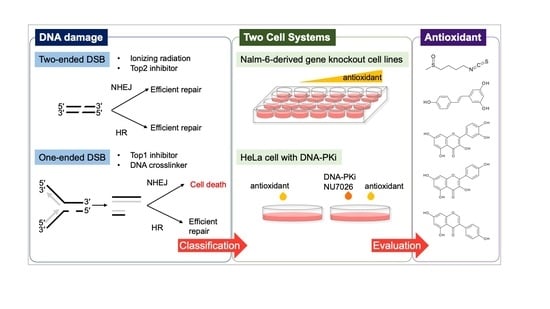
A Cell System-Assisted Strategy for Evaluating the Natural Antioxidant-Induced Double-Stranded DNA Break (DSB) Style
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Growth Inhibition Assay
2.3. Sensitivity Assay with NHEJ Inhibition
2.4. Immunofluorescence Staining
2.5. Statistical Analyses
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xu, D.P.; Li, Y.; Meng, X.; Zhou, T.; Zhou, Y.; Zheng, J.; Zhang, J.J.; Li, H.B. Natural Antioxidants in Foods and Medicinal Plants: Extraction, Assessment and Resources. Int. J. Mol. Sci. 2017, 18, 96. [Google Scholar] [CrossRef] [PubMed]
- Manach, C.; Scalbert, A.; Morand, C.; Rémésy, C.; Jiménez, L. Polyphenols: Food sources and bioavailability. Am. J. Clin. Nutr. 2004, 79, 727–747. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-J.; Gan, R.-Y.; Li, S.; Zhou, Y.; Li, A.-N.; Xu, D.-P.; Li, H.-B. Antioxidant Phytochemicals for the Prevention and Treatment of Chronic Diseases. Molecules 2015, 20, 21138–21156. [Google Scholar] [CrossRef]
- Pinto, C.; Cidade, H.; Pinto, M.; Tiritan, M.E. Chiral Flavonoids as Antitumor Agents. Pharmaceuticals 2021, 14, 1267. [Google Scholar] [CrossRef]
- Mangla, B.; Javed, S.; Sultan, M.H.; Kumar, P.; Kohli, K.; Najmi, A.; Alhazmi, H.A.; Al Bratty, M.; Ahsan, W. Sulforaphane: A review of its therapeutic potentials, advances in its nanodelivery, recent patents, and clinical trials. Phytother. Res. 2021, 35, 5440–5458. [Google Scholar] [CrossRef] [PubMed]
- Calcabrini, C.; Maffei, F.; Turrini, E.; Fimognari, C. Sulforaphane Potentiates Anticancer Effects of Doxorubicin and Cisplatin and Mitigates Their Toxic Effects. Front. Pharmacol. 2020, 11, 567. [Google Scholar] [CrossRef] [PubMed]
- Georgikou, C.; Buglioni, L.; Bremerich, M.; Roubicek, N.; Yin, L.; Gross, W.; Sticht, C.; Bolm, C.; Herr, I. Novel Broccoli Sulforaphane-Based Analogues Inhibit the Progression of Pancreatic Cancer without Side Effects. Biomolecules 2020, 10, 769. [Google Scholar] [CrossRef]
- Piberger, A.L.; Keil, C.; Platz, S.; Rohn, S.; Hartwig, A. Sulforaphane inhibits damage-induced poly (ADP-ribosyl)ation via direct interaction of its cellular metabolites with PARP-1. Mol. Nutr. Food Res. 2015, 59, 2231–2242. [Google Scholar] [CrossRef]
- Naumann, P.; Liermann, J.; Fortunato, F.; Schmid, T.E.; Weber, K.-J.; Debus, J.; Combs, S.E. Sulforaphane enhances irradiation effects in terms of perturbed cell cycle progression and increased DNA damage in pancreatic cancer cells. PLoS ONE 2017, 12, e0180940. [Google Scholar] [CrossRef]
- Jeggo, P.A.; Pearl, L.H.; Carr, A.M. DNA repair, genome stability and cancer: A historical perspective. Nat. Rev. Cancer 2015, 16, 35–42. [Google Scholar] [CrossRef]
- Terabayashi, T.; Hanada, K. Genome instability syndromes caused by impaired DNA repair and aberrant DNA damage responses. Cell Biol. Toxicol. 2018, 34, 337–350. [Google Scholar] [CrossRef] [PubMed]
- Tatin, X.; Muggiolu, G.; Sauvaigo, S.; Breton, J. Evaluation of DNA double-strand break repair capacity in human cells: Critical overview of current functional methods. Mutat. Res. Mol. Mech. Mutagen. 2021, 788, 108388. [Google Scholar] [CrossRef] [PubMed]
- Vodicka, P.; Vodenkova, S.; Horak, J.; Opattova, A.; Tomasova, K.; Vymetalkova, V.; Stetina, R.; Hemminki, K.; Vodickova, L. An investigation of DNA damage and DNA repair in chemical carcinogenesis triggered by small-molecule xenobiotics and in cancer: Thirty years with the comet assay. Mutat. Res. Toxicol. Environ. Mutagen. 2023, 885, 503564. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Zhang, D.; Yu, F.; Ma, Y.; Liu, Y.; Li, X.; Wang, H. Precise sequencing of single protected-DNA fragment molecules for profiling of protein distribution and assembly on DNA. Chem. Sci. 2021, 12, 2039–2049. [Google Scholar] [CrossRef]
