Unexpected Findings in Hereditary Breast and Ovarian Cancer Syndrome: Low-Level Constitutional Mosaicism in BRCA2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Case Report
2.2. Collection of Samples and Clinical Information
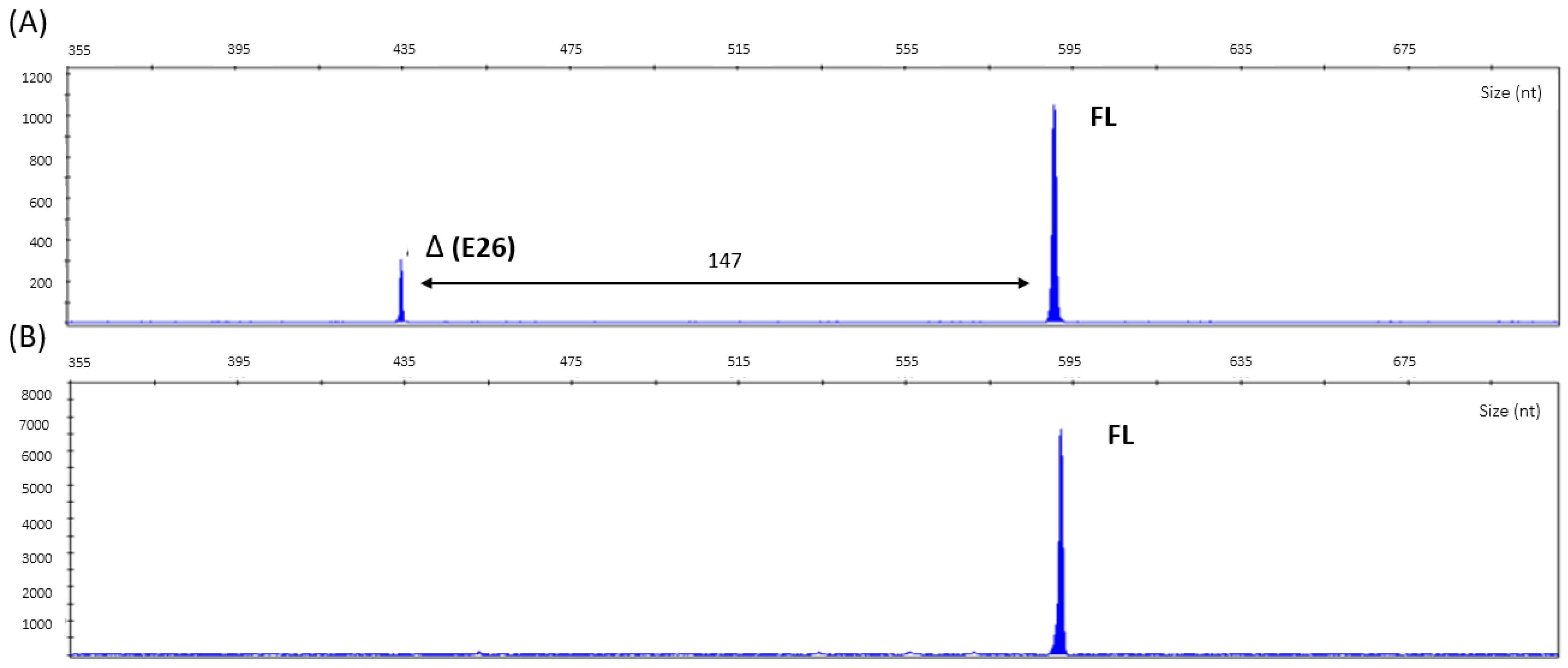
2.3. Molecular Genetics Studies
2.4. Confirmation Analysis
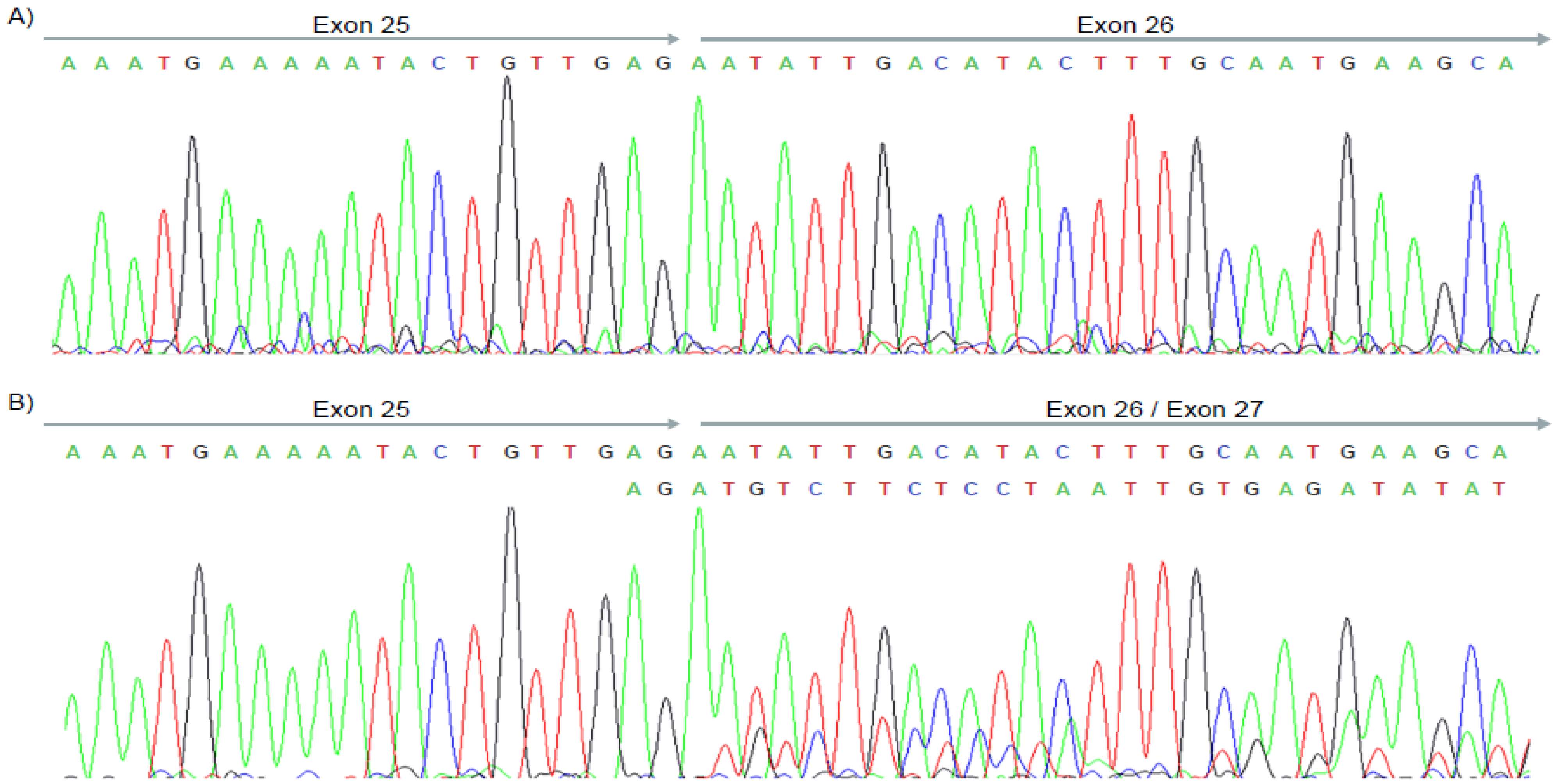
2.5. cDNA Analysis
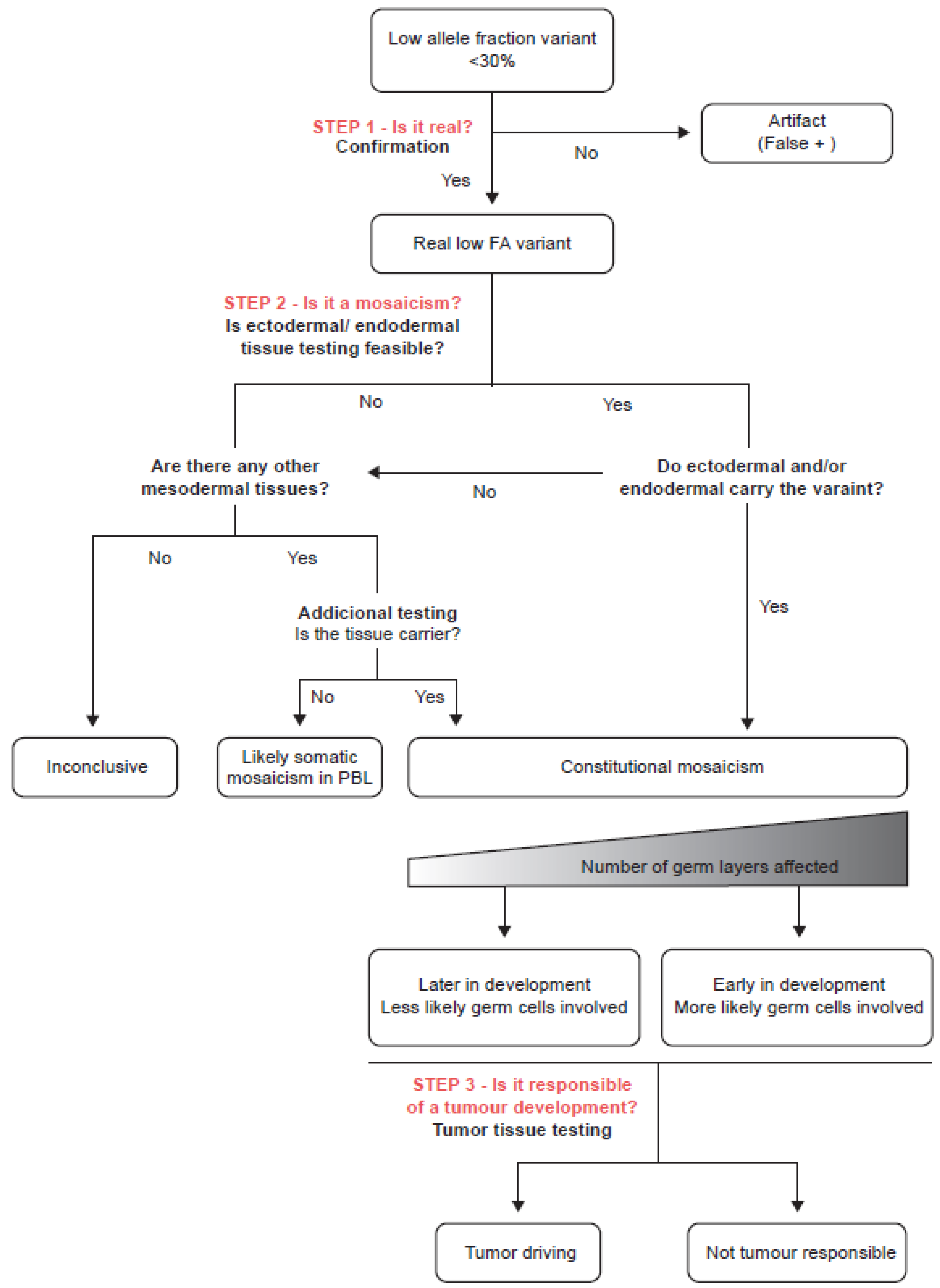
2.6. Diagnostic Algorithm Definition
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Srivastava, S.; Love-Nichols, J.A.; Dies, K.A.; Ledbetter, D.H.; Martin, C.L.; Chung, W.K.; Firth, H.V.; Frazier, T.; Hansen, R.L.; Prock, L.; et al. Meta-analysis and multidisciplinary consensus statement: Exome sequencing is a first-tier clinical diagnostic test for individuals with neurodevelopmental disorders. Genet. Med. 2019, 21, 2413–2421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedman, E.; Efrat, N.; Soussan-Gutman, L.; Dvir, A.; Kaplan, Y.; Ekstein, T.; Nykamp, K.; Powers, M.; Rabideau, M.; Sorenson, J.; et al. Low-level constitutional mosaicism of a de novoBRCA1 gene mutation. Br. J. Cancer 2015, 112, 765–768. [Google Scholar] [CrossRef] [Green Version]
- Alhopuro, P.; Vainionpää, R.; Anttonen, A.K.; Aittomäki, K.; Nevanlinna, H.; Pöyhönen, M. Constitutional mosaicism for a BRCA2 mutation as a cause of early-onset breast cancer. Fam. Cancer 2020, 19, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Glez, V.; Tenorio, J.; Nevado, J.; Gordo, G.; Rodríguez-Laguna, L.; Feito, M.; de Lucas, R.; Pérez-Jurado, L.A.; Ruiz Pérez, V.L.; Torrelo, A.; et al. A six-attribute classification of genetic mosaicism. Genet. Med. 2020, 22, 1743–1757. [Google Scholar] [CrossRef] [PubMed]
