tooth Development
4.2. Tooth Morphology and Number Are Affected in Several Double Mutants
4.3. Growth Rate Is Decreased in Wnt10a and Wnt10b Double Mutants
4.4. Wnt10a and Wnt10b Function Have a Great Effect on Repartition of Tooth Number
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bailleul-Forestier, I.; Molla, M.; Verloes, A.; Berdal, A. The genetic basis of inherited anomalies of the teeth. Part 1: Clinical and molecular aspects of non-syndromic dental disorders. Eur. J. Med. Genet. 2008, 51, 273–291. [Google Scholar] [CrossRef] [PubMed]
- Boeira, R.B., Jr.; Echeverrigaray, S. Novel missense mutation in PAX9 gene associated with familial tooth agenesis. J. Oral Pathol. Med. 2013, 42, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Nikopensius, T.; Annilo, T.; Jagomägi, T.; Gilissen, C.; Kals, M.; Krjutškov, K.; Mägi, R.; Eelmets, M.; Gerst-Talas, U.; Remm, M.; et al. Non-syndromic tooth agenesis associated with a nonsense mutation in ectodysplasin-A (EDA). J. Dent. Res. 2013, 92, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Vastardis, H.; Karimbux, N.; Guthua, S.W.; Seidman, J.; Seidman, C.E. A human MSX1 homeodomain missense mutation causes selective tooth agenesis. Nat. Genet. 1996, 13, 417–421. [Google Scholar] [CrossRef]
- Yu, M.; Wong, S.; Han, D.; Cai, T. Genetic analysis: Wnt and other pathways in nonsyndromic tooth agenesis. Oral Dis. 2019, 25, 646–651. [Google Scholar] [CrossRef] [Green Version]
- Arzoo, P.S.; Klar, J.; Bergendal, B.; Norderyd, J.; Dahl, N. WNT10A mutations account for ¼ of population-based isolated oligodontia and show phenotypic correlations. Am. J. Med. Genet A 2014, 164, 353–359. [Google Scholar] [CrossRef]
- Kantaputra, P.; Hutsadaloi, A.; Kaewgahya, M.; Intachai, W.; German, R.; Koparal, M.; Leethanakul, C.; Tolun, A.; Cairns, J.K. WNT10B mutations associated with isolated dental anomalies. Clin. Genet. 2018, 93, 992–999. [Google Scholar] [CrossRef]
- Ahn, Y.; Sanderson, B.W.; Klein, O.; Krumlauf, R. Inhibition of Wnt signaling by Wise (Sostdc1) and negative feedback from Shh controls tooth number and patterning. Development 2010, 137, 3221–3231. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Chu, E.Y.; Watt, B.; Zhang, Y.; Gallant, N.M.; Andl, T.; Yang, S.H.; Lu, M.-M.; Piccolo, S.; Schmidt-Ullrich, R.; et al. Wnt/β-catenin signaling directs multiple stages of tooth morphogenesis. Dev. Biol. 2008, 313, 210–224. [Google Scholar] [CrossRef] [Green Version]
- Thesleff, I.; Sharpe, P. Signalling networks regulating dental development. Mech. Dev. 1997, 67, 111–123. [Google Scholar] [CrossRef]
- He, H.; Han, D.; Feng, H.; Qu, H.; Song, S.; Bai, B.; Zhang, Z. Involvement of and Interaction between WNT10A and EDA mutations in tooth agenesis cases in the Chinese population. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [Green Version]
- Sidow, A. Diversification of the Wnt gene family on the ancestral line-age of vertebrates. Proc. Natl. Acad. Sci. USA 1992, 89, 5098–5102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nusse, R. An ancient cluster of Wnt paralogues. Trends Genet. 2001, 17, 443. [Google Scholar] [CrossRef] [PubMed]
- Dassule, H.R.; McMahon, A.P. Analysis of epithelial-mesenchymal interactions in the initial morphogenesis of the mammalian tooth. Dev. Biol. 1998, 202, 215–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satokata, I.; Maas, R. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat. Genet. 1994, 6, 348–356. [Google Scholar] [CrossRef]
