Whole-Genome Analysis Reveals the Dynamic Evolution of Holocentric Chromosomes in Satyrine Butterflies
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Detection of Macrosynteny and Chromosomal Rearrangements
3. Results
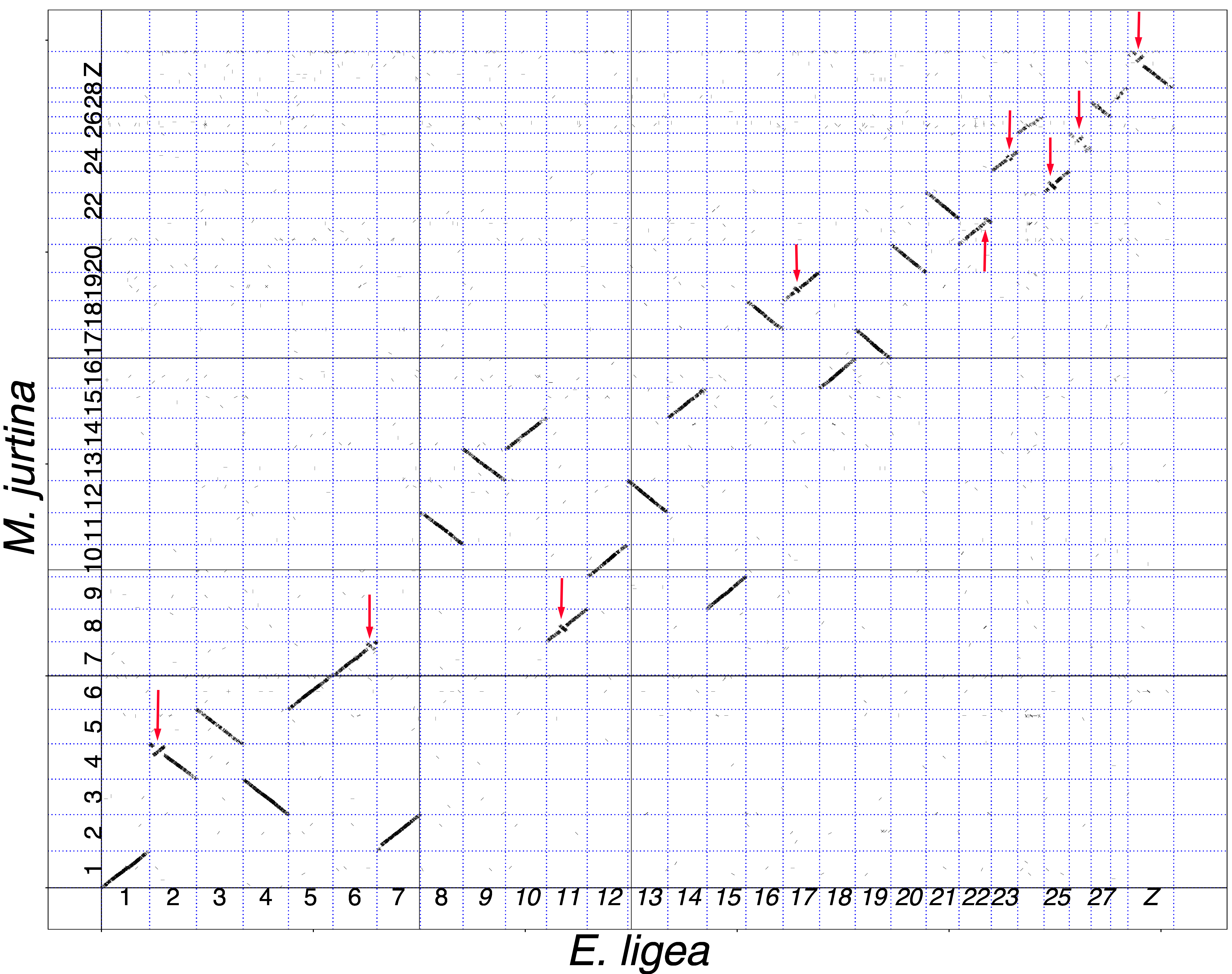
3.1. Analysis of Macrosynteny and Collinearity
3.2. Chromosomal Rearrangements: Inversions and Fusions
4. Discussion
4.1. Karyotypes of Maniola and Erebia Butterflies
4.2. Dynamic Evolution of Holocentric Chromosomes in Satyrine Butterflies
4.3. Inversions and Holocentric Model of Chromosomal Speciation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schrader, F. Notes on the mitotic behavior of long chromosomes. Cytologia 1935, 6, 422–430. [Google Scholar] [CrossRef]
- White, M.J.D. Animal Cytology and Evolution, 3rd ed.; Cambridge University Press: Cambridge, UK, 1973; p. 468. [Google Scholar]
- Mandrioli, M.; Manicardi, G.C. Holocentric chromosomes. PLoS Genet. 2020, 16, e1008918. [Google Scholar] [CrossRef]
- Hofstatter, P.G.; Thangavel, G.; Lux, T.; Neumann, P.; Vondrak, T.; Novak, P.; Zhang, M.; Costa, L.; Castellani, M.; Scott, A.; et al. Repeat-based holocentromeres influence genome architecture and karyotype evolution. Cell 2022, 185, 3153–3168.e18. [Google Scholar] [CrossRef] [PubMed]
- Melters, D.P.; Paliulis, L.V.; Korf, I.F.; Chan, S.W. Holocentric chromosomes: Convergent evolution, meiotic adaptations, and genomic analysis. Chromosome Res. 2012, 20, 579–593. [Google Scholar] [CrossRef]
- Mayrose, I.; Lysak, M.A. The evolution of chromosome numbers: Mechanistic models and experimental approaches. Genome Biol. Evol. 2022, 13, evaa220. [Google Scholar] [CrossRef]
- Jankowska, M.; Fuchs, J.; Klocke, E.; Fojtová, M.; Polanská, P.; Fajkus, J.; Schubert, V.; Houben, A. Holokinetic centromeres and efficient telomere healing enable rapid karyotype evolution. Chromosoma 2015, 124, 519–528. [Google Scholar] [CrossRef]
- Haber, J.E.; Thorburn, P.; Rogers, D. Meiotic and mitotic behavior of dicentric chromosomes in Saccharomyces cerevisiae. Genetics 1983, 106, 185–205. [Google Scholar] [CrossRef]
- Bongiorni, S.; Fiorenzo, P.; Pippoletti, D.; Prantera, G. Inverted meiosis and meiotic drive in mealybugs. Chromosoma 2004, 112, 331–341. [Google Scholar] [CrossRef]
- Cabral, G.; Marques, A.; Schubert, V.; Pedrosa-Harand, A.; Schlögelhofer, P. Chiasmatic and achiasmatic inverted meiosis of plants with holocentric chromosomes. Nat. Commun. 2014, 5, 5070. [Google Scholar] [CrossRef]
- Heckmann, S.; Jankowska, M.; Schubert, V.; Kumke, K.; Ma, W.; Houben, A. Alternative meiotic chromatid segregation in the holocentric plant Luzula elegans. Nat. Commun. 2014, 5, 4979. [Google Scholar] [CrossRef]