- Carlson, B. Human Embriology & Developmental Biology, 4th ed.; Elsevier: Philadelphia, PA, USA, 1999. [Google Scholar]
- Waldman, L.; Mahon, S.M. Mosaicism: Implications for genetic counseling and medical management. Clin. J. Oncol. Nurs. 2017, 21, 428–432. [Google Scholar] [PubMed]
- Chen, J.L.; Miller, D.T.; Schmidt, L.S.; Malkin, D.; Korf, B.R.; Eng, C.; Kwiatkowski, D.J.; Giannikou, K. Mosaicism in Tumor Suppressor Gene Syndromes: Prevalence, Diagnostic Strategies, and Transmission Risk. Annu. Rev. Genom. Hum. Genet. 2022, 23, 331–361. [Google Scholar] [CrossRef]
- Delon, I.; Taylor, A.; Molenda, A.; Drummond, J.; Oakhill, K.; Girling, A.; Liu, H.; Wthittaker, J.; Treacy, R.; Tischkowitz, M. A germline mosaic BRCA1 exon deletion in a woman with bilateral basal-like breast cancer. Clin Genet. 2013, 84, 297–299. [Google Scholar] [CrossRef]
- Golmard, L.; Delnatte, C.; Laugé, A.; Moncoutier, V.; Lefol, C.; Abidallah, K.; Tenreiro, H.; Copigny, F.; Giraudeau, M.; Guy, C.; et al. Breast and ovarian cancer predisposition due to de novo BRCA1 and BRCA2 mutations. Oncogene 2016, 35, 1324–1327. [Google Scholar] [CrossRef]
- Gráf, A.; Enyedi, M.Z.; Pintér, L.; Kriston-Pál, É.; Jaksa, G.; Bálind, Á.; Ezer, É.; Horváth, P.; Sükösd, F.; Kiss, E.; et al. The combination of single-cell and next-generation sequencing can reveal mosaicism for BRCA2 mutations and the fine molecular details of tumorigenesis. Cancers 2021, 13, 2354. [Google Scholar] [CrossRef]
