Nonsense-Mediated mRNA Decay as a Mediator of Tumorigenesis
Abstract
1. Introduction
2. Physiological Mechanisms of NMD
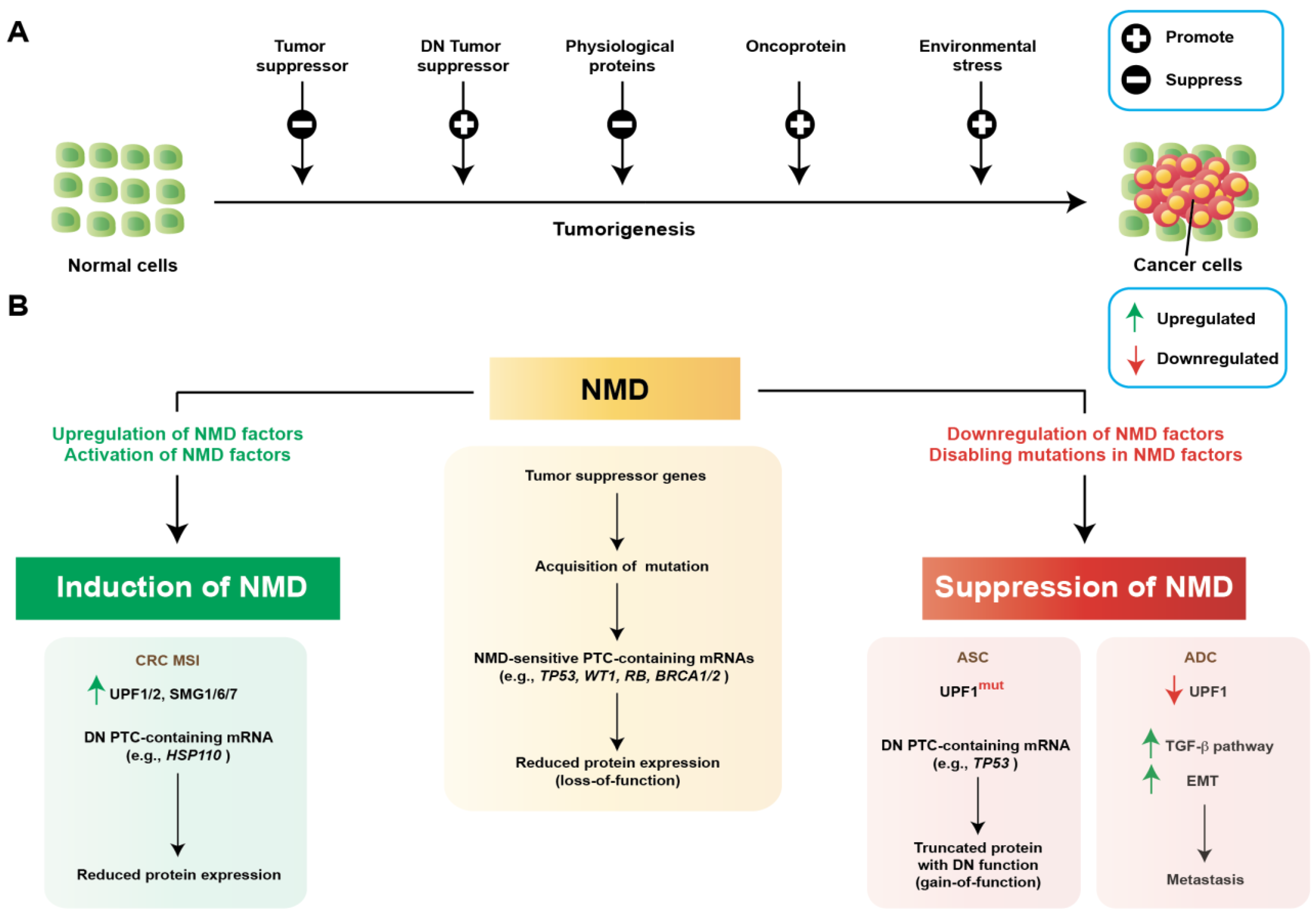
3. Regulation of NMD in Cancer
3.1. Activation of NMD to Promote Tumorigenesis
3.2. Suppression of NMD to Promote Tumorigenesis
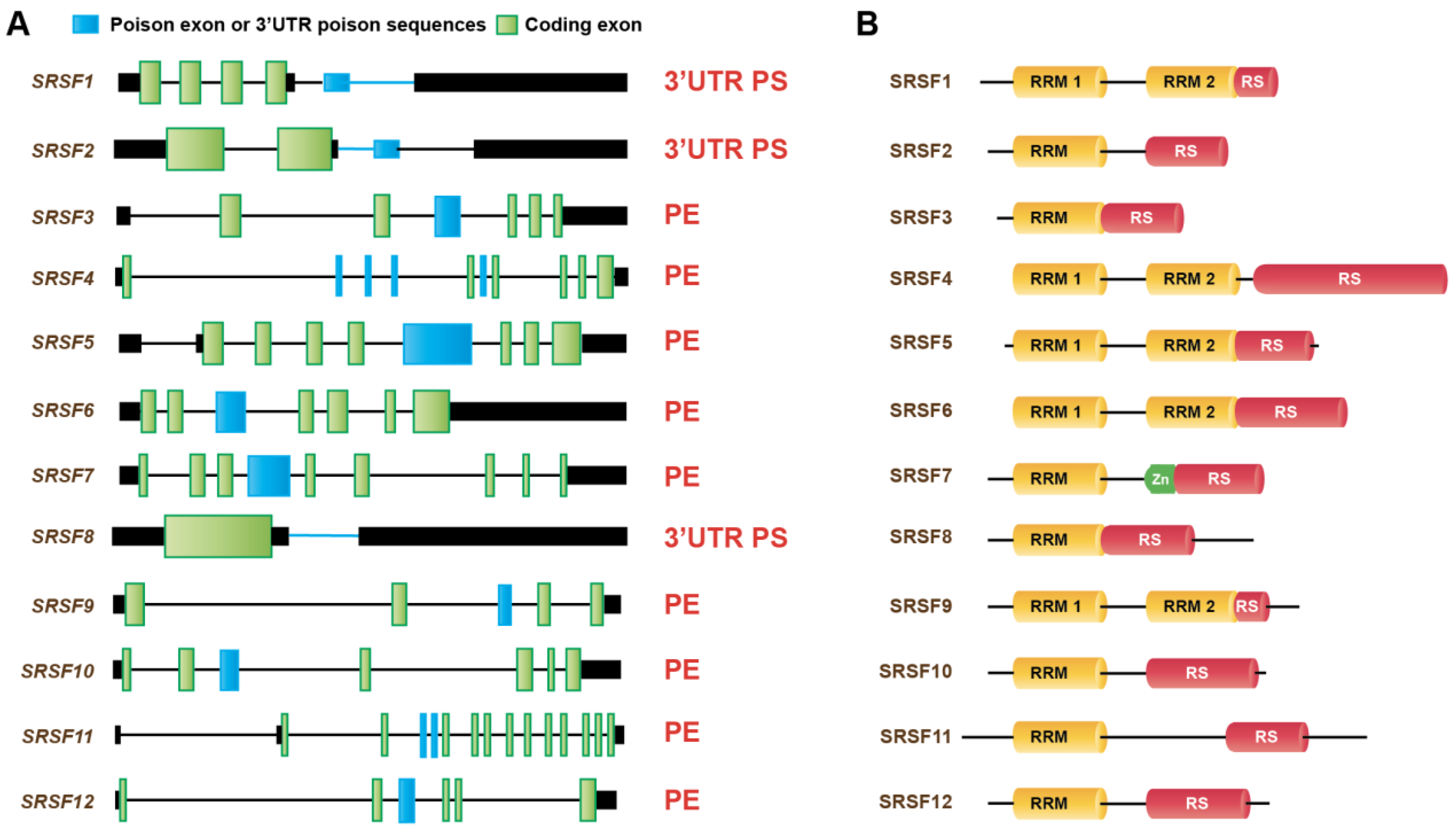
4. Regulation of Alternative Splicing Coupled to NMD in Cancer
4.1. Roles of SR Protein Splicing Factors Regulating AS-NMD in Cancer
4.2. Roles of Other Splicing Factors and RNA Binding Proteins Regulating AS-NMD in Cancer
5. NMD under Tumor Microenvironment
6. Perspectives and Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Popp, M.W.; Maquat, L.E. Nonsense-Mediated mRNA Decay and Cancer. Curr. Opin. Genet. Dev. 2018, 48, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Lykke-Andersen, S.; Jensen, T.H. Nonsense-Mediated mRNA Decay: An Intricate Machinery That Shapes Transcriptomes. Nat. Rev. Mol. Cell Biol. 2015, 16, 665–677. [Google Scholar] [CrossRef]
- Miller, J.N.; Pearce, D.A. Nonsense-Mediated Decay in Genetic Disease: Friend or Foe? Mutat. Res. Rev. Mutat. Res. 2014, 762, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.; Maquat, L.E. Nonsense-Mediated mRNA Decay (NMD) in Animal Embryogenesis: To Die or Not to Die, That Is the Question. Curr. Opin. Genet. Dev. 2011, 21, 422–430. [Google Scholar] [CrossRef]
- Brogna, S.; Wen, J. Nonsense-Mediated mRNA Decay (NMD) Mechanisms. Nat. Struct. Mol. Biol. 2009, 16, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Tani, H.; Imamachi, N.; Salam, K.A.; Mizutani, R.; Ijiri, K.; Irie, T.; Yada, T.; Suzuki, Y.; Akimitsu, N. Identification of Hundreds of Novel UPF1 Target Transcripts by Direct Determination of Whole Transcriptome Stability. RNA Biol. 2012, 9, 1370–1379. [Google Scholar] [CrossRef]
- Mendell, J.T.; Sharifi, N.A.; Meyers, J.L.; Martinez-Murillo, F.; Dietz, H.C. Nonsense Surveillance Regulates Expression of Diverse Classes of Mammalian Transcripts and Mutes Genomic Noise. Nat. Genet. 2004, 36, 1073–1078. [Google Scholar] [CrossRef]
- Nasif, S.; Contu, L.; Mühlemann, O. Beyond Quality Control: The Role of Nonsense-Mediated mRNA Decay (NMD) in Regulating Gene Expression. Semin. Cell Dev. Biol. 2018, 75, 78–87. [Google Scholar] [CrossRef]
- Leclair, N.K.; Brugiolo, M.; Urbanski, L.; Lawson, S.C.; Thakar, K.; Yurieva, M.; George, J.; Hinson, J.T.; Cheng, A.; Graveley, B.R.; et al. Poison Exon Splicing Regulates a Coordinated Network of SR Protein Expression during Differentiation and Tumorigenesis. Mol. Cell 2020, 80, 648–665.e9. [Google Scholar] [CrossRef]
- Chang, L.; Li, C.; Guo, T.; Wang, H.; Ma, W.; Yuan, Y.; Liu, Q.; Ye, Q.; Liu, Z. The Human RNA Surveillance Factor UPF1 Regulates Tumorigenesis by Targeting Smad7 in Hepatocellular Carcinoma. J. Exp. Clin. Cancer Res. CR 2016, 35, 8. [Google Scholar] [CrossRef] [PubMed]
- Bokhari, A.; Jonchere, V.; Lagrange, A.; Bertrand, R.; Svrcek, M.; Marisa, L.; Buhard, O.; Greene, M.; Demidova, A.; Jia, J.; et al. Targeting Nonsense-Mediated mRNA Decay in Colorectal Cancers with Microsatellite Instability. Oncogenesis 2018, 7, 70. [Google Scholar] [CrossRef] [PubMed]
- Metze, S.; Herzog, V.A.; Ruepp, M.-D.; Mühlemann, O. Comparison of EJC-Enhanced and EJC-Independent NMD in Human Cells Reveals Two Partially Redundant Degradation Pathways. RNA 2013, 19, 1432–1448. [Google Scholar] [CrossRef] [PubMed]
