Assessing Outlier Probabilities in Transcriptomics Data When Evaluating a Classifier
Abstract
:1. Introduction
2. Materials and Methods
2.1. Feature Selection, Classification Models, and Bootstrap Validation
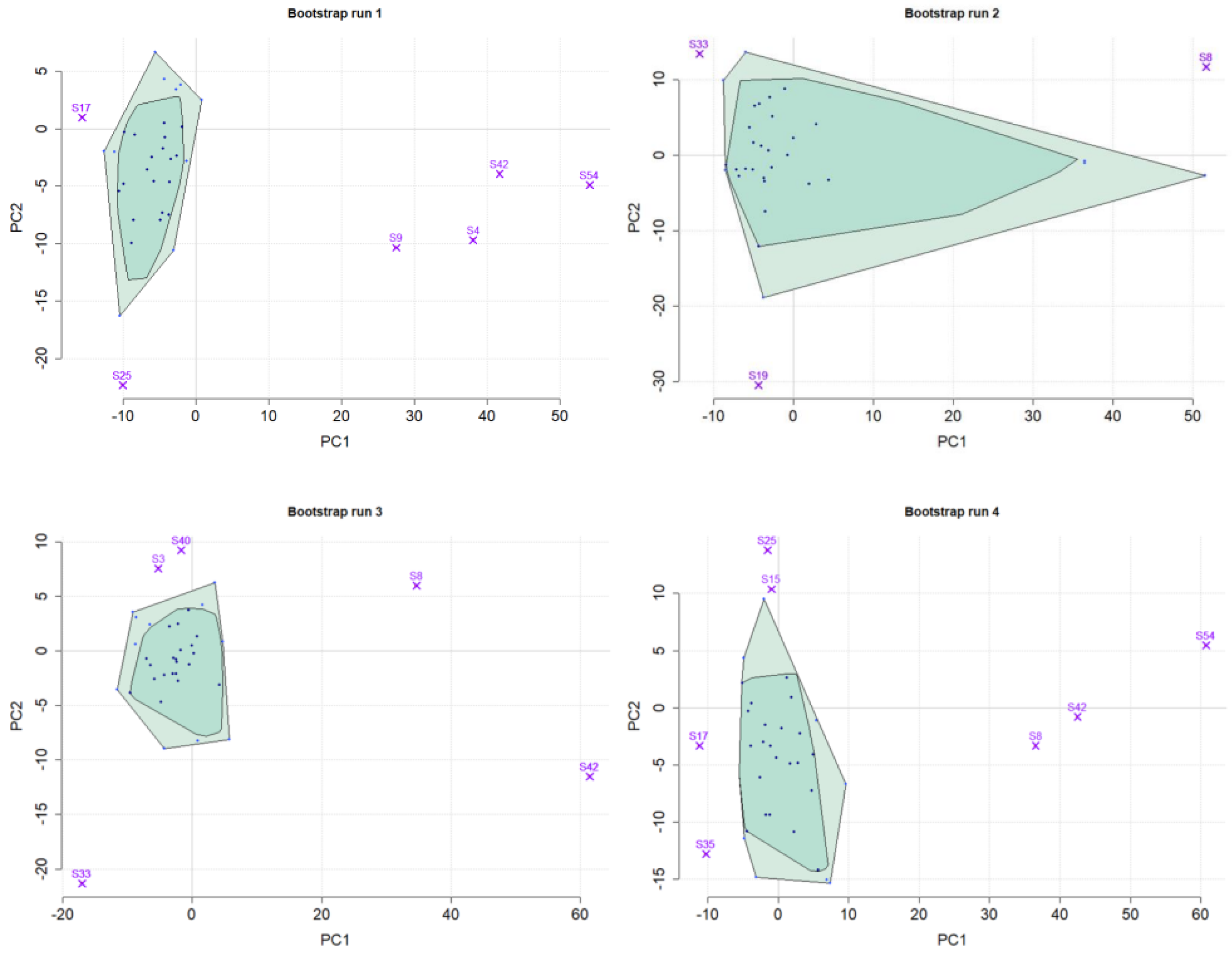
2.2. Assessment of Outlier Probabilities
| Algorithm 1 Pseudo-code of bootstrap–bagplot algorithm for outlier detection for two study groups. |
|
2.3. Simulated and Real-World Example Data
2.3.1. Simulated Data
2.3.2. SARS-CoV-2 versus Other Respiratory Viruses
2.3.3. West-Nile Infected versus Control Samples
3. Results
3.1. Classifier Evaluation in Simulated Data
3.2. Classifier Evaluation in SARS-CoV-2 Study
3.3. Classifier Evaluation in West-Nile Virus Study
3.4. Findings for Differentially Expressed Genes Analyses
3.5. Run Time Evaluation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ARI | Acute respiratory illness |
| DE | Differential expression |
| GEO | Gene Expression Omnibus |
| LDA | Linear discriminant analysis |
| ML | Machine learning |
| PBMC | Peripheral blood mononuclear cell |
| PCA | Principal component analysis |
| RF | Random forest |
| SVM | Support vector machine |
| WNV | West Nile virus |
References
- Bumgarner, R. Overview of DNA microarrays: Types, applications, and their future. Curr. Protoc. Mol. Biol. 2013, 101, 22-1. [Google Scholar] [CrossRef] [PubMed]
- Marguerat, S.; Bähler, J. RNA-seq: From technology to biology. Cell. Mol. Life Sci. 2010, 67, 569–579. [Google Scholar] [CrossRef]
- Bair, E.; Tibshirani, R. Machine learning methods applied to DNA microarray data can improve the diagnosis of cancer. ACM SIGKDD Explor. Newsl. 2003, 5, 48–55. [Google Scholar] [CrossRef]
- Zhang, W.; Yu, Y.; Hertwig, F.; Thierry-Mieg, J.; Zhang, W.; Thierry-Mieg, D.; Wang, J.; Furlanello, C.; Devanarayan, V.; Cheng, J.; et al. Comparison of RNA-seq and microarray-based models for clinical endpoint prediction. Genome Biol. 2015, 16, 133. [Google Scholar] [CrossRef]
- Huang, Z.; Johnson, T.S.; Han, Z.; Helm, B.; Cao, S.; Zhang, C.; Salama, P.; Rizkalla, M.; Yu, C.Y.; Cheng, J.; et al. Deep learning-based cancer survival prognosis from RNA-seq data: Approaches and evaluations. BMC Med. Genom. 2020, 13, 41. [Google Scholar] [CrossRef] [PubMed]
- Best, M.G.; Sol, N.; Kooi, I.; Tannous, J.; Westerman, B.A.; Rustenburg, F.; Schellen, P.; Verschueren, H.; Post, E.; Koster, J.; et al. RNA-Seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell 2015, 28, 666–676. [Google Scholar] [CrossRef] [PubMed]
- Fischer, N.; Indenbirken, D.; Meyer, T.; Lütgehetmann, M.; Lellek, H.; Spohn, M.; Aepfelbacher, M.; Alawi, M.; Grundhoff, A. Evaluation of unbiased next-generation sequencing of RNA (RNA-seq) as a diagnostic method in influenza virus-positive respiratory samples. J. Clin. Microbiol. 2015, 53, 2238–2250. [Google Scholar] [CrossRef]
