Telomere Fragility and MiDAS: Managing the Gaps at the End of the Road
Abstract
:1. Introduction
2. Telomere Fragility
2.1. Initial Discovery and Characterization
2.2. Telomere Fragility, DNA Unwinding, Fork Remodeling
2.3. POT1 and CST Roles in Telomere Fragility
2.4. Excision-Repair Proteins, Oxidative DNA Damage and Telomere Fragility
3. Mitotic DNA Synthesis (MiDAS)
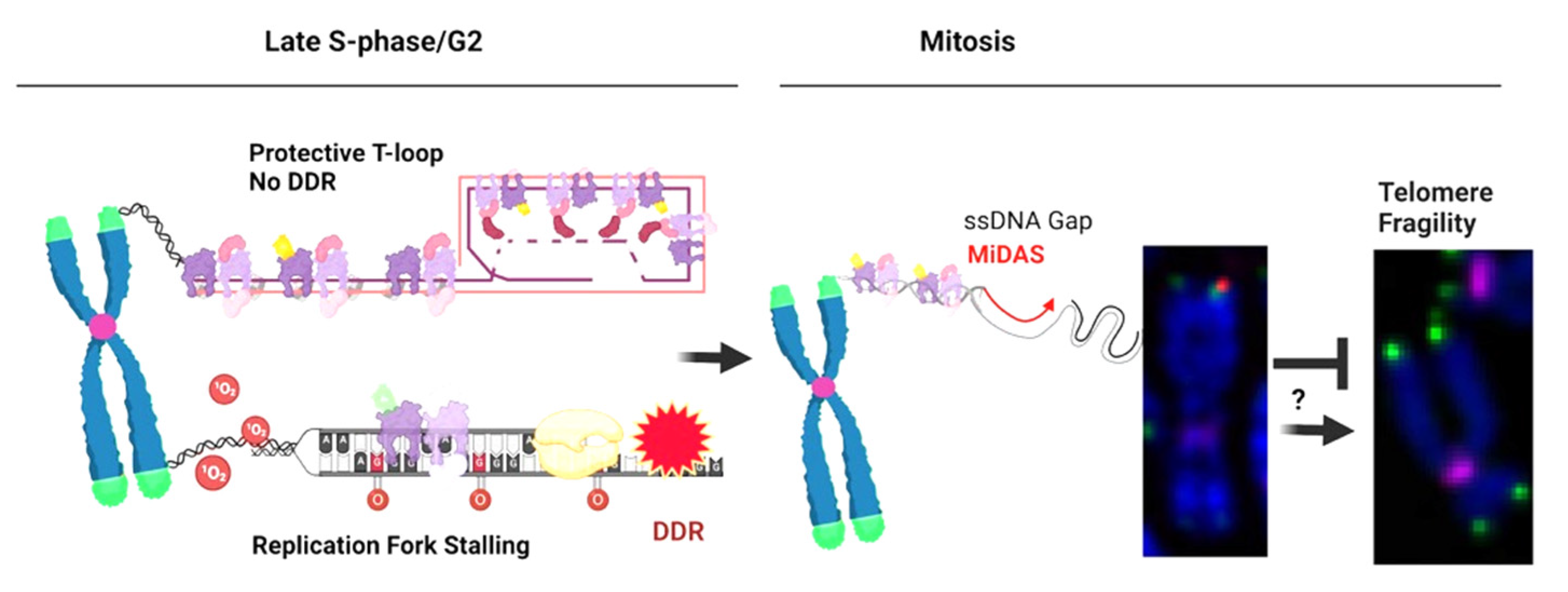
3.1. Initial Discovery and Characterization
3.2. Telomere MiDAS
3.3. BIR and MiDAS at ALT Telomeres
3.4. Shelterin, SUMO, and MiDAS
4. Replication Stress Enhances Telomere Fragility and MiDAS
4.1. TERRA, RNA/DNA Hybrids, and G4s
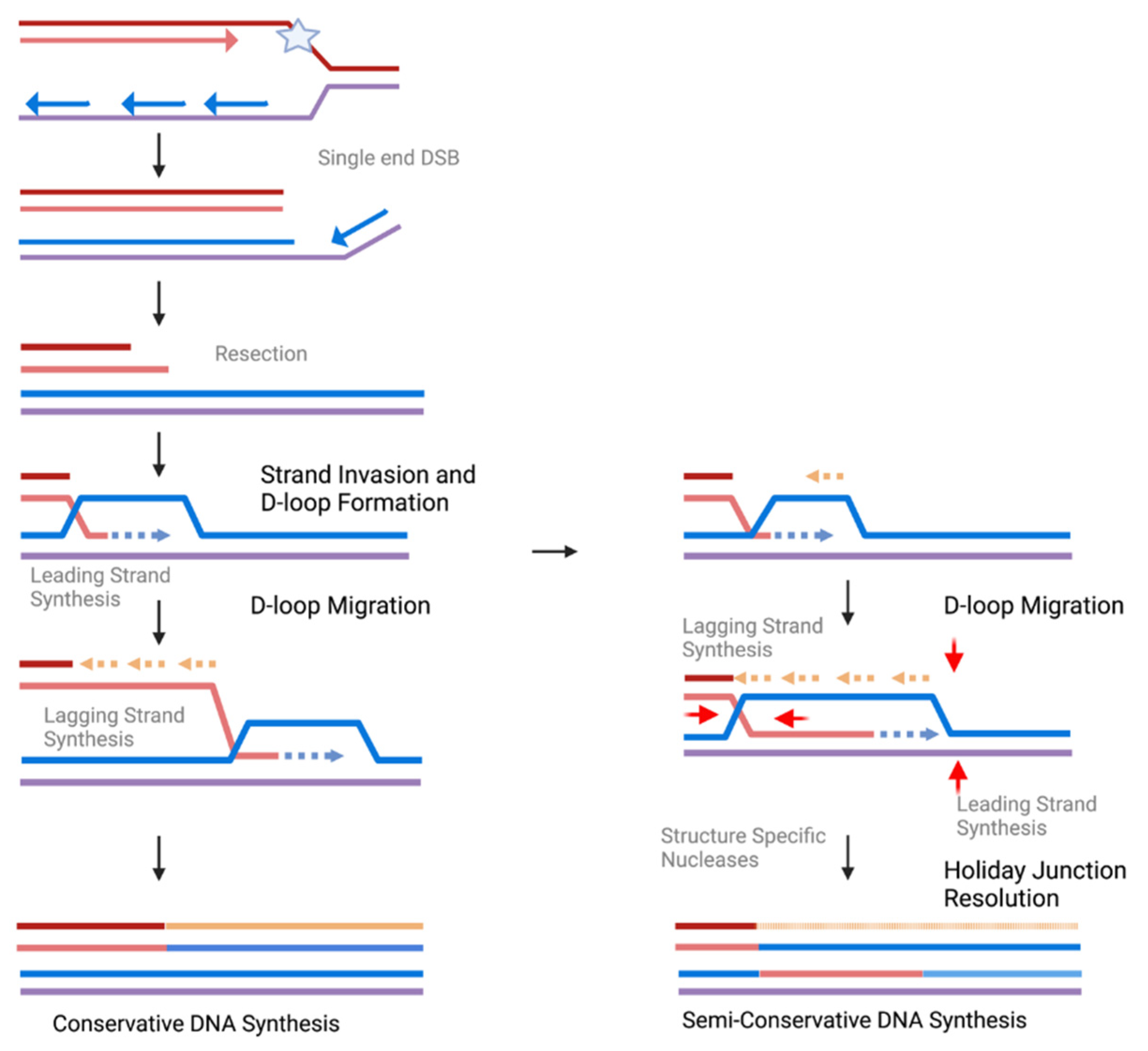
4.2. Stalled Fork Processing: HR, Nucleases, and Protection
5. Telomere Replication Stress as a Therapeutic Opportunity
6. How Are Telomere Fragility and MiDAS Connected?
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Lange, T. Shelterin-Mediated Telomere Protection. Annu. Rev. Genet. 2018, 52, 223–247. [Google Scholar] [CrossRef] [PubMed]
- Lormand, J.D.; Buncher, N.; Murphy, C.T.; Kaur, P.; Lee, M.Y.; Burgers, P.; Wang, H.; Kunkel, T.A.; Opresko, P.L. DNA polymerase delta stalls on telomeric lagging strand templates independently from G-quadruplex formation. Nucleic Acids Res. 2013, 41, 10323–10333. [Google Scholar] [CrossRef]
- Bryan, T.M. G-Quadruplexes at Telomeres: Friend or Foe? Molecules 2020, 25, 686. [Google Scholar] [CrossRef] [PubMed]
- Azzalin, C.M.; Reichenbach, P.; Khoriauli, L.; Giulotto, E.; Lingner, J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 2007, 318, 798–801. [Google Scholar] [CrossRef] [PubMed]
- Toubiana, S.; Selig, S. DNA:RNA hybrids at telomeres—When it is better to be out of the (R) loop. FEBS J. 2018, 285, 2552–2566. [Google Scholar] [CrossRef] [Green Version]
- Vannier, J.B.; Pavicic-Kaltenbrunner, V.; Petalcorin, M.I.; Ding, H.; Boulton, S.J. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell 2012, 149, 795–806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clynes, D.; Jelinska, C.; Xella, B.; Ayyub, H.; Scott, C.; Mitson, M.; Taylor, S.; Higgs, D.R.; Gibbons, R.J. Suppression of the alternative lengthening of telomere pathway by the chromatin remodelling factor ATRX. Nat. Commun. 2015, 6, 7538. [Google Scholar] [CrossRef] [Green Version]
- Lu, R.; Pickett, H.A. Telomeric replication stress: The beginning and the end for alternative lengthening of telomeres cancers. Open Biol. 2022, 12, 220011. [Google Scholar] [CrossRef] [PubMed]
- Brenner, K.A.; Nandakumar, J. Consequences of telomere replication failure: The other end-replication problem. Trends Biochem. Sci. 2022, 47, 506–517. [Google Scholar] [CrossRef]
- Bower, K.; Napier, C.E.; Cole, S.L.; Dagg, R.A.; Lau, L.M.; Duncan, E.L.; Moy, E.L.; Reddel, R.R. Loss of wild-type ATRX expression in somatic cell hybrids segregates with activation of Alternative Lengthening of Telomeres. PLoS ONE 2012, 7, e50062. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Deng, Z.; Zhang, L.; Wu, C.; Jin, Y.; Hwang, I.; Vladimirova, O.; Xu, L.; Yang, L.; Lu, B.; et al. ATRX loss induces telomere dysfunction and necessitates induction of alternative lengthening of telomeres during human cell immortalization. EMBO J. 2019, 38, e96659. [Google Scholar] [CrossRef]
- Zeman, M.K.; Cimprich, K.A. Causes and consequences of replication stress. Nat. Cell Biol. 2014, 16, 2–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, J.S. The Heritability of Replication Problems. Cells 2021, 10, 1464. [Google Scholar] [CrossRef] [PubMed]