- Le Hir, H.; Izaurralde, E.; Maquat, L.E.; Moore, M.J. The Spliceosome Deposits Multiple Proteins 20-24 Nucleotides Upstream of mRNA Exon-Exon Junctions. EMBO J. 2000, 19, 6860–6869. [Google Scholar] [CrossRef] [PubMed]
- Le Hir, H.; Gatfield, D.; Izaurralde, E.; Moore, M.J. The Exon-Exon Junction Complex Provides a Binding Platform for Factors Involved in mRNA Export and Nonsense-Mediated mRNA Decay. EMBO J. 2001, 20, 4987–4997. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.-K.; Huang, L.; Gudikote, J.P.; Chang, Y.-F.; Imam, J.S.; MacLean, J.A.; Wilkinson, M.F. An Alternative Branch of the Nonsense-Mediated Decay Pathway. EMBO J. 2007, 26, 1820–1830. [Google Scholar] [CrossRef]
- Alexandrov, A.; Colognori, D.; Shu, M.-D.; Steitz, J.A. Human Spliceosomal Protein CWC22 Plays a Role in Coupling Splicing to Exon Junction Complex Deposition and Nonsense-Mediated Decay. Proc. Natl. Acad. Sci. USA 2012, 109, 21313–21318. [Google Scholar] [CrossRef]
- Singh, G.; Kucukural, A.; Cenik, C.; Leszyk, J.D.; Shaffer, S.A.; Weng, Z.; Moore, M.J. The Cellular EJC Interactome Reveals Higher-Order mRNP Structure and an EJC-SR Protein Nexus. Cell 2012, 151, 750–764. [Google Scholar] [CrossRef]
- Saulière, J.; Murigneux, V.; Wang, Z.; Marquenet, E.; Barbosa, I.; Le Tonquèze, O.; Audic, Y.; Paillard, L.; Roest Crollius, H.; Le Hir, H. CLIP-Seq of eIF4AIII Reveals Transcriptome-Wide Mapping of the Human Exon Junction Complex. Nat. Struct. Mol. Biol. 2012, 19, 1124–1131. [Google Scholar] [CrossRef]
- Ishigaki, Y.; Li, X.; Serin, G.; Maquat, L.E. Evidence for a Pioneer Round of mRNA Translation: mRNAs Subject to Nonsense-Mediated Decay in Mammalian Cells Are Bound by CBP80 and CBP20. Cell 2001, 106, 607–617. [Google Scholar] [CrossRef]
- Lejeune, F.; Ishigaki, Y.; Li, X.; Maquat, L.E. The Exon Junction Complex Is Detected on CBP80-Bound but Not EIF4E-Bound mRNA in Mammalian Cells: Dynamics of mRNP Remodeling. EMBO J. 2002, 21, 3536–3545. [Google Scholar] [CrossRef]
- Gehring, N.H.; Lamprinaki, S.; Hentze, M.W.; Kulozik, A.E. The Hierarchy of Exon-Junction Complex Assembly by the Spliceosome Explains Key Features of Mammalian Nonsense-Mediated mRNA Decay. PLOS Biol. 2009, 7, e1000120. [Google Scholar] [CrossRef] [PubMed]
- Gehring, N.H.; Lamprinaki, S.; Kulozik, A.E.; Hentze, M.W. Disassembly of Exon Junction Complexes by PYM. Cell 2009, 137, 536–548. [Google Scholar] [CrossRef] [PubMed]
- Popp, M.W.-L.; Maquat, L.E. The Dharma of Nonsense-Mediated mRNA Decay in Mammalian Cells. Mol. Cells 2014, 37, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, A.; Izumi, N.; Kashima, I.; Ohnishi, T.; Saari, B.; Katsuhata, Y.; Muramatsu, R.; Morita, T.; Iwamatsu, A.; Hachiya, T.; et al. SMG-8 and SMG-9, Two Novel Subunits of the SMG-1 Complex, Regulate Remodeling of the mRNA Surveillance Complex during Nonsense-Mediated mRNA Decay. Genes Dev. 2009, 23, 1091–1105. [Google Scholar] [CrossRef]
- Shum, E.Y.; Jones, S.H.; Shao, A.; Dumdie, J.; Krause, M.D.; Chan, W.-K.; Lou, C.-H.; Espinoza, J.L.; Song, H.-W.; Phan, M.H.; et al. The Antagonistic Gene Paralogs Upf3a and Upf3b Govern Nonsense-Mediated RNA Decay. Cell 2016, 165, 382–395. [Google Scholar] [CrossRef]
- Kashima, I.; Yamashita, A.; Izumi, N.; Kataoka, N.; Morishita, R.; Hoshino, S.; Ohno, M.; Dreyfuss, G.; Ohno, S. Binding of a Novel SMG-1-Upf1-ERF1-ERF3 Complex (SURF) to the Exon Junction Complex Triggers Upf1 Phosphorylation and Nonsense-Mediated mRNA Decay. Genes Dev. 2006, 20, 355–367. [Google Scholar] [CrossRef]