- Manicardi, G.C.; Mandrioli, M.; Blackman, R.L. The cytogenetic architecture of the aphid genome. Biol. Rev. Camb. Philos. Soc. 2015, 90, 112–125. [Google Scholar] [CrossRef]
- Golub, N.V.; Golub, V.B.; Kuznetsova, V.G. New data on karyotypes of lace bugs (Tingidae, Cimicomorpha, Hemiptera) with analysis of the 18S rDNA clusters distribution. Comp. Cytogenet. 2018, 12, 515–528. [Google Scholar] [CrossRef]
- Lukhtanov, V.A.; Dantchenko, A.V.; Khakimov, F.R.; Sharafutdinov, D.; Pazhenkova, E.A. Karyotype evolution and flexible (conventional versus inverted) meiosis in insects with holocentric chromosomes: A case study based on Polyommatus butterflies. Biol. J. Linn. Soc. 2020, 130, 683–699. [Google Scholar] [CrossRef]
- Bogdanov, Y.F. Inverted meiosis and its place in the evolution of sexual reproduction pathways. Genetika 2016, 52, 541–560. [Google Scholar] [CrossRef]
- Lenormand, T.; Engelstaedter, J.; Johnston, S.E.; Wijnker, E.; Haag, C.R. Evolutionary mysteries in meiosis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016, 371, 20160001. [Google Scholar] [CrossRef]
- Loidl, J. Conservation and variability of meiosis across the eukaryotes. Ann. Rev. Genet. 2016, 50, 293–316. [Google Scholar] [CrossRef]
- Archetti, M. Inverted meiosis and the evolution of sex by loss of complementation. J. Evol. Biol. 2020, 33, 460–467. [Google Scholar] [CrossRef]
- Lukhtanov, V.A.; Dincă, V.; Friberg, M.; Šíchová, J.; Olofsson, M.; Vila, R.; Marec, F.; Wiklund, C. Versatility of multivalent orientation, inverted meiosis, and rescued fitness in holocentric chromosomal hybrids. Proc. Natl. Acad. Sci. USA 2018, 115, E9610–E9619. [Google Scholar] [CrossRef]
- Lukhtanov, V.A.; Dincă, V.; Friberg, M.; Vila, R.; Wiklund, C. Incomplete sterility of chromosomal hybrids: Implications for karyotype evolution and homoploid hybrid speciation. Front. Genet. 2020, 11, 583827. [Google Scholar] [CrossRef]
- Bureš, P.; Zedek, F.; Marková, M. Holocentric chromosomes. In Plant Genome Diversity; Greilhuber, J., Dolezel, J., Wendel, J.F., Eds.; Springer: Vienna, Austria, 2013; Volume 2, pp. 187–208. [Google Scholar]
- Ruckman, S.N.; Jonika, M.M.; Casola, C.; Blackmon, H. Chromosome number evolves at equal rates in holocentric and monocentric clades. PLoS Genet. 2020, 16, e1009076. [Google Scholar] [CrossRef]
- Talavera, G.; Lukhtanov, V.A.; Pierce, N.E.; Vila, R. DNA barcodes combined with multilocus data of representative taxa can generate reliable higher-level phylogenies. Syst. Biol. 2022, 71, 382–395. [Google Scholar] [CrossRef]