- Peters, H.; Neubüser, A.; Kratochwil, K.; Balling, R. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 1998, 12, 2735–2747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srivastava, A.K.; Pispa, J.; Hartung, A.J.; Du, Y.; Ezer, S.; Jenks, T.; Shimada, T.; Pekkanen, M.; Mikkola, M.L.; Ko, M.S.H.; et al. The Tabby phenotype is caused by mutation in a mouse homologue of the EDA gene that reveals novel mouse and human exons and encodes a protein (ectodysplasin-A) with collagenous domains. Proc. Natl. Acad. Sci. USA 1997, 94, 13069–13074. [Google Scholar] [CrossRef] [Green Version]
- Pispa, J.; Jung, H.S.; Jernvall, J.; Kettunen, P.; Mustonen, T.; Tabata, M.J.; Kere, J.; Thesleff, I. Cusp patterning defect in Tabby mouse teeth and its partial rescue by FGF. Dev. Biol. 1999, 15, 216. [Google Scholar] [CrossRef] [Green Version]
- Murphy, P.; Armit, C.; Hill, B.; Venkataraman, S.; Frankel, P.; Baldock, R.A.; Davidson, D.R. Integrated analysis of Wnt signalling system component gene expression. bioRxiv 2022. [Google Scholar] [CrossRef] [PubMed]
- Järvinen, E.; Salazar-Ciudad, I.; Birchmeier, W. Continuous tooth generation in mouse is induced by activated epithelial Wnt/β-catenin signaling. Proc. Natl. Acad. Sci. USA 2006, 103, 18627–18632. [Google Scholar] [CrossRef]
- Yamashiro, T.; Zheng, L.; Shitaku, Y.; Saito, M.; Tsubakimoto, T.; Takada, K.; Takano-Yamamoto, T.; Thesleff, I. Wnt10a regulates dentin sialophosphoprotein mRNA expression and possibly links odontoblast differentiation and tooth morphogenesis. Differentiation 2007, 75, 452–462. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Yang, W.; Han, D.; Wang, X.; Guo, S.; Li, J.; Li, F.; Zhang, X.; Wong, S.-W.; Bai, B.; et al. Mutations in WNT10B Are Identified in Individuals with Oligodontia. Am. J. Hum. Genet. 2016, 99, 195–201. [Google Scholar] [CrossRef] [Green Version]
- Stevens, J.R.; Miranda-Carboni, A.G.; Singer, A.M.; Brugger, S.M.; Lyons, K.M.; Lane, T.F. Wnt10b Deficiency Results in Age-Dependent Loss of Bone Mass and Progressive Reduction of Mesenchymal Progenitor Cells. J. Bone Miner. Res. 2010, 25, 2138–2147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrade, A.C.; Nilsson, O.; Barnes, K.M.; Baron, J. Wnt gene expression in the post-natal growth plate: Regulation with chondrocyte differentiation. Bone 2007, 40, 1361–1369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, H.; Mak, W.; Zheng, Y.; Dunstan, C.R.; Seibel, M.J. Osteoblasts Directly Control Lineage Commitment of Mesenchymal Progenitor Cells through Wnt Signaling. J. Biol. Chem. 2007, 283, 1936–1945. [Google Scholar] [CrossRef] [Green Version]
- Nawaz, S.; Klar, J.; Wajid, M.; Aslam, M.; Tariq, M.; Schuster, J.; Baig, S.M.; Dahl, N. WNT10A missense mutation associated with a complete Odonto-Onycho-Dermal Dysplasia syndrome. Eur. J. Hum. Genet. 2009, 17, 1600–1605. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.; Horrell, J.; Snitow, M.; Cui, J.; Gochnauer, H.; Syrett, C.M.; Kallish, S.; Seykora, J.T.; Liu, F.; Gaillard, D.; et al. WNT10A mutation causes ectodermal dysplasia by impairing progenitor cell proliferation and KLF4-mediated differentiation. Nat. Commun. 2017, 8, 15397. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Shih-Kai, W.; Choi, M.; Reid, B.M.; Hu, Y.; Lee, Y.-L.; Herzog, C.R.; Kim-Berman, H.; Lee, M.; Benke, P.J.; et al. Taurodontism, variations in tooth number, and misshapened crowns in Wnt10a null mice and human kindreds. Mol. Genet. Genom. Med. 2015, 3, 40–58. [Google Scholar] [CrossRef] [Green Version]
- Lyngstadaas, S.P.; Møinichen, C.B.; Risnes, S. Crown Morphology, Enamel Distribution, and Enamel Structure in Mouse Molars. Anat. Rec. 1998, 250, 268–290. [Google Scholar] [CrossRef]