- Lukas, C.; Savic, V.; Bekker-Jensen, S.; Doil, C.; Neumann, B.; Pedersen, R.S.; Grofte, M.; Chan, K.L.; Hickson, I.D.; Bartek, J.; et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat. Cell. Biol. 2011, 13, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Minocherhomji, S.; Ying, S.; Bjerregaard, V.A.; Bursomanno, S.; Aleliunaite, A.; Wu, W.; Mankouri, H.W.; Shen, H.; Liu, Y.; Hickson, I.D. Replication stress activates DNA repair synthesis in mitosis. Nature 2015, 528, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Dilley, R.L.; Verma, P.; Cho, N.W.; Winters, H.D.; Wondisford, A.R.; Greenberg, R.A. Break-induced telomere synthesis underlies alternative telomere maintenance. Nature 2016, 539, 54–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhowmick, R.; Minocherhomji, S.; Hickson, I.D. RAD52 Facilitates Mitotic DNA Synthesis Following Replication Stress. Mol. Cell. 2016, 64, 1117–1126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pennarun, G.; Hoffschir, F.; Revaud, D.; Granotier, C.; Gauthier, L.R.; Mailliet, P.; Biard, D.S.; Boussin, F.D. ATR contributes to telomere maintenance in human cells. Nucleic Acids Res. 2010, 38, 2955–2963. [Google Scholar] [CrossRef] [Green Version]
- Sfeir, A.; Kosiyatrakul, S.T.; Hockemeyer, D.; MacRae, S.L.; Karlseder, J.; Schildkraut, C.L.; de Lange, T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 2009, 138, 90–103. [Google Scholar] [CrossRef] [Green Version]
- Özer, Ö.; Bhowmick, R.; Liu, Y.; Hickson, I.D. Human cancer cells utilize mitotic DNA synthesis to resist replication stress at telomeres regardless of their telomere maintenance mechanism. Oncotarget 2018, 9, 15836–15846. [Google Scholar] [CrossRef]
- Min, J.; Wright, W.E.; Shay, J.W. Alternative Lengthening of Telomeres Mediated by Mitotic DNA Synthesis Engages Break-Induced Replication Processes. Mol. Cell. Biol. 2017, 37, e00226-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Wang, B.; Li, T.; Liu, R.; Xiao, Y.; Geng, X.; Li, G.; Liu, Q.; Price, C.M.; Liu, Y.; et al. Mammalian CST averts replication failure by preventing G-quadruplex accumulation. Nucleic Acids Res. 2019, 47, 5243–5259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnes, R.P.; de Rosa, M.; Thosar, S.A.; Detwiler, A.C.; Roginskaya, V.; Van Houten, B.; Bruchez, M.P.; Stewart-Ornstein, J.; Opresko, P.L. Telomeric 8-oxo-guanine drives rapid premature senescence in the absence of telomere shortening. Nat. Struct. Mol. Biol. 2022, 29, 639–652. [Google Scholar] [CrossRef] [PubMed]
- Fouquerel, E.; Barnes, R.P.; Uttam, S.; Watkins, S.C.; Bruchez, M.P.; Opresko, P.L. Targeted and Persistent 8-Oxoguanine Base Damage at Telomeres Promotes Telomere Loss and Crisis. Mol. Cell. 2019, 75, 117–130.e6. [Google Scholar] [CrossRef]
- Yang, Z.; Takai, K.K.; Lovejoy, C.A.; de Lange, T. Break-induced replication promotes fragile telomere formation. Genes Dev. 2020, 34, 1392–1405. [Google Scholar] [CrossRef] [PubMed]
- Suram, A.; Kaplunov, J.; Patel, P.L.; Ruan, H.; Cerutti, A.; Boccardi, V.; Fumagalli, M.; Di Micco, R.; Mirani, N.; Gurung, R.L.; et al. Oncogene-induced telomere dysfunction enforces cellular senescence in human cancer precursor lesions. EMBO J. 2012, 31, 2839–2851. [Google Scholar] [CrossRef]
- Boccardi, V.; Razdan, N.; Kaplunov, J.; Mundra, J.J.; Kimura, M.; Aviv, A.; Herbig, U. Stn1 is critical for telomere maintenance and long-term viability of somatic human cells. Aging Cell. 2015, 14, 372–381. [Google Scholar] [CrossRef] [Green Version]
- Stout, G.J.; Blasco, M.A. Telomere length and telomerase activity impact the UV sensitivity syndrome xeroderma pigmentosum C. Cancer Res. 2013, 73, 1844–1854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Margalef, P.; Kotsantis, P.; Borel, V.; Bellelli, R.; Panier, S.; Boulton, S.J. Stabilization of Reversed Replication Forks by Telomerase Drives Telomere Catastrophe. Cell 2018, 172, 439–453.e14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porreca, R.M.; Herrera-Moyano, E.; Skourti, E.; Law, P.P.; Gonzalez Franco, R.; Montoya, A.; Faull, P.; Kramer, H.; Vannier, J.-B. TRF1 averts chromatin remodelling, recombination and replication dependent-break induced replication at mouse telomeres. eLife 2020, 9, e49817. [Google Scholar] [CrossRef]