- Lukhtanov, V.A.; Kandul, N.P.; Plotkin, J.B.; Dantchenko, A.V.; Haig, D.; Pierce, N.E. Reinforcement of pre-zygotic isolation and karyotype evolution in Agrodiaetus butterflies. Nature 2005, 436, 385–389. [Google Scholar] [CrossRef]
- Talavera, G.; Lukhtanov, V.A.; Rieppel, L.; Pierce, N.E.; Vila, R. In the shadow of phylogenetic uncertainty: The recent diversification of Lysandra butterflies through chromosomal change. Mol. Phylogenet. Evol. 2013, 69, 469–478. [Google Scholar] [CrossRef]
- Lukhtanov, V.A. The blue butterfly Polyommatus (Plebicula) atlanticus (Lepidoptera, Lycaenidae) holds the record of the highest number of chromosomes in the non-polyploid eukaryotic organisms. Comp. Cytogenet. 2015, 9, 683–690. [Google Scholar] [CrossRef]
- de Lesse, H. Spéciation et variation chromosomique chez les Lépidoptères Rhopalocères. Annls Sci. Nat. Zool. Sér. 1960, 2, 1–223. [Google Scholar]
- Robinson, R. Lepidoptera Genetics; Pergamon Press: Oxford, UK, 1971. [Google Scholar]
- Brown, K.S.; von Schoultz, B.; Suomalainen, E. Chromosome evolution in Neotropical Danainae and Ithomiinae (Lepidoptera). Hereditas 2004, 141, 216–236. [Google Scholar] [CrossRef]
- Beliajeff, N.K. Die Chromosomenkomplexe und ihre Beziehung zur Phylogenie bei den Lepidopteren. Z. Für Inductive Abstamm. Und Verer. 1930, 14, 369–399. [Google Scholar] [CrossRef]
- Suomalainen, E. Chromosome evolution in Lepidoptera. Chromosome Today 1969, 2, 132–138. [Google Scholar]
- Lukhtanov, V.A. Sex chromatin and sex chromosome systems in nonditrysian Lepidoptera (Insecta). J. Zool. Syst. Evol. Res. 2000, 38, 73–79. [Google Scholar] [CrossRef]
- Ahola, V.; Lehtonen, R.; Somervuo, P.; Salmela, L.; Koskinen, P.; Rastas, P.; Välimäki, N.; Paulin, L.; Kvist, J.; Wahlberg, N.; et al. The Glanville fritillary genome retains an ancient karyotype and reveals selective chromosomal fusions in Lepidoptera. Nat. Commun. 2014, 5, 4737. [Google Scholar] [CrossRef]
- Šíchová, J.; Voleníková, A.; Dincă, V.; Nguyen, P.; Vila, R.; Sahara, K.; Marec, F. Dynamic karyotype evolution and unique sex determination systems in Leptidea wood white butterflies. BMC Evol. Biol. 2015, 15, 89. [Google Scholar] [CrossRef] [PubMed]
- Šíchová, J.; Ohno, M.; Dincă, V.; Watanabe, M.; Sahara, K.; Marec, F. Fissions, fusions, and translocations shaped the karyotype and multiple sex chromosome constitution of the northeast-Asian wood white butterfly, Leptidea amurensis. Biol. J. Linn. Soc. Lond. 2016, 118, 457–471. [Google Scholar] [CrossRef]
- Sahara, K.; Yoshido, A.; Marec, F.; Fuková, I.; Zhang, H.B.; Wu, C.C.; Goldsmith, M.R.; Yasukochi, Y. Conserved synteny of genes between chromosome 15 of Bombyx mori and a chromosome of Manduca sexta shown by five-color BAC-FISH. Genome 2007, 50, 1061–1065. [Google Scholar] [CrossRef] [PubMed]