- Bhattacharya, I.; Bhatia, M.P.S. SVM classification to distinguish Parkinson disease patients. In Proceedings of the 1st Amrita ACM-W Celebration on Women in Computing in India, Tamilnadu, India, 16–17 September 2010; pp. 1–6. [Google Scholar]
- Wong, T.T. Performance evaluation of classification algorithms by k-fold and leave-one-out cross validation. Pattern Recognit. 2015, 48, 2839–2846. [Google Scholar] [CrossRef]
- Efron, B.; Tibshirani, R. Improvements on cross-validation: The 632+ bootstrap method. J. Am. Stat. Assoc. 1997, 92, 548–560. [Google Scholar]
- Dupuy, A.; Simon, R.M. Critical review of published microarray studies for cancer outcome and guidelines on statistical analysis and reporting. J. Natl. Cancer Inst. 2007, 99, 147–157. [Google Scholar] [CrossRef]
- Subramanian, J.; Simon, R. Gene expression–based prognostic signatures in lung cancer: Ready for clinical use? J. Natl. Cancer Inst. 2010, 102, 464–474. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Parmigiani, G.; Johnson, W.E. ComBat-seq: Batch effect adjustment for RNA-seq count data. NAR Genom. Bioinform. 2020, 2, lqaa078. [Google Scholar] [CrossRef] [PubMed]
- Rau, A.; Marot, G.; Jaffrézic, F. Differential meta-analysis of RNA-seq data from multiple studies. BMC Bioinform. 2014, 15, 91. [Google Scholar] [CrossRef]
- Krepel, J.; Kircher, M.; Kohls, M.; Jung, K. Comparison of merging strategies for building machine learning models on multiple independent gene expression data sets. Stat. Anal. Data Min. ASA Data Sci. J. 2022, 15, 112–124. [Google Scholar] [CrossRef]
- Wu, C.; Ma, S. A selective review of robust variable selection with applications in bioinformatics. Briefings Bioinform. 2015, 16, 873–883. [Google Scholar] [CrossRef] [PubMed]
- Singhania, A.; Wilkinson, R.J.; Rodrigue, M.; Haldar, P.; O’Garra, A. The value of transcriptomics in advancing knowledge of the immune response and diagnosis in tuberculosis. Nat. Immunol. 2018, 19, 1159–1168. [Google Scholar] [CrossRef] [PubMed]
- Van Laere, S.; Van der Auwera, I.; Van den Eynden, G.G.; Fox, S.B.; Bianchi, F.; Harris, A.L.; Van Dam, P.; Van Marck, E.A.; Vermeulen, P.B.; Dirix, L.Y. Distinct molecular signature of inflammatory breast cancer by cDNA microarray analysis. Breast Cancer Res. Treat. 2005, 93, 237–246. [Google Scholar] [CrossRef]
- Westermann, A.J.; Barquist, L.; Vogel, J. Resolving host–pathogen interactions by dual RNA-seq. PLoS Pathog. 2017, 13, e1006033. [Google Scholar] [CrossRef]