- Min, J.; Wright, W.E.; Shay, J.W. Clustered telomeres in phase-separated nuclear condensates engage mitotic DNA synthesis through BLM and RAD52. Genes Dev. 2019, 33, 814–827. [Google Scholar] [CrossRef] [PubMed]
- Pinzaru, M.; Alexandra Hom, A.; Robert Beal, A.; Phillips, F.; Aaron Ni, E.; Cardozo, T.; Nair, N.; Choi, J.; Wuttke, S.; Deborah Sfeir, A.; et al. Telomere Replication Stress Induced by POT1 Inactivation Accelerates Tumorigenesis. Cell Rep. 2016, 15, 2170–2184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinzaru, A.M.; Kareh, M.; Lamm, N.; Lazzerini-Denchi, E.; Cesare, A.J.; Sfeir, A. Replication stress conferred by POT1 dysfunction promotes telomere relocalization to the nuclear pore. Genes Dev. 2020, 34, 1619–1636. [Google Scholar] [CrossRef] [PubMed]
- Verma, P.; Dilley, R.L.; Zhang, T.; Gyparaki, M.T.; Li, Y.; Greenberg, R.A. RAD52 and SLX4 act nonepistatically to ensure telomere stability during alternative telomere lengthening. Genes Dev. 2019, 33, 221–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotsantis, P.; Segura-Bayona, S.; Margalef, P.; Marzec, P.; Ruis, P.; Hewitt, G.; Bellelli, R.; Patel, H.; Goldstone, R.; Poetsch, A.R.; et al. RTEL1 Regulates G4/R-Loops to Avert Replication-Transcription Collisions. Cell Rep. 2020, 33, 108546. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Rossi, M.L.; Singh, D.K.; Dunn, C.; Ramamoorthy, M.; Croteau, D.L.; Liu, Y.; Bohr, V.A. RECQL4, the protein mutated in Rothmund-Thomson syndrome, functions in telomere maintenance. J. Biol. Chem. 2012, 287, 196–209. [Google Scholar] [CrossRef] [Green Version]
- Lin, W.; Sampathi, S.; Dai, H.; Liu, C.; Zhou, M.; Hu, J.; Huang, Q.; Campbell, J.; Shin-Ya, K.; Zheng, L.; et al. Mammalian DNA2 helicase/nuclease cleaves G-quadruplex DNA and is required for telomere integrity. EMBO J. 2013, 32, 1425–1439. [Google Scholar] [CrossRef] [Green Version]
- Kwon, M.S.; Lee, J.J.; Min, J.; Hwang, K.; Park, S.G.; Kim, E.H.; Kim, B.C.; Bhak, J.; Lee, H. Brca2 abrogation engages with the alternative lengthening of telomeres via break-induced replication. FEBS J. 2019, 286, 1841–1858. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.S.; Tejera, A.M.; Castor, D.; Toth, R.; Blasco, M.A.; Rouse, J. Localization-dependent and -independent roles of SLX4 in regulating telomeres. Cell Rep. 2013, 4, 853–860. [Google Scholar] [CrossRef] [Green Version]
- Guh, C.Y.; Shen, H.J.; Chen, L.W.; Chiu, P.C.; Liao, I.H.; Lo, C.C.; Chen, Y.; Hsieh, Y.H.; Chang, T.C.; Yen, C.P.; et al. XPF activates break-induced telomere synthesis. Nat. Commun. 2022, 13, 5781. [Google Scholar] [CrossRef]
- Van Overbeek, M.; de Lange, T. Apollo, an Artemis-related nuclease, interacts with TRF2 and protects human telomeres in S phase. Curr. Biol. 2006, 16, 1295–1302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petti, E.; Buemi, V.; Zappone, A.; Schillaci, O.; Broccia, P.V.; Dinami, R.; Matteoni, S.; Benetti, R.; Schoeftner, S. SFPQ and NONO suppress RNA:DNA-hybrid-related telomere instability. Nat. Commun. 2019, 10, 1001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le, P.N.; Maranon, D.G.; Altina, N.H.; Battaglia, C.L.; Bailey, S.M. TERRA, hnRNP A1, and DNA-PKcs Interactions at Human Telomeres. Front. Oncol. 2013, 3, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vohhodina, J.; Goehring, L.J.; Liu, B.; Kong, Q.; Botchkarev, V.V., Jr.; Huynh, M.; Liu, Z.; Abderazzaq, F.O.; Clark, A.P.; Ficarro, S.B.; et al. BRCA1 binds TERRA RNA and suppresses R-Loop-based telomeric DNA damage. Nat. Commun. 2021, 12, 3542. [Google Scholar] [CrossRef]