- Sahara, K.; Yoshido, A.; Shibata, F.; Fujikawa-Kojima, N.; Okabe, T.; Tanaka-Okuyama, M.; Yasukochi, Y. FISH identification of Helicoverpa armigera and Mamestra brassicae chromosomes by BAC and fosmid probes. Insect Biochem. Mol. Biol. 2013, 43, 644–653. [Google Scholar] [CrossRef]
- Hill, J.; Rastas, P.; Hornett, E.A.; Neethiraj, R.; Clark, N.; Morehouse, N.; de la Paz Celorio-Mancera, M.; Cols, J.C.; Dircksen, H.; Meslin, C.; et al. Unprecedented reorganization of holocentric chromosomes provides insights into the enigma of lepidopteran chromosome evolution. Sci. Adv. 2019, 5, eaau3648. [Google Scholar] [CrossRef]
- Cicconardi, F.; Lewis, J.J.; Martin, S.H.; Reed, R.D.; Danko, C.G.; Montgomery, S.H. Chromosome fusion affects genetic diversity and evolutionary turnover of functional loci but consistently depends on chromosome size. Mol. Biol. Evol. 2021, 38, 4449–4462. [Google Scholar] [CrossRef]
- Mackintosh, A.; Laetsch, D.R.; Baril, T.; Foster, R.; Dincă, V.; Vila, R.; Hayward, A.; Lohse, K. The genome sequence of the lesser marbled fritillary, Brenthis ino, and evidence for a segregating neo-Z chromosome. G3-Genes Genom. Genet. 2022, 12, jkac069. [Google Scholar] [CrossRef]
- Höök, L.; Näsvall, K.; Vila, R.; Wiklund, C.; Backström, N. High-density linkage maps and chromosome level genome assemblies unveil direction and frequency of extensive structural rearrangements in wood white butterflies (Leptidea spp.). Chromosome Res. 2023, 31, 2. [Google Scholar] [CrossRef]
- Pringle, E.G.; Baxter, S.W.; Webster, C.L.; Papanicolaou, A.; Lee, S.F.; Jiggins, C.D. Synteny and chromosome evolution in the Lepidoptera: Evidence from mapping in Heliconius melpomene. Genetics 2007, 177, 417–426. [Google Scholar] [CrossRef]
- Papa, R.; Morrison, C.M.; Walters, J.R.; Counterman, B.A.; Chen, R.; Halder, G.; Ferguson, L.; Chamberlain, N.; Ffrench-Constant, R.; Kapan, D.D.; et al. Highly conserved gene order and numerous novel repetitive elements in genomic regions linked to wing pattern variation in Heliconius butterflies. BMC Genom. 2008, 9, 345. [Google Scholar] [CrossRef]
- Beldade, P.; Saenko, S.V.; Pul, N.; Long, A.D. A gene-based linkage map for Bicyclus anynana butterflies allows for a comprehensive analysis of synteny with the lepidopteran reference genome. PLoS Genet. 2009, 5, e1000366. [Google Scholar] [CrossRef]
- d’Alençon, E.; Sezutsu, H.; Legeai, F.; Permal, E.; Bernard-Samain, S.; Gimenez, S.; Gagneur, C.; Cousserans, F.; Shimomura, M.; Brun-Barale, A.; et al. Extensive synteny conservation of holocentric chromosomes in Lepidoptera despite high rates of local genome rearrangements. Proc. Natl. Acad. Sci. USA 2010, 107, 7680–7685. [Google Scholar] [CrossRef] [PubMed]