- Speight, B.; Colvin, E.; Epurescu, E.D.; Drummond, J.; Verhoef, S.; Pereira, M.; Evans, D.G. Low—level constitutional mosaicism of BRCA1 in two women with young onset ovarian cancer. Hered. Cancer Clin. Pract. 2022, 20, 1–7. [Google Scholar] [CrossRef]
- Taylor, A.; Brady, A.F.; Frayling, I.M.; Hanson, H.; Tischkowitz, M.; Turnbull, C.; Side, L. Consensus for genes to be included on cancer panel tests offered by UK genetics services: Guidelines of the UK Cancer Genetics Group. J. Med. Genet. 2018, 55, 372–377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sim, N.L.; Kumar, P.; Hu, J.; Henikoff, S.; Schneider, G.; Ng, P.C. SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012, 40, 452–457. [Google Scholar] [CrossRef] [PubMed]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarz, J.M.; Cooper, D.N.; Schuelke, M.; Seelow, D. Mutationtaster2: Mutation prediction for the deep-sequencing age. Nat. Methods 2014, 11, 361–362. [Google Scholar] [CrossRef] [PubMed]
- Pollard, K.S.; Hubisz, M.J.; Rosenbloom, K.R.; Siepel, A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010, 20, 110–121. [Google Scholar] [CrossRef] [Green Version]
- Brewer, C.J.; Gillespie, M.; Fierro, J.; Scaringe, W.A.; Li, J.; Lee, C.Y.; Yen, H.Y.; Gao, H.; Strom, S.P. The Value of Parental Testing by Next-Generation Sequencing Includes the Detection of Germline Mosaicism. J. Mol. Diagn. 2020, 22, 670–678. [Google Scholar] [CrossRef] [Green Version]
- Mu, W.; Lu, H.M.; Chen, J.; Li, S.; Elliott, A.M. Sanger Confirmation Is Required to Achieve Optimal Sensitivity and Specificity in Next-Generation Sequencing Panel Testing. J. Mol. Diagn. 2016, 18, 923–932. [Google Scholar] [CrossRef] [Green Version]