- Badie, S.; Escandell, J.M.; Bouwman, P.; Carlos, A.R.; Thanasoula, M.; Gallardo, M.M.; Suram, A.; Jaco, I.; Benitez, J.; Herbig, U.; et al. BRCA2 acts as a RAD51 loader to facilitate telomere replication and capping. Nat. Struct. Mol. Biol. 2010, 17, 1461–1469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Min, J.; Choi, E.S.; Hwang, K.; Kim, J.; Sampath, S.; Venkitaraman, A.R.; Lee, H. The breast cancer susceptibility gene BRCA2 is required for the maintenance of telomere homeostasis. J. Biol. Chem. 2012, 287, 5091–5101. [Google Scholar] [CrossRef] [Green Version]
- Barroso-Gonzalez, J.; Garcia-Exposito, L.; Hoang, S.M.; Lynskey, M.L.; Roncaioli, J.L.; Ghosh, A.; Wallace, C.T.; Modesti, M.; Bernstein, K.A.; Sarkar, S.N.; et al. RAD51AP1 Is an Essential Mediator of Alternative Lengthening of Telomeres. Mol. Cell. 2019, 76, 217. [Google Scholar] [CrossRef] [Green Version]
- Root, H.; Larsen, A.; Komosa, M.; Al-Azri, F.; Li, R.; Bazett-Jones, D.P.; Stephen Meyn, M. FANCD2 limits BLM-dependent telomere instability in the alternative lengthening of telomeres pathway. Hum. Mol. Genet. 2016, 25, 3255–3268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joksic, I.; Vujic, D.; Guc-Scekic, M.; Leskovac, A.; Petrovic, S.; Ojani, M.; Trujillo, J.P.; Surralles, J.; Zivkovic, M.; Stankovic, A.; et al. Dysfunctional telomeres in primary cells from Fanconi anemia FANCD2 patients. Genome Integr. 2012, 3, 6. [Google Scholar] [CrossRef] [Green Version]
- Lu, R.; O’Rourke, J.J.; Sobinoff, A.P.; Allen, J.A.M.; Nelson, C.B.; Tomlinson, C.G.; Lee, M.; Reddel, R.R.; Deans, A.J.; Pickett, H.A. The FANCM-BLM-TOP3A-RMI complex suppresses alternative lengthening of telomeres (ALT). Nat. Commun. 2019, 10, 2252. [Google Scholar] [CrossRef]
- Silva, B.; Pentz, R.; Figueira, A.M.; Arora, R.; Lee, Y.W.; Hodson, C.; Wischnewski, H.; Deans, A.J.; Azzalin, C.M. FANCM limits ALT activity by restricting telomeric replication stress induced by deregulated BLM and R-loops. Nat. Commun. 2019, 10, 2253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leman, A.R.; Dheekollu, J.; Deng, Z.; Lee, S.W.; Das, M.M.; Lieberman, P.M.; Noguchi, E. Timeless preserves telomere length by promoting efficient DNA replication through human telomeres. Cell Cycle 2012, 11, 2337–2347. [Google Scholar] [CrossRef] [Green Version]
- Shastrula, P.K.; Rice, C.T.; Wang, Z.; Lieberman, P.M.; Skordalakes, E. Structural and functional analysis of an OB-fold in human Ctc1 implicated in telomere maintenance and bone marrow syndromes. Nucleic Acids Res. 2018, 46, 972–984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, E.; Batenburg, N.L.; Walker, J.R.; Ho, A.; Mitchell, T.R.H.; Qin, J.; Zhu, X.D. CSB cooperates with SMARCAL1 to maintain telomere stability in ALT cells. J. Cell. Sci. 2020, 133, jcs234914. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Exposito, L.; Bournique, E.; Bergoglio, V.; Bose, A.; Barroso-Gonzalez, J.; Zhang, S.; Roncaioli, J.L.; Lee, M.; Wallace, C.T.; Watkins, S.C.; et al. Proteomic Profiling Reveals a Specific Role for Translesion DNA Polymerase eta in the Alternative Lengthening of Telomeres. Cell Rep. 2016, 17, 1858–1871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saharia, A.; Teasley, D.C.; Duxin, J.P.; Dao, B.; Chiappinelli, K.B.; Stewart, S.A. FEN1 ensures telomere stability by facilitating replication fork re-initiation. J. Biol. Chem. 2010, 285, 27057–27066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teasley, D.C.; Parajuli, S.; Nguyen, M.; Moore, H.R.; Alspach, E.; Lock, Y.J.; Honaker, Y.; Saharia, A.; Piwnica-Worms, H.; Stewart, S.A. Flap Endonuclease 1 Limits Telomere Fragility on the Leading Strand. J. Biol. Chem. 2015, 290, 15133–15145. [Google Scholar] [CrossRef] [Green Version]
- Barroso-Gonzalez, J.; Garcia-Exposito, L.; Galaviz, P.; Lynskey, M.L.; Allen, J.A.M.; Hoang, S.; Watkins, S.C.; Pickett, H.A.; O’Sullivan, R.J. Anti-recombination function of MutSalpha restricts telomere extension by ALT-associated homology-directed repair. Cell Rep. 2021, 37, 110088. [Google Scholar] [CrossRef]