- Nicholson, P.; Gkratsou, A.; Josi, C.; Colombo, M.; Mühlemann, O. Dissecting the Functions of SMG5, SMG7, and PNRC2 in Nonsense-Mediated mRNA Decay of Human Cells. RNA 2018, 24, 557–573. [Google Scholar] [CrossRef]
- Okada-Katsuhata, Y.; Yamashita, A.; Kutsuzawa, K.; Izumi, N.; Hirahara, F.; Ohno, S. N- and C-Terminal Upf1 Phosphorylations Create Binding Platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res. 2012, 40, 1251–1266. [Google Scholar] [CrossRef]
- Cho, H.; Kim, K.M.; Kim, Y.K. Human Proline-Rich Nuclear Receptor Coregulatory Protein 2 Mediates an Interaction between mRNA Surveillance Machinery and Decapping Complex. Mol. Cell 2009, 33, 75–86. [Google Scholar] [CrossRef]
- Kashima, I.; Jonas, S.; Jayachandran, U.; Buchwald, G.; Conti, E.; Lupas, A.N.; Izaurralde, E. SMG6 Interacts with the Exon Junction Complex via Two Conserved EJC-Binding Motifs (EBMs) Required for Nonsense-Mediated mRNA Decay. Genes Dev. 2010, 24, 2440. [Google Scholar] [CrossRef]
- Huntzinger, E.; Kashima, I.; Fauser, M.; Saulière, J.; Izaurralde, E. SMG6 Is the Catalytic Endonuclease That Cleaves mRNAs Containing Nonsense Codons in Metazoan. RNA 2008, 14, 2609–2617. [Google Scholar] [CrossRef] [PubMed]
- Eberle, A.B.; Lykke-Andersen, S.; Mühlemann, O.; Jensen, T.H. SMG6 Promotes Endonucleolytic Cleavage of Nonsense mRNA in Human Cells. Nat. Struct. Mol. Biol. 2009, 16, 49–55. [Google Scholar] [CrossRef]
- Lejeune, F.; Li, X.; Maquat, L.E. Nonsense-Mediated mRNA Decay in Mammalian Cells Involves Decapping, Deadenylating, and Exonucleolytic Activities. Mol. Cell 2003, 12, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Unterholzner, L.; Izaurralde, E. SMG7 Acts as a Molecular Link between mRNA Surveillance and mRNA Decay. Mol. Cell 2004, 16, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Boehm, V.; Kueckelmann, S.; Gerbracht, J.V.; Kallabis, S.; Britto-Borges, T.; Altmüller, J.; Krüger, M.; Dieterich, C.; Gehring, N.H. SMG5-SMG7 Authorize Nonsense-Mediated mRNA Decay by Enabling SMG6 Endonucleolytic Activity. Nat. Commun. 2021, 12, 3965. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Karousis, E.D.; Bourquin, J.; Bruggmann, R.; Mühlemann, O. Transcriptome-Wide Identification of NMD-Targeted Human mRNAs Reveals Extensive Redundancy between SMG6- and SMG7-Mediated Degradation Pathways. RNA 2017, 23, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Jonas, S.; Weichenrieder, O.; Izaurralde, E. An Unusual Arrangement of Two 14-3-3-like Domains in the SMG5-SMG7 Heterodimer Is Required for Efficient Nonsense-Mediated mRNA Decay. Genes Dev. 2013, 27, 211–225. [Google Scholar] [CrossRef]
- Boehm, V.; Gerbracht, J.V.; Marx, M.-C.; Gehring, N.H. Interrogating the Degradation Pathways of Unstable mRNAs with XRN1-Resistant Sequences. Nat. Commun. 2016, 7, 13691. [Google Scholar] [CrossRef]
- Toma, K.G.; Rebbapragada, I.; Durand, S.; Lykke-Andersen, J. Identification of Elements in Human Long 3′ UTRs That Inhibit Nonsense-Mediated Decay. RNA N. Y. N 2015, 21, 887–897. [Google Scholar] [CrossRef]
- Ge, Z.; Quek, B.L.; Beemon, K.L.; Hogg, J.R. Polypyrimidine Tract Binding Protein 1 Protects mRNAs from Recognition by the Nonsense-Mediated mRNA Decay Pathway. eLife 2016, 5, e11155. [Google Scholar] [CrossRef]