- Mpindi, J.P.; Sara, H.; Haapa-Paananen, S.; Kilpinen, S.; Pisto, T.; Bucher, E.; Ojala, K.; Iljin, K.; Vainio, P.; Björkman, M.; et al. GTI: A novel algorithm for identifying outlier gene expression profiles from integrated microarray datasets. PLoS ONE 2011, 6, e17259. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, B.; Wang, T.; Bonni, A.; Zhao, G. Robust principal component analysis for accurate outlier sample detection in RNA-Seq data. BMC Bioinform. 2020, 21, 269. [Google Scholar] [CrossRef]
- Hubert, M.; Rousseeuw, P.; Verdonck, T. Robust PCA for skewed data and its outlier map. Comput. Stat. Data Anal. 2009, 53, 2264–2274. [Google Scholar] [CrossRef]
- Kruppa, J.; Jung, K. Automated multigroup outlier identification in molecular high-throughput data using bagplots and gemplots. BMC Bioinform. 2017, 18, 232. [Google Scholar] [CrossRef] [PubMed]
- Filzmoser, P.; Hron, K.; Reimann, C. Principal component analysis for compositional data with outliers. Environmetr. Off. J. Int. Environmetr. Soc. 2009, 20, 621–632. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Efron, B.; Halloran, E.; Holmes, S. Bootstrap confidence levels for phylogenetic trees. Proc. Natl. Acad. Sci. USA 1996, 93, 13429. [Google Scholar] [CrossRef]
- Saremi, B.; Kohls, M.; Liebig, P.; Siebert, U.; Jung, K. Measuring reproducibility of virus metagenomics analyses using bootstrap samples from FASTQ-files. Bioinformatics 2021, 37, 1068–1075. [Google Scholar] [CrossRef]
- Yu, J.; Xue, A.; Redei, E.; Bagheri, N. A support vector machine model provides an accurate transcript-level-based diagnostic for major depressive disorder. Transl. Psychiatry 2016, 6, e931. [Google Scholar] [CrossRef]
- Acharjee, A.; Kloosterman, B.; Visser, R.G.; Maliepaard, C. Integration of multi-omics data for prediction of phenotypic traits using random forest. BMC Bioinform. 2016, 17, 180. [Google Scholar] [CrossRef]
- Giger, T.; Excoffier, L.; Amstutz, U.; Day, P.J.; Champigneulle, A.; Hansen, M.M.; Kelso, J.; Largiadèr, C.R. Population transcriptomics of life-history variation in the genus Salmo. Mol. Ecol. 2008, 17, 3095–3108. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Venables, W.N.; Ripley, B.D. Modern Applied Statistics with S-PLUS; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Brier, G.W. Verification of forecasts expressed in terms of probability. Mon. Weather Rev. 1950, 78, 1–3. [Google Scholar] [CrossRef]
- Rousseeuw, P.J.; Ruts, I.; Tukey, J.W. The bagplot: A bivariate boxplot. Am. Stat. 1999, 53, 382–387. [Google Scholar]
- Wolf, H.P.; Bielefeld, U.; Wolf, M.P. Package ‘aplpack’. 2019. Available online: https://cran.rproject.org/web/packages/aplpack/aplpack.pdf (accessed on 13 June 2022).