- Davey, J.W.; Chouteau, M.; Barker, S.L.; Maroja, L.; Baxter, S.W.; Simpson, F.; Merrill, R.M.; Joron, M.; Mallet, J.; Dasmahapatra, K.K.; et al. Major improvements to the Heliconius melpomene genome assembly used to confirm 10 chromosome fusion events in 6 million years of butterfly evolution. G3-Genes Genom. Genet. 2016, 6, 695–708. [Google Scholar] [CrossRef]
- Yasukochi, Y.; Tanaka-Okuyama, M.; Shibata, F.; Yoshido, A.; Marec, F.; Wu, C.; Zhang, H.; Goldsmith, M.R.; Sahara, K. Extensive conserved synteny of genes between the karyotypes of Manduca sexta and Bombyx mori revealed by BAC-FISH mapping. PLoS ONE 2009, 4, e7465. [Google Scholar] [CrossRef]
- Mongue, A.J.; Kawahara, A.Y. Population differentiation and structural variation in the Manduca sexta genome across the United States. G3-Genes Genom. Genet. 2022, 12, jkac047. [Google Scholar] [CrossRef]
- de Lesse, H. Caractères et repartition en France d’Erebia aethiopellus Hoffmsg. et E. mnestra Hb. Alexanor 1959, 1, 72–81. [Google Scholar]
- The Darwin Tree of Life Project Consortium. Sequence locally, think globally: The Darwin tree of life project. Proc. Natl. Acad. Sci. USA 2022, 119, e2115642118. [Google Scholar] [CrossRef]
- Lohse, K.; Weir, J.; Darwin Tree of Life Barcoding Collective; Wellcome Sanger Institute Tree of Life Programme; Wellcome Sanger Institute Scientific Operations: DNA Pipelines Collective; Tree of Life Core Informatics Collective; Darwin Tree of Life Consortium. The genome sequence of the meadow brown, Maniola jurtina (Linnaeus, 1758). Wellcome Open. Res. 2021, 6, 296. [Google Scholar] [CrossRef]
- Lohse, K.; Hayward, A.; Laetsch, D.R.; Vila, R.; Lucek, K.; Wellcome Sanger Institute Tree of Life Programme; Wellcome Sanger Institute Scientific Operations: DNA Pipelines Collective; Tree of Life Core Informatics Collective; Darwin Tree of Life Consortium. The genome sequence of the Arran brown, Erebia ligea (Linnaeus, 1758) [version 1; peer review: Awaiting peer review]. Wellcome Open. Res. 2022, 7, 259. [Google Scholar] [CrossRef]
- Lohse, O.; Lohse, K.; Wellcome Sanger Institute Tree of Life programme; Wellcome Sanger Institute Scientific Operations: DNA Pipelines collective; Tree of Life Core Informatics c ollective; Darwin Tree of Life Consortium. The genome sequence of the scotch argus butterfly, Erebia aethiops (Esper, 1777). Wellcome Open. Res. 2022, 7, 217. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Winter, D.; Lee, K.; Cox, M. 2020 pafr: Read, Manipulate and Visualize Pairwise mApping Format. Available online: https://dwinter.github.io/pafr/ (accessed on 18 December 2022).