- Matsuda, D.; Hosoda, N.; Kim, Y.K.; Maquat, L.E. Failsafe Nonsense-Mediated mRNA Decay Does Not Detectably Target eIF4E-Bound mRNA. Nat. Struct. Mol. Biol. 2007, 14, 974–979. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.L.; Ribeiro, P.; Inácio, A.; Liebhaber, S.A.; Romão, L. Proximity of the Poly(A)-Binding Protein to a Premature Termination Codon Inhibits Mammalian Nonsense-Mediated mRNA Decay. RNA 2008, 14, 563–576. [Google Scholar] [CrossRef] [PubMed]
- Kurosaki, T.; Popp, M.W.; Maquat, L.E. Quality and Quantity Control of Gene Expression by Nonsense-Mediated mRNA Decay. Nat. Rev. Mol. Cell Biol. 2019, 20, 406–420. [Google Scholar] [CrossRef]
- Lindeboom, R.G.H.; Supek, F.; Lehner, B. The Rules and Impact of Nonsense-Mediated mRNA Decay in Human Cancers. Nat. Genet. 2016, 48, 1112–1118. [Google Scholar] [CrossRef]
- Rhee, J.-K.; Lee, S.; Park, W.-Y.; Kim, Y.-H.; Kim, T.-M. Allelic Imbalance of Somatic Mutations in Cancer Genomes and Transcriptomes. Sci. Rep. 2017, 7, 1653. [Google Scholar] [CrossRef]
- Hoek, T.A.; Khuperkar, D.; Lindeboom, R.G.H.; Sonneveld, S.; Verhagen, B.M.P.; Boersma, S.; Vermeulen, M.; Tanenbaum, M.E. Single-Molecule Imaging Uncovers Rules Governing Nonsense-Mediated mRNA Decay. Mol. Cell 2019, 75, 324–339.e11. [Google Scholar] [CrossRef]
- Mort, M.; Ivanov, D.; Cooper, D.N.; Chuzhanova, N.A. A Meta-Analysis of Nonsense Mutations Causing Human Genetic Disease. Hum. Mutat. 2008, 29, 1037–1047. [Google Scholar] [CrossRef]
- Coban-Akdemir, Z.; White, J.J.; Song, X.; Jhangiani, S.N.; Fatih, J.M.; Gambin, T.; Bayram, Y.; Chinn, I.K.; Karaca, E.; Punetha, J.; et al. Identifying Genes Whose Mutant Transcripts Cause Dominant Disease Traits by Potential Gain-of-Function Alleles. Am. J. Hum. Genet. 2018, 103, 171–187. [Google Scholar] [CrossRef]
- Lindeboom, R.G.H.; Vermeulen, M.; Lehner, B.; Supek, F. The Impact of Nonsense-Mediated mRNA Decay on Genetic Disease, Gene Editing and Cancer Immunotherapy. Nat. Genet. 2019, 51, 1645–1651. [Google Scholar] [CrossRef]
- Gardner, L.B. Nonsense-Mediated RNA Decay Regulation by Cellular Stress: Implications for Tumorigenesis. Mol. Cancer Res. MCR 2010, 8, 295–308. [Google Scholar] [CrossRef]
- Nogueira, G.; Fernandes, R.; García-Moreno, J.F.; Romão, L. Nonsense-Mediated RNA Decay and Its Bipolar Function in Cancer. Mol. Cancer 2021, 20, 72. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.; Stupack, D.G.; Wilkinson, M.F. Nonsense-Mediated RNA Decay: An Emerging Modulator of Malignancy. Nat. Rev. Cancer 2022, 22, 437–451. [Google Scholar] [CrossRef] [PubMed]
- Reddy, J.C.; Morris, J.C.; Wang, J.; English, M.A.; Haber, D.A.; Shi, Y.; Licht, J.D. WT1-Mediated Transcriptional Activation Is Inhibited by Dominant Negative Mutant Proteins. J. Biol. Chem. 1995, 270, 10878–10884. [Google Scholar] [CrossRef]
- Ware, M.D.; DeSilva, D.; Sinilnikova, O.M.; Stoppa-Lyonnet, D.; Tavtigian, S.V.; Mazoyer, S. Does Nonsense-Mediated mRNA Decay Explain the Ovarian Cancer Cluster Region of the BRCA2 Gene? Oncogene 2006, 25, 323–328. [Google Scholar] [CrossRef]
- Perrin-Vidoz, L.; Sinilnikova, O.M.; Stoppa-Lyonnet, D.; Lenoir, G.M.; Mazoyer, S. The Nonsense-Mediated mRNA Decay Pathway Triggers Degradation of Most BRCA1 mRNAs Bearing Premature Termination Codons. Hum. Mol. Genet. 2002, 11, 2805–2814. [Google Scholar] [CrossRef] [PubMed]
- Anczuków, O.; Ware, M.D.; Buisson, M.; Zetoune, A.B.; Stoppa-Lyonnet, D.; Sinilnikova, O.M.; Mazoyer, S. Does the Nonsense-Mediated mRNA Decay Mechanism Prevent the Synthesis of Truncated BRCA1, CHK2, and P53 Proteins? Hum. Mutat. 2008, 29, 65–73. [Google Scholar] [CrossRef]