- Chang, H.H.Y.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017, 18, 495–506. [Google Scholar] [CrossRef]
- Ensminger, M.; Lobrich, M. One end to rule them all: Non-homologous end-joining and homologous recombination at DNA double-strand breaks. Br. J. Radiol. 2020, 93, 20191054. [Google Scholar] [CrossRef]
- Cortez, D. Replication-Coupled DNA Repair. Mol. Cell 2019, 74, 866–876. [Google Scholar] [CrossRef]
- Kurosawa, A.; Saito, S.; So, S.; Hashimoto, M.; Iwabuchi, K.; Watabe, H.; Adachi, N. DNA Ligase IV and Artemis Act Cooperatively to Suppress Homologous Recombination in Human Cells: Implications for DNA Double-Strand Break Repair. PLoS ONE 2013, 8, e72253. [Google Scholar] [CrossRef]
- Zhao, B.; Zhang, D.; Li, C.; Yuan, Z.; Yu, F.; Zhong, S.; Jiang, G.; Yang, Y.-G.; Le, X.C.; Weinfeld, M.; et al. ATPase activity tightly regulates RecA nucleofilaments to promote homologous recombination. Cell Discov. 2017, 3, 16053. [Google Scholar] [CrossRef]
- Shibata, A.; Jeggo, P.A. ATM’s Role in the Repair of DNA Double-Strand Breaks. Genes 2021, 12, 1370. [Google Scholar] [CrossRef]
- Kurosawa, A.; Kuboshima, H.; Adachi, N. Complex genetic interactions between DNA polymerase β and the NHEJ ligase. FEBS J. 2020, 287, 377–385. [Google Scholar] [CrossRef]
- Llorens-Agost, M.; Ensminger, M.; Le, H.P.; Gawai, A.; Liu, J.; Cruz-Garcia, A.; Bhetawal, S.; Wood, R.D.; Heyer, W.D.; Lobrich, M. POLtheta-mediated end joining is restricted by RAD52 and BRCA2 until the onset of mitosis. Nat. Cell Biol. 2021, 23, 1095–1104. [Google Scholar] [CrossRef]
- Enoiu, M.; Jiricny, J.; Schärer, O.D. Repair of cisplatin-induced DNA interstrand crosslinks by a replication-independent pathway involving transcription-coupled repair and translesion synthesis. Nucleic Acids Res. 2012, 40, 8953–8964. [Google Scholar] [CrossRef] [PubMed]
- Baddock, H.T.; Yosaatmadja, Y.; Newman, J.A.; Schofield, C.J.; Gileadi, O.; McHugh, P.J. The SNM1A DNA repair nuclease. DNA Repair. (Amst.) 2020, 95, 102941. [Google Scholar] [CrossRef] [PubMed]
- Bunting, S.F.; Callen, E.; Kozak, M.L.; Kim, J.M.; Wong, N.; Lopez-Contreras, A.J.; Ludwig, T.; Baer, R.; Faryabi, R.B.; Malhowski, A.; et al. BRCA1 Functions Independently of Homologous Recombination in DNA Interstrand Crosslink Repair. Mol. Cell 2012, 46, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Lundin, C.; North, M.; Erixon, K.; Walters, K.; Jenssen, D.; Goldman, A.S.H.; Helleday, T. Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 2012, 40, 5794. [Google Scholar] [CrossRef]
- Ensminger, M.; Iloff, L.; Ebel, C.; Nikolova, T.; Kaina, B.; Löbrich, M. DNA breaks and chromosomal aberrations arise when replication meets base excision repair. J. Cell Biol. 2014, 206, 29–43. [Google Scholar] [CrossRef]
- Toyoda, E.; Kagaya, S.; Cowell, I.G.; Kurosawa, A.; Kamoshita, K.; Nishikawa, K.; Iiizumi, S.; Koyama, H.; Austin, C.A.; Adachi, N. NK314, a Topoisomerase II Inhibitor That Specifically Targets the α Isoform. J. Biol. Chem. 2008, 283, 23711–23720. [Google Scholar] [CrossRef]
- Nomura, Y.; Adachi, N.; Koyama, H. Human Mus81 and FANCB independently contribute to repair of DNA damage during replication. Genes Cells 2007, 12, 1111–1122. [Google Scholar] [CrossRef]