- Filzmoser, P.; Fritz, H.; Kalcher, K.; Todorov, M.V. Package ‘pcaPP’. J. Am. Stat. Assoc. 2022, 314, 436–439. [Google Scholar]
- Todorov, V.; Filzmoser, P. An Object-Oriented Framework for Robust Multivariate Analysis. J. Stat. Softw. 2010, 32, 1–47. [Google Scholar] [CrossRef]
- Frazee, A.C.; Jaffe, A.E.; Langmead, B.; Leek, J.T. Polyester: Simulating RNA-seq datasets with differential transcript expression. Bioinformatics 2015, 31, 2778–2784. [Google Scholar] [CrossRef]
- Ng, D.L.; Granados, A.C.; Santos, Y.A.; Servellita, V.; Goldgof, G.M.; Meydan, C.; Sotomayor-Gonzalez, A.; Levine, A.G.; Balcerek, J.; Han, L.M.; et al. A diagnostic host response biosignature for COVID-19 from RNA profiling of nasal swabs and blood. Sci. Adv. 2021, 7, eabe5984. [Google Scholar] [CrossRef]
- Qian, F.; Goel, G.; Meng, H.; Wang, X.; You, F.; Devine, L.; Raddassi, K.; Garcia, M.N.; Murray, K.O.; Bolen, C.R.; et al. Systems immunology reveals markers of susceptibility to West Nile virus infection. Clin. Vaccine Immunol. 2015, 22, 6–16. [Google Scholar] [CrossRef]
- Domany, E. Using high-throughput transcriptomic data for prognosis: A critical overview and perspectives. Cancer Res. 2014, 74, 4612–4621. [Google Scholar] [CrossRef]
- Lee, H.S.; Cleynen, I. Molecular profiling of inflammatory bowel disease: Is it ready for use in clinical decision-making? Cells 2019, 8, 535. [Google Scholar] [CrossRef]
- Haywood, S.C.; Gupta, S.; Heemers, H.V. PAM50 and beyond: When will tissue transcriptomics guide clinical decision-making? Eur. Urol. Focus 2022, 8, 916–918. [Google Scholar] [CrossRef]
- Pepe, M.S.; Longton, G.; Anderson, G.L.; Schummer, M. Selecting differentially expressed genes from microarray experiments. Biometrics 2003, 59, 133–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]








| Dataset | Number of Samples | Number of Transcripts | Study Design | |
|---|---|---|---|---|
| Simulated dataset | D = 1000 | Group 1: | 55 samples including 5 outliers | |
| Group 2: | 55 samples including 5 outliers | |||
| Subset of SARS-CoV-2 study (GSE163151) | D = 26,485 | Group ‘COVID’: | Nasopharyngeal swabs from 138 humans with ARI caused by SARS-CoV-2 | |
| Group ‘Other virus’: | Nasopharyngeal swabs from 120 humans with ARI caused by other viruses | |||
| Subset of WNV study (GSE46681) | D = 47,323 | Group ‘Mock’: | PBMCs from 39 humans incubated in medium alone | |
| Group ‘WNV’: | PBMCs from 39 humans infected with WNV ex vivo | |||
| Sample ID | Occurrences in Bootstrapping | Detected as Outlier | Outlier Probability | 95%-CI | p-Value |