- Zhao, B.; Pritchard, J.R. Evolution of the Nonsense-Mediated Decay Pathway Is Associated with Decreased Cytolytic Immune Infiltration. PLoS Comput. Biol. 2019, 15, e1007467. [Google Scholar] [CrossRef]
- Karam, R.; Carvalho, J.; Bruno, I.; Graziadio, C.; Senz, J.; Huntsman, D.; Carneiro, F.; Seruca, R.; Wilkinson, M.F.; Oliveira, C. The NMD mRNA Surveillance Pathway Downregulates Aberrant E-Cadherin Transcripts in Gastric Cancer Cells and in CDH1 Mutation Carriers. Oncogene 2008, 27, 4255–4260. [Google Scholar] [CrossRef]
- Wang, D.; Zavadil, J.; Martin, L.; Parisi, F.; Friedman, E.; Levy, D.; Harding, H.; Ron, D.; Gardner, L.B. Inhibition of Nonsense-Mediated RNA Decay by the Tumor Microenvironment Promotes Tumorigenesis. Mol. Cell. Biol. 2011, 31, 3670–3680. [Google Scholar] [CrossRef]
- Zhu, C.; Zhang, L.; Zhao, S.; Dai, W.; Xu, Y.; Zhang, Y.; Zheng, H.; Sheng, W.; Xu, Y. UPF1 Promotes Chemoresistance to Oxaliplatin through Regulation of TOP2A Activity and Maintenance of Stemness in Colorectal Cancer. Cell Death Dis. 2021, 12, 1–15. [Google Scholar] [CrossRef]
- Hu, Z.; Yau, C.; Ahmed, A.A. A Pan-Cancer Genome-Wide Analysis Reveals Tumour Dependencies by Induction of Nonsense-Mediated Decay. Nat. Commun. 2017, 8, 15943. [Google Scholar] [CrossRef]
- Pestova, T.V.; Lomakin, I.B.; Lee, J.H.; Choi, S.K.; Dever, T.E.; Hellen, C.U. The Joining of Ribosomal Subunits in Eukaryotes Requires eIF5B. Nature 2000, 403, 332–335. [Google Scholar] [CrossRef] [PubMed]
- Schäffler, K.; Schulz, K.; Hirmer, A.; Wiesner, J.; Grimm, M.; Sickmann, A.; Fischer, U. A Stimulatory Role for the La-Related Protein 4B in Translation. RNA 2010, 16, 1488–1499. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Gaidamakov, S.A.; Xie, J.; Lee, J.; Martino, L.; Kozlov, G.; Crawford, A.K.; Russo, A.N.; Conte, M.R.; Gehring, K.; et al. La-Related Protein 4 Binds Poly(A), Interacts with the Poly(A)-Binding Protein MLLE Domain via a Variant PAM2w Motif, and Can Promote mRNA Stability. Mol. Cell. Biol. 2011, 31, 542–556. [Google Scholar] [CrossRef] [PubMed]
- Nitulescu, G.M.; Van De Venter, M.; Nitulescu, G.; Ungurianu, A.; Juzenas, P.; Peng, Q.; Olaru, O.T.; Grădinaru, D.; Tsatsakis, A.; Tsoukalas, D.; et al. The Akt Pathway in Oncology Therapy and beyond (Review). Int. J. Oncol. 2018, 53, 2319–2331. [Google Scholar] [CrossRef] [PubMed]
- Khezri, M.R.; Jafari, R.; Yousefi, K.; Zolbanin, N.M. The PI3K/AKT Signaling Pathway in Cancer: Molecular Mechanisms and Possible Therapeutic Interventions. Exp. Mol. Pathol. 2022, 127, 104787. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Bode, A.M.; Dong, Z.; Lee, M.-H. AKT as a Therapeutic Target for Cancer. Cancer Res. 2019, 79, 1019–1031. [Google Scholar] [CrossRef]
- Cho, H.; Abshire, E.T.; Popp, M.W.; Pröschel, C.; Schwartz, J.L.; Yeo, G.W.; Maquat, L.E. AKT Constitutes a Signal-Promoted Alternative Exon-Junction Complex That Regulates Nonsense-Mediated mRNA Decay. Mol. Cell 2022, 82, 2779–2796.e10. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Karam, R.; Zhou, Y.; Su, F.; Ji, Y.; Li, G.; Xu, G.; Lu, L.; Wang, C.; Song, M.; et al. The UPF1 RNA Surveillance Gene Is Commonly Mutated in Pancreatic Adenosquamous Carcinoma. Nat. Med. 2014, 20, 596–598. [Google Scholar] [CrossRef]
- Lu, J.; Plank, T.-D.; Su, F.; Shi, X.; Liu, C.; Ji, Y.; Li, S.; Huynh, A.; Shi, C.; Zhu, B.; et al. The Nonsense-Mediated RNA Decay Pathway Is Disrupted in Inflammatory Myofibroblastic Tumors. J. Clin. Investig. 2016, 126, 3058–3062. [Google Scholar] [CrossRef]