- Yeo, G.; Burge, C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004, 11, 377–394. [Google Scholar] [CrossRef]
- Reese, M.G.; Eeckman, F.H.; Kulp, D.; Haussler, D. Improved Splice Site Detection in Genie. J. Comput. Biol. 1997, 4, 311–323. [Google Scholar] [CrossRef]
- Pertea, M.; Lin, X.; Salzberg, S.L. GeneSplicer: A new computational method for splice site prediction. Nucleic Acids Res. 2001, 29, 1185–1190. [Google Scholar] [CrossRef] [Green Version]
- Jaganathan, K.; Kyriazopoulou Panagiotopoulou, S.; McRae, J.F.; Darbandi, S.F.; Knowles, D.; Li, Y.I.; Kosmicki, J.A.; Arbelaez, J.; Cui, W.; Schwartz, G.B.; et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell 2019, 176, 535–548.e24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bamford, S.; Dawson, E.; Forbes, S.; Clements, J.; Pettett, R.; Dogan, A.; Flanagan, A.; Teague, J.; Futreal, P.A.; Stratton, M.R.; et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br. J. Cancer 2004, 91, 355–358. [Google Scholar] [CrossRef] [PubMed]
- Behjati, S.; Maschietto, M.; Williams, R.D.; Side, L.; Hubank, M.; West, R.; Pearson, K.; Sebire, N.; Tarpey, P.; Futreal, A.; et al. A pathogenic mosaic TP53 mutation in two germ layers detected by next generation sequencing. PLoS ONE 2014, 9, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Moran, K.; Richards-Yutz, J.; Toorens, E.; Gerhart, D.; Ganguly, T.; Shields, C.L.; Ganguly, A. Enhanced sensitivity for detection of low-level germline mosaic RB1 mutations in sporadic retinoblastoma cases using deep semiconductor sequencing. Hum. Mutat. 2014, 35, 384–391. [Google Scholar] [CrossRef] [Green Version]