- Hao, Z.; Lv, D.; Ge, Y.; Shi, J.; Weijers, D.; Yu, G.; Chen, J. RIdeogram: Drawing SVG graphics to visualize and map genome-wide data on the idiograms. PeerJ Comput. Sci. 2020, 6, e251. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. circlize implements and enhances circular visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef] [PubMed]
- Federley, H. Chromosomenzahlen finnländischer Lepidopteren I. Rhopalocera. Hereditas 1938, 24, 397–464. [Google Scholar] [CrossRef]
- Lorković, Z. Die Chromosomezahlen in der Spermatogenese der Tagfalter. Chromosoma 1941, 2, 155–191. [Google Scholar] [CrossRef]
- Bigger, T.R.L. Chromosome numbers of Lepidoptera. Part I. Ent. Gaz. 1960, 11, 149–152. [Google Scholar]
- Saitoh, K.; Abe, A. The chromosomes of Erebia ligea rishirizana (Nymphalidae, Satyrinae). Nota lepid. 1997, 20, 326–329. [Google Scholar]
- Larsen, T.B. Chromosome numbers and notes on testicular morphology of some Lebanese Rhopalocera (Insecta Lepidoptera). Entomol. Scand. 1975, 6, 253–260. [Google Scholar] [CrossRef]
- Lorković, Z. The butterfly chromosomes and their application in systematics and phylogeny. In Butterflies of Europe; Kudrna, O., Ed.; Aula Verlag: Wiesbaden, Germany, 1990; Volume 2, pp. 332–396. [Google Scholar]
- Brown, K.; Freitas, A.V.L.; von Schoultz, B.; Saura, A.O.; Saura, A. Chromosomal evolution of South American frugivorous butterflies in the Satyroid clade (Nymphalidae: Charaxinae, Morphinae and Satyrinae). Biol. J. Linn. Soc. 2007, 92, 467–481. [Google Scholar] [CrossRef]
- de Lesse, H. Formules chromosomiques de Lépidoptères Rhopalocères d’Afrique du Nord. Bull. Soc. Entomol. Fr. 1967, 72, 20–25. [Google Scholar] [CrossRef]
- de Lesse, H. Formules chromosomiques de Lépidoptères Rhopalocères d’Uganda et du Kenya. Ann. Soc. Entomol. Fr. 1968, 4, 581–599. [Google Scholar]
- de Lesse, H.; Condamin, M. Formules chromosomiques de quelques Lépidoptères Rhopalocères du Sénégal. Bull. IFAN Ser. A 1962, 24, 464–473. [Google Scholar]
- de Lesse, H.; Condamin, M. Formules chromosomiques de quelques Lépidoptères Rhopalocères du Sénégal et de Côte d’Ivoire. Bull. IFAN Ser. A 1965, 27, 1089–1094. [Google Scholar]
- Espeland, M.; Breinholt, J.; Willmott, K.R.; Warren, A.D.; Vila, R.; Toussaint, E.F.A.; Maunsell, S.C.; Aduse-Poku, K.; Talavera, G.; Eastwood, R.; et al. A comprehensive and dated phylogenomic analysis of butterflies. Curr. Biol. 2018, 28, 770–778. [Google Scholar] [CrossRef]
- Lukhtanov, V.; Sourakov, A.; Tikhonov, V.; Zakharov, E. Taxonomic rearrangement of the Erebia tyndarus species group (Lepidoptera, Nymphalidae, Satyrinae) based on an analysis of COI barcodes, morphology and geographic distribution. Folia Biol. 2019, 67, 149–157. [Google Scholar] [CrossRef]
- Lukhtanov, V.A.; Dinca, V.; Talavera, G.; Vila, R. Unprecedented within-species chromosome number cline in the Wood White butterfly Leptidea sinapis and its significance for karyotype evolution and speciation. BMC Evol. Biol. 2011, 11, 109. [Google Scholar] [CrossRef]