- Kassai, Y.; Munne, P.; Hotta, Y.; Penttilä, E.; Kavanagh, K.; Ohbayashi, N.; Takada, S.; Thesleff, I.; Jernvall, J.; Itoh, N. Regulation of Mammalian Tooth Cusp Patterning by Ectodin. Science 2005, 309, 2067–2069. [Google Scholar] [CrossRef]
- Ohazama, A.; Johnson, E.B.; Ota, M.S.; Choi, H.J.; Porntaveetus, T.; Oommen, S.; Itoh, N.; Eto, K.; Gritli-Linde, A.; Herz, J.; et al. Lrp4 modulates extracellular integration of cell signaling pathways in development. PLoS ONE 2008, 3, e4092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahn, Y.; Sims, C.; Murray, M.J.; Kuhlmann, P.K.; Fuentes-Antrás, J.; Weatherbee, S.D.; Krumlauf, R. Multiple modes of Lrp4 function in modulation of Wnt/β-catenin signaling during tooth development. Development 2017, 144, 2824–2836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cawthorn, W.P.; Bree, A.J.; Yao, Y.; Du, B.; Hemati, N.; Martinez-Santibañez, G.; MacDougald, O.A. Wnt6, Wnt10a and Wnt10b inhibit adipogenesis and stimulate osteoblastogenesis through a β-catenin-dependent mechanism. Bone 2012, 50, 477–489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsukamoto, M.; Wang, K.; Tasaki, T.; Murata, Y.; Okada, Y.; Yamanaka, Y.; Nakamura, E.; Yamada, S.; Izumi, H.; Zhou, Q.; et al. Findings as a starting point to unravel the underlying mechanisms of in vivo interactions involving Wnt10a in bone, fat and muscle. Bone 2019, 120, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Li, L.; Liu, C.; Liu, H.; He, F.; Yan, F.; Zhang, Y.; Chen, Y. Wnt5a regulates growth, patterning, and odontoblast differentiation of developing mouse tooth. Dev. Dyn. 2011, 240, 432–440. [Google Scholar] [CrossRef] [Green Version]
- Cai, J.; Cho, S.-W.; Kim, J.-Y.; Lee, M.-J.; Cha, Y.-G.; Jung, H.-S. Patterning the size and number of tooth and its cusps. Dev. Biol. 2007, 304, 499–507. [Google Scholar] [CrossRef] [Green Version]
- Cho, S.-W.; Kwak, S.; Woolley, T.; Lee, M.-J.; Kim, E.-J.; Baker, R.E.; Kim, H.-J.; Shin, J.-S.; Tickle, C.; Maini, P.K.; et al. Interactions between Shh, Sostdc1 and Wnt signaling and a new feedback loop for spatial patterning of the teeth. Development 2011, 138, 1807–1816. [Google Scholar] [CrossRef] [Green Version]
- Laurikkala, J.; Kassai, Y.; Pakkasjärvi, L.; Thesleff, I.; Itoh, N. Identification of a secreted BMP antagonist, ectodin, integrating BMP, FGF, and SHH signals from the tooth enamel knot. Dev. Biol. 2003, 264, 91–105. [Google Scholar] [CrossRef]

| Genotypes | Phenotypes | Ratio |
|---|---|---|
| Wnt10a−/− | M4 appearance | 3/5 |
| Wnt10b−/− | No change | 5/5 |
| Wnt10a+/−;Wnt10b+/− | No change | 10/10 |
| Wnt10a−/−;Wnt10b+/− | M4 appearance | 4/8 |
| Wnt10a+/−;Wnt10b−/− | A supernumerary tooth appearance | 8/10 |
| Wnt10a−/−;Wnt10b−/− | M3 defect | 3/5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoshinaga, K.; Yasue, A.; Mitsui, S.N.; Minegishi, Y.; Oyadomari, S.; Imoto, I.; Tanaka, E. Effects of Wnt10a and Wnt10b Double Mutations on Tooth Development. Genes 2023, 14, 340. https://doi.org/10.3390/genes14020340
Yoshinaga K, Yasue A, Mitsui SN, Minegishi Y, Oyadomari S, Imoto I, Tanaka E. Effects of Wnt10a and Wnt10b Double Mutations on Tooth Development. Genes. 2023; 14(2):340. https://doi.org/10.3390/genes14020340
Chicago/Turabian StyleYoshinaga, Kaoru, Akihiro Yasue, Silvia Naomi Mitsui, Yoshiyuki Minegishi, Seiichi Oyadomari, Issei Imoto, and Eiji Tanaka. 2023. "Effects of Wnt10a and Wnt10b Double Mutations on Tooth Development" Genes 14, no. 2: 340. https://doi.org/10.3390/genes14020340
APA StyleYoshinaga, K., Yasue, A., Mitsui, S. N., Minegishi, Y., Oyadomari, S., Imoto, I., & Tanaka, E. (2023). Effects of Wnt10a and Wnt10b Double Mutations on Tooth Development. Genes, 14(2), 340. https://doi.org/10.3390/genes14020340