|---|---|---|---|---|---|
| GSM4972835 | 68 | 68 | 1.00 | [0.96, 1.00] | <0.001 |
| GSM4972888 | 64 | 64 | 1.00 | [0.95, 1.00] | <0.001 |
| GSM4972929 | 68 | 68 | 1.00 | [0.96, 1.00] | <0.001 |
| GSM4972877 | 67 | 66 | 0.99 | [0.93, 1.00] | <0.001 |
| GSM4972927 | 66 | 65 | 0.98 | [0.93, 1.00] | <0.001 |
| GSM4972944 | 66 | 63 | 0.95 | [0.89, 1.00] | <0.001 |
| GSM4973094 | 60 | 57 | 0.95 | [0.88, 1.00] | <0.001 |
| GSM4972878 | 63 | 59 | 0.94 | [0.86, 1.00] | <0.001 |
| GSM4973123 | 57 | 53 | 0.93 | [0.85, 1.00] | <0.001 |
| GSM4972907 | 63 | 45 | 0.71 | [0.61, 1.00] | <0.001 |
| GSM4973051 | 61 | 43 | 0.70 | [0.59, 1.00] | 0.001 |
| GSM4972892 | 87 | 59 | 0.68 | [0.59, 1.00] | 0.001 |
| GSM4972872 | 72 | 43 | 0.60 | [0.49, 1.00] | 0.062 |
| GSM4972919 | 66 | 38 | 0.58 | [0.47, 1.00] | 0.134 |
| GSM4972891 | 58 | 33 | 0.57 | [0.45, 1.00] | 0.179 |
| GSM4972875 | 67 | 37 | 0.55 | [0.44, 1.00] | 0.232 |
| GSM4972971 | 59 | 31 | 0.53 | [0.41, 1.00] | 0.397 |
| GSM4972973 | 66 | 33 | 0.50 | [0.39, 1.00] | 0.549 |
| GSM4972946 | 60 | 29 | 0.48 | [0.37, 1.00] | 0.651 |
| Sample ID | Occurrences in Bootstrapping | Detected as Outlier | Outlier Probability | 95%-CI | p-Value |
|---|---|---|---|---|---|
| GSM1133935 | 71 | 55 | 0.77 | [0.68, 1.00] | <0.001 |
| GSM1058065 | 66 | 23 | 0.35 | [0.25, 1.00] | 0.995 |
| GSM1133954 | 68 | 21 | 0.31 | [0.22, 1.00] | 1.00 |
| GSM1058069 | 69 | 18 | 0.26 | [0.18, 1.00] | 1.00 |
| GSM1133940 | 62 | 12 | 0.19 | [0.12, 1.00] | 1.00 |
| GSM1058085 | 62 | 11 | 0.18 | [0.10, 1.00] | 1.00 |
| GSM1058062 | 64 | 11 | 0.17 | [0.10, 1.00] | 1.00 |
| GSM1133950 | 62 | 10 | 0.16 | [0.09, 1.00] | 1.00 |
| GSM1058064 | 61 | 9 | 0.15 | [0.08, 1.00] | 1.00 |
| GSM1058082 | 64 | 8 | 0.12 | [0.06, 1.00] | 1.00 |
| GSM1133931 | 64 | 8 | 0.12 | [0.06, 1.00] | 1.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kircher, M.; Säurich, J.; Selle, M.; Jung, K. Assessing Outlier Probabilities in Transcriptomics Data When Evaluating a Classifier. Genes 2023, 14, 387. https://doi.org/10.3390/genes14020387
Kircher M, Säurich J, Selle M, Jung K. Assessing Outlier Probabilities in Transcriptomics Data When Evaluating a Classifier. Genes. 2023; 14(2):387. https://doi.org/10.3390/genes14020387
Chicago/Turabian StyleKircher, Magdalena, Josefin Säurich, Michael Selle, and Klaus Jung. 2023. "Assessing Outlier Probabilities in Transcriptomics Data When Evaluating a Classifier" Genes 14, no. 2: 387. https://doi.org/10.3390/genes14020387