- Hejníčková, M.; Dalíková, M.; Potocký, P.; Tammaru, T.; Trehubenko, M.; Kubíčková, S.; Marec, F.; Zrzavá, M. Degenerated, undifferentiated, rearranged, lost: High variability of sex chromosomes in Geometridae (Lepidoptera) identified by sex chromatin. Cells 2021, 10, 2230. [Google Scholar] [CrossRef]
- Lucek, K. Evolutionary mechanisms of varying chromosome numbers in the radiation of Erebia butterflies. Genes 2018, 9, 166. [Google Scholar] [CrossRef]
- de Vos, J.M.; Augustijnen, H.; Batscher, L.; Lucek, K. Speciation through chromosomal fusion and fission in Lepidoptera. Philos. Trans. R. Soc. B Biol. Sci. 2020, 375, 20190539. [Google Scholar] [CrossRef]
- Kandul, N.P.; Lukhtanov, V.A.; Pierce, N.E. Karyotypic diversity and speciation in Agrodiaetus butterflies. Evolution 2007, 61, 546–559. [Google Scholar] [CrossRef] [PubMed]
- Vershinina, A.O.; Lukhtanov, V.A. Evolutionary mechanisms of runaway chromosome number change in Agrodiaetus butterflies. Sci. Rep. 2017, 7, 8199. [Google Scholar] [CrossRef] [PubMed]
- Peña, C.; Nylin, S.; Wahlberg, N. The radiation of Satyrini butterflies (Nymphalidae: Satyrinae): A challenge for phylogenetic methods. Zool. J. Linn. Soc. 2011, 161, 64–87. [Google Scholar] [CrossRef]
- Ruggieri, A.A.; Livraghi, L.; Lewis, J.J.; Evans, E.; Cicconardi, F.; Hebberecht, L.; Ortiz-Ruiz, Y.; Montgomery, S.H.; Ghezzi, A.; Rodriguez-Martinez, J.A.; et al. A butterfly pan-genome reveals a large amount of structural variation underlies the evolution of chromatin accessibility. Genome Res. 2022, 32, 1862–1875. [Google Scholar] [CrossRef]
- Peña, C.; Witthauer, H.; Klečková, I.; Fric, Z.; Wahlberg, N. Adaptive radiations in butterflies: Evolutionary history of the genus Erebia (Nymphalidae: Satyrinae). Biol. J. Linn. Soc. 2015, 116, 449–467. [Google Scholar] [CrossRef]
- Nilsson, N.O.; Löfstedt, C.; Dävring, L. Unusual sex-chromosome inheritance in six species of small ermine moths (Yponomeuta, Yponomeutidae, Lepidoptera). Hereditas 2008, 108, 259–265. [Google Scholar] [CrossRef]
- Nguyen, P.; Sýkorová, M.; Šichová, J.; Kůta, V.; Daliková, M.; Frydrychová, R.Č.; Neven, L.G.; Sahara, K.; Marec, F. Neo-sex chromosomes and adaptive potential in tortricid pests. Proc. Natl. Acad. Sci. USA 2013, 110, 6931–6936. [Google Scholar] [CrossRef]
- Šíchová, J.; Nguyen, P.; Dalíková, M.; Marec, F. Chromosomal evolution in tortricid moths: Conserved karyotypes with diverged features. PLoS ONE 2013, 8, e64520. [Google Scholar] [CrossRef]
- Van’t Hof, A.; Nguyen, P.; Dalíková, M.; Edmonds, N.; Marec, F.; Saccheri, I.J. Linkage map of the peppered moth, Biston betularia (Lepidoptera, Geometridae): A model of industrial melanism. Heredity 2013, 110, 283–295. [Google Scholar] [CrossRef]
- Dalíková, M.; Zrzavá, M.; Hladová, I.; Nguyen, P.; Šonský, I.; Flegrová, M.; Kubíčková, S.; Voleníková, A.; Kawahara, A.Y.; Peters, R.S.; et al. New insights into the evolution of the W chromosome in Lepidoptera. J. Hered. 2017, 108, 709–719. [Google Scholar] [CrossRef]
- Mongue, A.J.; Nguyen, P.; Voleníková, A.; Walters, J.R. Neo-sex chromosomes in the monarch butterfly, Danaus plexippus. G3-Genes Genom. Genet. 2017, 7, 3281–3294. [Google Scholar] [CrossRef] [PubMed]