- Cao, L.; Qi, L.; Zhang, L.; Song, W.; Yu, Y.; Xu, C.; Li, L.; Guo, Y.; Yang, L.; Liu, C.; et al. Human Nonsense-Mediated RNA Decay Regulates EMT by Targeting the TGF-ß Signaling Pathway in Lung Adenocarcinoma. Cancer Lett. 2017, 403, 246–259. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Shai, O.; Lee, L.J.; Frey, B.J.; Blencowe, B.J. Deep Surveying of Alternative Splicing Complexity in the Human Transcriptome by High-Throughput Sequencing. Nat. Genet. 2008, 40, 1413–1415. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.A.; Krainer, A.R.; Abdel-Wahab, O. SnapShot: Splicing Alterations in Cancer. Cell 2020, 180, 208–208.e1. [Google Scholar] [CrossRef]
- Rahman, M.A.; Nasrin, F.; Bhattacharjee, S.; Nandi, S. Hallmarks of Splicing Defects in Cancer: Clinical Applications in the Era of Personalized Medicine. Cancers 2020, 12, 1381. [Google Scholar] [CrossRef]
- Hillman, R.T.; Green, R.E.; Brenner, S.E. An Unappreciated Role for RNA Surveillance. Genome Biol. 2004, 5, R8. [Google Scholar] [CrossRef] [PubMed]
- Long, J.C.; Caceres, J.F. The SR Protein Family of Splicing Factors: Master Regulators of Gene Expression. Biochem. J. 2009, 417, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Lareau, L.F.; Inada, M.; Green, R.E.; Wengrod, J.C.; Brenner, S.E. Unproductive Splicing of SR Genes Associated with Highly Conserved and Ultraconserved DNA Elements. Nature 2007, 446, 926–929. [Google Scholar] [CrossRef]
- Sureau, A.; Gattoni, R.; Dooghe, Y.; Stévenin, J.; Soret, J. SC35 Autoregulates Its Expression by Promoting Splicing Events That Destabilize Its mRNAs. EMBO J. 2001, 20, 1785–1796. [Google Scholar] [CrossRef]
- Sun, S.; Zhang, Z.; Sinha, R.; Karni, R.; Krainer, A.R. SF2/ASF Autoregulation Involves Multiple Layers of Post-Transcriptional and Translational Control. Nat. Struct. Mol. Biol. 2010, 17, 306–312. [Google Scholar] [CrossRef]
- Jumaa, H.; Nielsen, P.J. The Splicing Factor SRp20 Modifies Splicing of Its Own mRNA and ASF/SF2 Antagonizes This Regulation. EMBO J. 1997, 16, 5077–5085. [Google Scholar] [CrossRef]
- Pervouchine, D.; Popov, Y.; Berry, A.; Borsari, B.; Frankish, A.; Guigó, R. Integrative Transcriptomic Analysis Suggests New Autoregulatory Splicing Events Coupled with Nonsense-Mediated mRNA Decay. Nucleic Acids Res. 2019, 47, 5293–5306. [Google Scholar] [CrossRef]
- Yang, S.; Jia, R.; Bian, Z. SRSF5 Functions as a Novel Oncogenic Splicing Factor and Is Upregulated by Oncogene SRSF3 in Oral Squamous Cell Carcinoma. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 1161–1172. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.Z.; Grate, L.; Donohue, J.P.; Preston, C.; Nobida, N.; O’Brien, G.; Shiue, L.; Clark, T.A.; Blume, J.E.; Ares, M. Ultraconserved Elements Are Associated with Homeostatic Control of Splicing Regulators by Alternative Splicing and Nonsense-Mediated Decay. Genes Dev. 2007, 21, 708–718. [Google Scholar] [CrossRef] [PubMed]
- Änkö, M.-L.; Müller-McNicoll, M.; Brandl, H.; Curk, T.; Gorup, C.; Henry, I.; Ule, J.; Neugebauer, K.M. The RNA-Binding Landscapes of Two SR Proteins Reveal Unique Functions and Binding to Diverse RNA Classes. Genome Biol. 2012, 13, R17. [Google Scholar] [CrossRef] [PubMed]
- Königs, V.; de Oliveira Freitas Machado, C.; Arnold, B.; Blümel, N.; Solovyeva, A.; Löbbert, S.; Schafranek, M.; Ruiz De Los Mozos, I.; Wittig, I.; McNicoll, F.; et al. SRSF7 Maintains Its Homeostasis through the Expression of Split-ORFs and Nuclear Body Assembly. Nat. Struct. Mol. Biol. 2020, 27, 260–273. [Google Scholar] [CrossRef]