- Sakellariou, D.; Bak, S.T.; Isik, E.; Barroso, S.I.; Porro, A.; Aguilera, A.; Bartek, J.; Janscak, P.; Pena-Diaz, J. MutSbeta regulates G4-associated telomeric R-loops to maintain telomere integrity in ALT cancer cells. Cell Rep. 2022, 39, 110602. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Sharma, K.; de Lange, T. TRF1 uses a noncanonical function of TFIIH to promote telomere replication. Genes Dev. 2022, 36, 956–969. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Kim, H.; Ji, Z.; Zhang, T.; Chen, B.; Ge, Y.; Hu, Y.; Feng, X.; Han, X.; Xu, H.; et al. The BUB3-BUB1 Complex Promotes Telomere DNA Replication. Mol. Cell. 2018, 70, 395–407.e4. [Google Scholar] [CrossRef] [Green Version]
- Chan, F.L.; Vinod, B.; Novy, K.; Schittenhelm, R.B.; Huang, C.; Udugama, M.; Nunez-Iglesias, J.; Lin, J.I.; Hii, L.; Chan, J.; et al. Aurora Kinase B, a novel regulator of TERF1 binding and telomeric integrity. Nucleic Acids Res. 2017, 45, 12340–12353. [Google Scholar] [CrossRef] [Green Version]
- D’Alcontres, M.S.; Palacios, J.A.; Mejias, D.; Blasco, M.A. TopoIIalpha prevents telomere fragility and formation of ultra thin DNA bridges during mitosis through TRF1-dependent binding to telomeres. Cell Cycle 2014, 13, 1463–1481. [Google Scholar] [CrossRef] [Green Version]
- Min, J.N.; Tian, Y.; Xiao, Y.; Wu, L.; Li, L.; Chang, S. The mINO80 chromatin remodeling complex is required for efficient telomere replication and maintenance of genome stability. Cell Res. 2013, 23, 1396–1413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez, P.; Thanasoula, M.; Munoz, P.; Liao, C.; Tejera, A.; Mcnees, C.; Flores, J.M.; Fernandez-Capetillo, O.; Tarsounas, M.; Blasco, M.A. Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev. 2009, 23, 2060–2075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmermann, M.; Kibe, T.; Kabir, S.; de Lange, T. TRF1 negotiates TTAGGG repeat-associated replication problems by recruiting the BLM helicase and the TPP1/POT1 repressor of ATR signaling. Genes Dev. 2014, 28, 2477–2491. [Google Scholar] [CrossRef] [Green Version]
- Mendoza, O.; Bourdoncle, A.; Boule, J.B.; Brosh, R.M., Jr.; Mergny, J.L. G-quadruplexes and helicases. Nucleic Acids Res. 2016, 44, 1989–2006. [Google Scholar] [CrossRef] [Green Version]
- Lim, C.J.; Cech, T.R. Shaping human telomeres: From shelterin and CST complexes to telomeric chromatin organization. Nat. Rev. Mol. Cell Biol. 2021, 22, 283–298. [Google Scholar] [CrossRef] [PubMed]
- Sarek, G.; Vannier, J.B.; Panier, S.; Petrini, J.H.; Boulton, S.J. TRF2 recruits RTEL1 to telomeres in S phase to promote t-loop unwinding. Mol. Cell. 2015, 57, 622–635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergoglio, V.; Boyer, A.-S.; Walsh, E.; Naim, V.; Legube, G.; Lee, M.Y.W.T.; Rey, L.; Rosselli, F.; Cazaux, C.; Eckert, K.A.; et al. DNA synthesis by Pol η promotes fragile site stability by preventing under-replicated DNA in mitosis. J. Cell Biol. 2013, 201, 395–408. [Google Scholar] [CrossRef] [PubMed]
- Sonneville, R.; Bhowmick, R.; Hoffmann, S.; Mailand, N.; Hickson, I.D.; Labib, K. TRAIP drives replisome disassembly and mitotic DNA repair synthesis at sites of incomplete DNA replication. eLife 2019, 8, e48686. [Google Scholar] [CrossRef] [PubMed]
- Villa, F.; Fujisawa, R.; Ainsworth, J.; Nishimura, K.; Lie, A.L.M.; Lacaud, G.; Labib, K.P. CUL2(LRR1), TRAIP and p97 control CMG helicase disassembly in the mammalian cell cycle. EMBO Rep. 2021, 22, e52164. [Google Scholar] [CrossRef] [PubMed]
- Graber-Feesl, C.L.; Pederson, K.D.; Aney, K.J.; Shima, N. Mitotic DNA Synthesis Is Differentially Regulated between Cancer and Noncancerous Cells. Mol. Cancer Res. 2019, 17, 1687–1698. [Google Scholar] [CrossRef]