- Pazhenkova, E.A.; Lukhtanov, V.A. Chromosomal conservatism vs. chromosomal megaevolution: Enigma of karyotypic evolution in Lepidoptera. BioRxiv 2022, 5, 494852. [Google Scholar] [CrossRef]
- Coyne, J.A.; Orr, H.A. Speciation; Sinauer Associates: Sunderland, MA, USA, 2004; p. 545. [Google Scholar]
- Presgraves, D.C. Sex chromosomes and speciation in Drosophila. Trends Genet. 2008, 24, 336–343. [Google Scholar] [CrossRef]
- Charlesworth, B.; Campos, J.L.; Jackson, B.C. Faster-X evolution: Theory and evidence from Drosophila. Mol. Ecol. 2018, 27, 3753–3771. [Google Scholar] [CrossRef] [PubMed]
- Mongue, A.J.; Hansen, M.E.; Walters, J.R. Support for faster and more adaptive Z chromosome evolution in two divergent lepidopteran lineages. Evolution 2022, 76, 332–345. [Google Scholar] [CrossRef] [PubMed]
- King, M. Species Evolution: The Role of Chromosomal Change; Cambridge University Press: Cambridge, UK, 1993; p. 360. [Google Scholar]
- Lucek, K.; Augustijnen, H.; Escudero, M. A holocentric twist to chromosomal speciation? Trends Ecol. Evol. 2022, 37, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Faria, R.; Navarro, A. Chromosomal speciation revisited: Rearranging theory with pieces of evidence. Trends Ecol. Evol. 2010, 25, 660–669. [Google Scholar] [CrossRef] [PubMed]
- Kirkpatrick, M.; Barton, N. Chromosome inversions, local adaptation and speciation. Genetics 2006, 173, 419–434. [Google Scholar] [CrossRef]




| Species | Assembly ID | Diploid Chromosome Number and Sex Chromosomes in Females | Diploid Chromosome Number and Sex Chromosomes in Males | Total Genome Size |
|---|---|---|---|---|
| M. jurtina | ilManJurt1.1 | 2n = 56 + ZW | 2n = 56 + ZZ | 402.0 Mb |
| E. ligea | ilEreLige1.2 | unstudied | 2n = 56 + ZZ | 506.4 Mb |
| E. aethiops | ilEreAeth2.2 | 2n = 36 + ZW | 2n = 36 + ZZ | 473.4 Mb |
| Comparison | Maximum Block Length, bp | Sum of Block Lengths, bp | Number of Blocks |
|---|---|---|---|
| M. jurtina–E. ligea | 13,739 | 32,722,763 | 15,870 |
| E. ligea–E. aethiops | 52,045 | 67,667,013 | 30,566 |
| M. jurtina–E. aethiops | 15,305 | 16,426,952 | 16,357 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pazhenkova, E.A.; Lukhtanov, V.A. Whole-Genome Analysis Reveals the Dynamic Evolution of Holocentric Chromosomes in Satyrine Butterflies. Genes 2023, 14, 437. https://doi.org/10.3390/genes14020437
Pazhenkova EA, Lukhtanov VA. Whole-Genome Analysis Reveals the Dynamic Evolution of Holocentric Chromosomes in Satyrine Butterflies. Genes. 2023; 14(2):437. https://doi.org/10.3390/genes14020437
Chicago/Turabian StylePazhenkova, Elena A., and Vladimir A. Lukhtanov. 2023. "Whole-Genome Analysis Reveals the Dynamic Evolution of Holocentric Chromosomes in Satyrine Butterflies" Genes 14, no. 2: 437. https://doi.org/10.3390/genes14020437
APA StylePazhenkova, E. A., & Lukhtanov, V. A. (2023). Whole-Genome Analysis Reveals the Dynamic Evolution of Holocentric Chromosomes in Satyrine Butterflies. Genes, 14(2), 437. https://doi.org/10.3390/genes14020437