- Dvinge, H.; Kim, E.; Abdel-Wahab, O.; Bradley, R.K. RNA Splicing Factors as Oncoproteins and Tumour Suppressors. Nat. Rev. Cancer 2016, 16, 413–430. [Google Scholar] [CrossRef]
- Urbanski, L.M.; Leclair, N.; Anczuków, O. Alternative-Splicing Defects in Cancer: Splicing Regulators and Their Downstream Targets, Guiding the Way to Novel Cancer Therapeutics. Wiley Interdiscip. Rev. RNA 2018, 9, e1476. [Google Scholar] [CrossRef]
- Karni, R.; de Stanchina, E.; Lowe, S.W.; Sinha, R.; Mu, D.; Krainer, A.R. The Gene Encoding the Splicing Factor SF2/ASF Is a Proto-Oncogene. Nat. Struct. Mol. Biol. 2007, 14, 185–193. [Google Scholar] [CrossRef]
- Anczuków, O.; Rosenberg, A.Z.; Akerman, M.; Das, S.; Zhan, L.; Karni, R.; Muthuswamy, S.K.; Krainer, A.R. The Splicing Factor SRSF1 Regulates Apoptosis and Proliferation to Promote Mammary Epithelial Cell Transformation. Nat. Struct. Mol. Biol. 2012, 19, 220–228. [Google Scholar] [CrossRef]
- Ghigna, C.; Giordano, S.; Shen, H.; Benvenuto, F.; Castiglioni, F.; Comoglio, P.M.; Green, M.R.; Riva, S.; Biamonti, G. Cell Motility Is Controlled by SF2/ASF through Alternative Splicing of the Ron Protooncogene. Mol. Cell 2005, 20, 881–890. [Google Scholar] [CrossRef]
- Aznarez, I.; Nomakuchi, T.T.; Tetenbaum-Novatt, J.; Rahman, M.A.; Fregoso, O.; Rees, H.; Krainer, A.R. Mechanism of Nonsense-Mediated mRNA Decay Stimulation by Splicing Factor SRSF1. Cell Rep. 2018, 23, 2186–2198. [Google Scholar] [CrossRef]
- Rahman, M.A.; Lin, K.-T.; Bradley, R.K.; Abdel-Wahab, O.; Krainer, A.R. Recurrent SRSF2 Mutations in MDS Affect Both Splicing and NMD. Genes Dev. 2020, 34, 413–427. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Sanada, M.; Shiraishi, Y.; Nowak, D.; Nagata, Y.; Yamamoto, R.; Sato, Y.; Sato-Otsubo, A.; Kon, A.; Nagasaki, M.; et al. Frequent Pathway Mutations of Splicing Machinery in Myelodysplasia. Nature 2011, 478, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Ilagan, J.O.; Liang, Y.; Daubner, G.M.; Lee, S.C.-W.; Ramakrishnan, A.; Li, Y.; Chung, Y.R.; Micol, J.-B.; Murphy, M.E.; et al. SRSF2 Mutations Contribute to Myelodysplasia by Mutant-Specific Effects on Exon Recognition. Cancer Cell 2015, 27, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Yoshimi, A.; Lin, K.-T.; Wiseman, D.H.; Rahman, M.A.; Pastore, A.; Wang, B.; Lee, S.C.-W.; Micol, J.-B.; Zhang, X.J.; de Botton, S.; et al. Coordinated Alterations in RNA Splicing and Epigenetic Regulation Drive Leukaemogenesis. Nature 2019, 574, 273–277. [Google Scholar] [CrossRef]
- Gardini, A.; Baillat, D.; Cesaroni, M.; Hu, D.; Marinis, J.M.; Wagner, E.J.; Lazar, M.A.; Shilatifard, A.; Shiekhattar, R. Integrator Regulates Transcriptional Initiation and Pause Release Following Activation. Mol. Cell 2014, 56, 128–139. [Google Scholar] [CrossRef]
- Inoue, D.; Chew, G.-L.; Liu, B.; Michel, B.C.; Pangallo, J.; D’Avino, A.R.; Hitchman, T.; North, K.; Lee, S.C.-W.; Bitner, L.; et al. Spliceosomal Disruption of the Non-Canonical BAF Complex in Cancer. Nature 2019, 574, 432–436. [Google Scholar] [CrossRef]
- Li, F.; Zhao, C.; Diao, Y.; Wang, Z.; Peng, J.; Yang, N.; Qiu, C.; Kong, B.; Li, Y. MEX3A Promotes the Malignant Progression of Ovarian Cancer by Regulating Intron Retention in TIMELESS. Cell Death Dis. 2022, 13, 553. [Google Scholar] [CrossRef]