- Chan, K.L.; Palmai-Pallag, T.; Ying, S.; Hickson, I.D. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat. Cell Biol. 2009, 11, 753–760. [Google Scholar] [CrossRef] [PubMed]
- Donnianni, R.A.; Symington, L.S. Break-induced replication occurs by conservative DNA synthesis. Proc. Natl. Acad. Sci. USA 2013, 110, 13475–13480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kramara, J.; Osia, B.; Malkova, A. Break-Induced Replication: The Where, The Why, and The How. Trends Genet. 2018, 34, 518–531. [Google Scholar] [CrossRef]
- Ying, S.; Minocherhomji, S.; Chan, K.L.; Palmai-Pallag, T.; Chu, W.K.; Wass, T.; Mankouri, H.W.; Liu, Y.; Hickson, I.D. MUS81 promotes common fragile site expression. Nat. Cell Biol. 2013, 15, 1001–1007. [Google Scholar] [CrossRef]
- Lundblad, V.; Blackburn, E.H. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell 1993, 73, 347–360. [Google Scholar] [CrossRef] [PubMed]
- Lydeard, J.R.; Jain, S.; Yamaguchi, M.; Haber, J.E. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature 2007, 448, 820–823. [Google Scholar] [CrossRef] [PubMed]
- Stavropoulos, D.J.; Bradshaw, P.S.; Li, X.; Pasic, I.; Truong, K.; Ikura, M.; Ungrin, M.; Meyn, M.S. The Bloom syndrome helicase BLM interacts with TRF2 in ALT cells and promotes telomeric DNA synthesis. Hum. Mol. Genet. 2002, 11, 3135–3144. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Drosopoulos, W.C.; Sethi, L.; Madireddy, A.; Schildkraut, C.L.; Zhang, D. FANCM, BRCA1, and BLM cooperatively resolve the replication stress at the ALT telomeres. Proc. Natl. Acad. Sci. USA 2017, 114, E5940–E5949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conomos, D.; Reddel, R.R.; Pickett, H.A. NuRD-ZNF827 recruitment to telomeres creates a molecular scaffold for homologous recombination. Nat. Struct. Mol. Biol. 2014, 21, 760–770. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.H.; Jiang, W.Q.; Cesare, A.J.; Neumann, A.A.; Wadhwa, R.; Reddel, R.R. Disruption of telomere maintenance by depletion of the MRE11/RAD50/NBS1 complex in cells that use alternative lengthening of telomeres. J. Biol. Chem. 2007, 282, 29314–29322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, B.; Yin, J.; Horvath, K.; Sarkar, J.; Chen, Y.; Wu, J.; Wan, K.; Lu, J.; Gu, P.; Yu, E.Y.; et al. SLX4 assembles a telomere maintenance toolkit by bridging multiple endonucleases with telomeres. Cell. Rep. 2013, 4, 861–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Potts, P.R.; Yu, H. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat. Struct. Mol. Biol. 2007, 14, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Hockemeyer, D.; Sfeir, A.J.; Shay, J.W.; Wright, W.E.; De Lange, T. POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J. 2005, 24, 2667–2678. [Google Scholar] [CrossRef] [Green Version]
- Bettin, N.; Oss Pegorar, C.; Cusanelli, E. The Emerging Roles of TERRA in Telomere Maintenance and Genome Stability. Cells 2019, 8, 246. [Google Scholar] [CrossRef] [Green Version]
- Moravec, M.; Wischnewski, H.; Bah, A.; Hu, Y.; Liu, N.; Lafranchi, L.; King, M.C.; Azzalin, C.M. TERRA promotes telomerase-mediated telomere elongation in Schizosaccharomyces pombe. EMBO Rep. 2016, 17, 999–1012. [Google Scholar] [CrossRef] [Green Version]
- Deng, Z.; Norseen, J.; Wiedmer, A.; Riethman, H.; Lieberman, P.M. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell. 2009, 35, 403–413. [Google Scholar] [CrossRef] [Green Version]
- Porro, A.; Feuerhahn, S.; Lingner, J. TERRA-reinforced association of LSD1 with MRE11 promotes processing of uncapped telomeres. Cell. Rep. 2014, 6, 765–776. [Google Scholar] [CrossRef] [PubMed]