- Lagunas-Rangel, F.A.; Bermúdez-Cruz, R.M. Natural Compounds That Target DNA Repair Pathways and Their Therapeutic Potential to Counteract Cancer Cells. Front. Oncol. 2020, 10, 2567. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, S.; Mei, H.; Xuan, J.; Guo, X.; Couch, L.; Dobrovolsky, V.N.; Guo, L.; Mei, N. Ginkgo biloba leaf extract induces DNA damage by inhibiting topoisomerase II activity in human hepatic cells. Sci. Rep. 2015, 5, 14633. [Google Scholar] [CrossRef]
- Sudan, S.; Rupasinghe, H.P. Quercetin-3-O-glucoside induces human DNA topoisomerase II inhibition, cell cycle arrest and apoptosis in hepatocellular carcinoma cells. Anticancer Res. 2014, 34, 1691–1699. [Google Scholar]
- Mizushina, Y.; Shiomi, K.; Kuriyama, I.; Takahashi, Y.; Yoshida, H. Inhibitory effects of a major soy isoflavone, genistein, on human DNA topoisomerase II activity and cancer cell proliferation. Int. J. Oncol. 2013, 43, 1117–1124. [Google Scholar] [CrossRef]
- Kelm, J.M.; Samarbakhsh, A.; Pillai, A.; VanderVere-Carozza, P.S.; Aruri, H.; Pandey, D.S.; Pawelczak, K.S.; Turchi, J.J.; Gavande, N.S. Recent advances in the development of non-PIKKs targeting small molecule inhibitors of DNA double-strand break repair. Front. Oncol. 2022, 12, 850833. [Google Scholar] [CrossRef]
- Matsumoto, Y. Development and Evolution of DNA-Dependent Protein Kinase Inhibitors toward Cancer Therapy. Int. J. Mol. Sci. 2022, 23, 4264. [Google Scholar] [CrossRef]
- Riballo, E.; Kuhne, M.; Rief, N.; Doherty, A.; Smith, G.C.; Recio, M.J.; Reis, C.; Dahm, K.; Fricke, A.; Krempler, A.; et al. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to γ-H2AX foci. Mol. Cell 2004, 16, 715–724. [Google Scholar] [CrossRef]
- Adachi, N.; Suzuki, H.; Iiizumi, S.; Koyama, H. Hypersensitivity of nonhomologous DNA end-joining mutants to VP-16 and ICRF-193: Implications for the repair of topoisomerase II-mediated DNA damage. J. Biol. Chem. 2003, 278, 35897–35902. [Google Scholar] [CrossRef]





| DNA Damage Inducer | Sensitivity | Type of DSB | Reference | |
|---|---|---|---|---|
| NHEJ-Deficient Cells | HR-Deficient Cells | |||
| X-ray, radiomimetic agents | More sensitive | More sensitive | Two-ended DSB | [18] |
| Top2 inhibitor | Hypersensitive | More sensitive | Two-ended DSB | This study, [18,28] |
| Alkylating agent | More sensitive | More sensitive | Two-ended DSB | This study, [21] |
| Top1 inhibitor | More resistant | More sensitive | One-ended DSB | This study, [18] |
| DNA interstrand crosslinking agent | More resistant | More sensitive | One-ended DSB | This study, [29] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Someya, Y.; Kobayashi, S.; Toriumi, K.; Takeda, S.; Adachi, N.; Kurosawa, A. A Cell System-Assisted Strategy for Evaluating the Natural Antioxidant-Induced Double-Stranded DNA Break (DSB) Style. Genes 2023, 14, 420. https://doi.org/10.3390/genes14020420
Someya Y, Kobayashi S, Toriumi K, Takeda S, Adachi N, Kurosawa A. A Cell System-Assisted Strategy for Evaluating the Natural Antioxidant-Induced Double-Stranded DNA Break (DSB) Style. Genes. 2023; 14(2):420. https://doi.org/10.3390/genes14020420
Chicago/Turabian StyleSomeya, Yuduki, Sakine Kobayashi, Kazuya Toriumi, Shigeki Takeda, Noritaka Adachi, and Aya Kurosawa. 2023. "A Cell System-Assisted Strategy for Evaluating the Natural Antioxidant-Induced Double-Stranded DNA Break (DSB) Style" Genes 14, no. 2: 420. https://doi.org/10.3390/genes14020420
APA StyleSomeya, Y., Kobayashi, S., Toriumi, K., Takeda, S., Adachi, N., & Kurosawa, A. (2023). A Cell System-Assisted Strategy for Evaluating the Natural Antioxidant-Induced Double-Stranded DNA Break (DSB) Style. Genes, 14(2), 420. https://doi.org/10.3390/genes14020420