- Zhang, P.; Su, T.; Zhang, S. Comprehensive Analysis of Prognostic Value of MEX3A and Its Relationship with Immune Infiltrates in Ovarian Cancer. J. Immunol. Res. 2021, 2021, 5574176. [Google Scholar] [CrossRef]
- Xiang, X.; Liu, Y.; Kang, Y.; Lu, X.; Xu, K. MEX3A Promotes Nasopharyngeal Carcinoma Progression via the MiR-3163/SCIN Axis by Regulating NF-ΚB Signaling Pathway. Cell Death Dis. 2022, 13, 420. [Google Scholar] [CrossRef]
- Gardner, L.B. Hypoxic Inhibition of Nonsense-Mediated RNA Decay Regulates Gene Expression and the Integrated Stress Response. Mol. Cell. Biol. 2008, 28, 3729–3741. [Google Scholar] [CrossRef] [PubMed]
- Usuki, F.; Yamashita, A.; Fujimura, M. Environmental Stresses Suppress Nonsense-Mediated mRNA Decay (NMD) and Affect Cells by Stabilizing NMD-Targeted Gene Expression. Sci. Rep. 2019, 9, 1279. [Google Scholar] [CrossRef] [PubMed]
- Harding, H.P.; Zhang, Y.; Zeng, H.; Novoa, I.; Lu, P.D.; Calfon, M.; Sadri, N.; Yun, C.; Popko, B.; Paules, R.; et al. An Integrated Stress Response Regulates Amino Acid Metabolism and Resistance to Oxidative Stress. Mol. Cell 2003, 11, 619–633. [Google Scholar] [CrossRef]
- Martin, L.; Gardner, L.B. Stress-Induced Inhibition of Nonsense-Mediated RNA Decay Regulates Intracellular Cystine Transport and Intracellular Glutathione through Regulation of the Cystine/Glutamate Exchanger SLC7A11. Oncogene 2015, 34, 4211–4218. [Google Scholar] [CrossRef] [PubMed]
- Pastor, F.; Kolonias, D.; Giangrande, P.H.; Gilboa, E. Induction of Tumour Immunity by Targeted Inhibition of Nonsense-Mediated mRNA Decay. Nature 2010, 465, 227–230. [Google Scholar] [CrossRef]
- Litchfield, K.; Reading, J.L.; Lim, E.L.; Xu, H.; Liu, P.; Al-Bakir, M.; Wong, Y.N.S.; Rowan, A.; Funt, S.A.; Merghoub, T.; et al. Escape from Nonsense-Mediated Decay Associates with Anti-Tumor Immunogenicity. Nat. Commun. 2020, 11, 3800. [Google Scholar] [CrossRef]
- Hurt, J.A.; Robertson, A.D.; Burge, C.B. Global Analyses of UPF1 Binding and Function Reveal Expanded Scope of Nonsense-Mediated mRNA Decay. Genome Res. 2013, 23, 1636–1650. [Google Scholar] [CrossRef]
- Nomakuchi, T.T.; Rigo, F.; Aznarez, I.; Krainer, A.R. Antisense Oligonucleotide-Directed Inhibition of Nonsense-Mediated mRNA Decay. Nat. Biotechnol. 2016, 34, 164–166. [Google Scholar] [CrossRef]
- Goldmann, T.; Overlack, N.; Möller, F.; Belakhov, V.; van Wyk, M.; Baasov, T.; Wolfrum, U.; Nagel-Wolfrum, K. A Comparative Evaluation of NB30, NB54 and PTC124 in Translational Read-through Efficacy for Treatment of an USH1C Nonsense Mutation. EMBO Mol. Med. 2012, 4, 1186–1199. [Google Scholar] [CrossRef]
- Rigo, F.; Hua, Y.; Krainer, A.R.; Bennett, C.F. Antisense-Based Therapy for the Treatment of Spinal Muscular Atrophy. J. Cell Biol. 2012, 199, 21–25. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nagar, P.; Islam, M.R.; Rahman, M.A. Nonsense-Mediated mRNA Decay as a Mediator of Tumorigenesis. Genes 2023, 14, 357. https://doi.org/10.3390/genes14020357
Nagar P, Islam MR, Rahman MA. Nonsense-Mediated mRNA Decay as a Mediator of Tumorigenesis. Genes. 2023; 14(2):357. https://doi.org/10.3390/genes14020357
Chicago/Turabian StyleNagar, Preeti, Md Rafikul Islam, and Mohammad Alinoor Rahman. 2023. "Nonsense-Mediated mRNA Decay as a Mediator of Tumorigenesis" Genes 14, no. 2: 357. https://doi.org/10.3390/genes14020357
APA StyleNagar, P., Islam, M. R., & Rahman, M. A. (2023). Nonsense-Mediated mRNA Decay as a Mediator of Tumorigenesis. Genes, 14(2), 357. https://doi.org/10.3390/genes14020357