- Porro, A.; Feuerhahn, S.; Reichenbach, P.; Lingner, J. Molecular dissection of telomeric repeat-containing RNA biogenesis unveils the presence of distinct and multiple regulatory pathways. Mol. Cell Biol. 2010, 30, 4808–4817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flynn, R.L.; Centore, R.C.; O’Sullivan, R.J.; Rai, R.; Tse, A.; Songyang, Z.; Chang, S.; Karlseder, J.; Zou, L. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature 2011, 471, 532–536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arora, R.; Lee, Y.; Wischnewski, H.; Brun, C.M.; Schwarz, T.; Azzalin, C.M. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat. Commun. 2014, 5, 5220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verdun, R.E.; Karlseder, J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell 2006, 127, 709–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferguson, B.M.; Brewer, B.J.; Reynolds, A.E.; Fangman, W.L. A yeast origin of replication is activated late in S phase. Cell 1991, 65, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Unsal-Kacmaz, K.; Chastain, P.D.; Qu, P.P.; Minoo, P.; Cordeiro-Stone, M.; Sancar, A.; Kaufmann, W.K. The human Tim/Tipin complex coordinates an Intra-S checkpoint response to UV that slows replication fork displacement. Mol. Cell Biol. 2007, 27, 3131–3142. [Google Scholar] [CrossRef] [Green Version]
- Madireddy, A.; Kosiyatrakul, S.T.; Boisvert, R.A.; Herrera-Moyano, E.; Garcia-Rubio, M.L.; Gerhardt, J.; Vuono, E.A.; Owen, N.; Yan, Z.; Olson, S.; et al. FANCD2 Facilitates Replication through Common Fragile Sites. Mol Cell. 2016, 64, 388–404. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.; Qin, J.; Wang, L.; Lee, H.J.; Kao, C.Y.; Liu, D.; Songyang, Z.; Chen, J.; Tsai, M.J.; Tsai, S.Y. Nuclear receptors regulate alternative lengthening of telomeres through a novel noncanonical FANCD2 pathway. Sci. Adv. 2019, 5, eaax6366. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Pickett, H.A. Targeting telomeres: Advances in telomere maintenance mechanism-specific cancer therapies. Nat. Rev. Cancer 2022, 22, 515–532. [Google Scholar] [CrossRef] [PubMed]
- Koneru, B.; Farooqi, A.; Nguyen, T.H.; Chen, W.H.; Hindle, A.; Eslinger, C.; Makena, M.R.; Burrow, T.A.; Wilson, J.; Smith, A.; et al. ALT neuroblastoma chemoresistance due to telomere dysfunction-induced ATM activation is reversible with ATM inhibitor AZD0156. Sci. Transl. Med. 2021, 13, eabd5750. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Eberhart, C.G.; Pratilas, C.A.; Blakeley, J.O.; Davis, C.; Stojanova, M.; Reilly, K.; Meeker, A.K.; Heaphy, C.M.; Rodriguez, F.J. Therapeutic Vulnerability to ATR Inhibition in Concurrent NF1 and ATRX-Deficient/ALT-Positive High-Grade Solid Tumors. Cancers 2022, 14, 3015. [Google Scholar] [CrossRef]
- Flynn, R.L.; Cox, K.E.; Jeitany, M.; Wakimoto, H.; Bryll, A.R.; Ganem, N.J.; Bersani, F.; Pineda, J.R.; Suva, M.L.; Benes, C.H.; et al. Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science 2015, 347, 273–277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laroche-Clary, A.; Chaire, V.; Verbeke, S.; Algeo, M.P.; Malykh, A.; Le Loarer, F.; Italiano, A. ATR Inhibition Broadly Sensitizes Soft-Tissue Sarcoma Cells to Chemotherapy Independent of Alternative Lengthening Telomere (ALT) Status. Sci. Rep. 2020, 10, 7488. [Google Scholar] [CrossRef] [PubMed]
- Deeg, K.I.; Chung, I.; Bauer, C.; Rippe, K. Cancer Cells with Alternative Lengthening of Telomeres Do Not Display a General Hypersensitivity to ATR Inhibition. Front. Oncol. 2016, 6, 186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruis, P.; Boulton, S.J. The end protection problem—An unexpected twist in the tail. Genes Dev. 2021, 35, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Cong, K.; Cantor, S.B. Exploiting replication gaps for cancer therapy. Mol. Cell. 2022, 82, 2363–2369. [Google Scholar] [CrossRef]

| Effects of Protein Depletion or Reagent Addition * | ||||
|---|---|---|---|---|
| Fragility | Mitotic DNA Synthesis | Spontaneous G2 Synthesis | BITS (FOKI-TRF1) | |
| Reagent | ||||
| ATRi/ATR-deficiency | Increase [18,19] | Increase [20] | Increase [16] | |
| ATMi | No Change [21] | |||
| Aphidicolin | Increase [19] | Increase [20] | ||
| G4 ligand | Increase b [21,22] | Increase [21] | ||
| FAP-TRF1 8oxoG | Increase [23] | Increase [23,24] | ||
| FokI-TRF1 break | Increase [25] | Increase [16] | ||
| Oncogene OE a | Increase [21,26] | Increase [21] | ||
| Oxidative stress | Increase [27,28] | |||
| PARPi | Decrease g [29] Increase c [25] | |||
| Shelterin | ||||
| TRF1 | Increase [19,30] | Increase [30] | ||
| TRF2 | No Change [19] | No Change d [31] | ||
| POT1 | Increase [32] | Increase [33] | ||
| Helicase | ||||
| BLM | Increase [19] | OE Increases [31] | Decrease [34] | |
| WRN | No Change [19] | |||
| RTEL1 | Increase b [6] | Decrease [35] | ||
| RECQL4 | Increase b [36] | |||
| Nuclease | ||||
| DNA2 | Increase b [37] | |||
| MRE11 | Increase [38] | Decrease [21] | ||
| SLX1/SLX4 | Increase [34,39] Decrease c [25] | Decrease [20] | No Change [34] | |
| MUS81 | No Change c [25] | No Change [20] | No Change [34] | |
| XPF | Increase c [25] | Decrease [40] | ||
| Apollo | Increase [41] | |||
| RNA Metabolism | ||||
| RNaseH1 | Increase [21] | Increase [21] | ||
| NONO | Increase, Leading Strand [42] | |||
| hnRNP1A | Increase [43] | |||
| HR Factor | ||||
| BRCA1 | Increase: U2OS [44] No Change: MEF [45] | Increase [21] | ||
| BRCA2 | Increase b [38,45,46] | Increase [38] | ||
| RAD51 | Increase [21] | Increase [21] | Increase [16] | Increase [16] |
| RAD51AP1 | Increase or Decrease f [25,47] | Decrease [47] | ||
| RAD52 | No Change [21] | Decrease [21] | Decrease [34] | Increase [34] |
| Fork Remodeler /Protector | ||||
| FANCD2 | Increase [48,49] | |||
| FANCM | OE Decreases [50] | Increase [40,51] | ||
| Timeless/Tipin | Increase [52] | Increase [21] | ||
| STN1/CTC1 | Increase [22,27,32,53] | |||
| SMARCAL1 | Increase [54] | |||
| ZRANB3 | Decrease g [29] | |||
| PARP1 | Decrease g [29] | |||
| DNA Replication Factor | ||||
| POLD1,3,4 | No Change TRF1ko [30], Decrease c [25] | Decrease [30] | Decrease [16] | Decrease [16] |
| POLE1,2 | Increase [16] | Increase [16] | ||
| POLA1,2 | Increase [16] | Increase [16] | ||
| Prim1 | No Change [16] | |||
| MCM2/7 | Increase [16] | |||
| PCNA | Decrease [16] | Decrease [16] | ||
| RFC1 | Decrease [16] | Decrease [16] | ||
| POLH (Pol eta) | Increase With APH [55] | Increase With APH [55] | No Change [16] | |
| REV3L (Pol zeta) | Increase [16] | Increase [16] | ||
| FEN1 | Increase b [56,57] | |||
| DNA Excision-repair Protein | ||||
| OGG1 | Increase e [24] | Increase e [24] | ||
| CSB | Increase b [54] | |||
| MSH6 | Increase [58] | |||
| MSH2 | Decrease [59] | |||
| MSH3 | Increase [59] | |||
| XPC | Increase [28] | |||
| XPD, XPB, TFIIH | Increase [60] | |||
| Mitotic Checkpoint | ||||
| BUB1/BUB3 | Increase [61] | |||
| Aurora Kinase B | Increase [62] | |||
| Chromatin Structure and Cohesion | ||||
| TopIIα | Increase [63] | |||
| SMC5/6 | No change [30] | Decrease (ALT) [21], No change (TRF1ko) [30] | ||
| INO80 | Increase [64] | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barnes, R.P.; Thosar, S.A.; Opresko, P.L. Telomere Fragility and MiDAS: Managing the Gaps at the End of the Road. Genes 2023, 14, 348. https://doi.org/10.3390/genes14020348
Barnes RP, Thosar SA, Opresko PL. Telomere Fragility and MiDAS: Managing the Gaps at the End of the Road. Genes. 2023; 14(2):348. https://doi.org/10.3390/genes14020348
Chicago/Turabian StyleBarnes, Ryan P., Sanjana A. Thosar, and Patricia L. Opresko. 2023. "Telomere Fragility and MiDAS: Managing the Gaps at the End of the Road" Genes 14, no. 2: 348. https://doi.org/10.3390/genes14020348
APA StyleBarnes, R. P., Thosar, S. A., & Opresko, P. L. (2023). Telomere Fragility and MiDAS: Managing the Gaps at the End of the Road. Genes, 14(2), 348. https://doi.org/10.3390/genes14020348