- Newman, H.; Long, J.; Zelley, K.; Baldino, S.; Li, M.M.; Maxwell, K.N.; MacFarland, S.P. Looking closely at overgrowth: Constitutional mosaicism in PTEN hamartoma tumor syndrome. Clin. Genet. 2022, 102, 557–559. [Google Scholar] [CrossRef]
- Stormorken, A.T.; Berg, T.; Norum, O.J.; Hølmebakk, T.; Aaberg, K.; Steigen, S.E.; Grindedal, E.M. APC mosaicism in a young woman with desmoid type fibromatosis and familial adenomatous polyposis. Fam. Cancer 2018, 17, 539–543. [Google Scholar] [CrossRef] [Green Version]
- Momozawa, Y.; Iwasaki, Y.; Parsons, M.T.; Kamatani, Y.; Takahashi, A.; Tamura, C.; Katagiri, T.; Yoshida, T.; Nakamura, S.; Sugano, K.; et al. Germline pathogenic variants of 11 breast cancer genes in 7,051 Japanese patients and 11,241 controls. Nat. Commun. 2018, 9, 4–6. [Google Scholar] [CrossRef]
- Andreassen, P.R.; Seo, J.; Wiek, C.; Hanenberg, H. Understanding BRCA2 function as a tumor suppressor based on domain-specific activities in DNA damage responses. Genes 2021, 12, 1034. [Google Scholar] [CrossRef]
- Yang, H.; Jeffrey, P.D.; Miller, J.; Kinnucan, E.; Sun, Y.; Thomä, N.H.; Zheng, N.; Chen, P.L.; Lee, W.H.; Pavletich, N.P. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science 2002, 297, 1837–1848. [Google Scholar] [CrossRef] [Green Version]
- Mesman, R.L.S.; Calléja, F.M.G.R.; de la Hoya, M.; Devilee, P.; van Asperen, C.J.; Vrieling, H.; Vreeswijk, M.P.G. Alternative mRNA splicing can attenuate the pathogenicity of presumed loss-of-function variants in BRCA2. Genet. Med. 2020, 22, 1355–1365. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abou Tayoun, A.N.; Pesaran, T.; DiStefano, M.T.; Oza, A.; Rehm, H.L.; Biesecker, L.G.; Harrison, S.M.; Abou Tayoun, A.; Jalila Children, A.; Hospital, S.; et al. Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion ClinGen Sequence Variant Interpretation Working Group (ClinGen SVI). Hum. Mutat. 2018, 39, 1517–1524. [Google Scholar] [CrossRef] [PubMed]
- Tavtigian, S.V.; Harrison, S.M.; Boucher, K.M.; Biesecker, L.G.; City, S.L.; City, S.L.; City, S.L.; Branch, M.G.; Human, N. Fitting a naturally scaled point system to the ACMG/AMP variant classification guidelines. Hum. Mutat. 2021, 41, 1734–1737. [Google Scholar] [CrossRef] [PubMed]
- Ju, Y.S.; Martincorena, I.; Gerstung, M.; Petljak, M.; Ludmil, B.; Rahbari, R.; Wedge, D.C.; Davies, H.R.; Fullam, A.; Martin, S.; et al. Somatic mutations reveal asymmetric cellular dynamics in the early human embryo. Nature 2017, 543, 714–718. [Google Scholar] [CrossRef]
- Campbell, I.M.; Shaw, C.A.; Pawel, S.; Lupski, J.R. Somatic mosaicism: Implications for disease and transmission genetics. Trends Genet. 2015, 31, 382–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-Santiago, S.; Ramón y Cajal, T.; Aguirre, E.; Alés-Martínez, J.E.; Andrés, R.; Balmaña, J.; Graña, B.; Herrero, A.; Llort, G.; González-del-Alba, A.; et al. SEOM clinical guidelines in hereditary breast and ovarian cancer (2019). Clin. Transl. Oncol. 2020, 22, 193–200. [Google Scholar] [CrossRef] [Green Version]
- Biesecker, L.G.; Spinner, N.B. A genomic view of mosaicism and human disease. Nat. Rev. Genet. 2013, 14, 307–320. [Google Scholar] [CrossRef]

| Tissue Type | Read Depth | VAF (%) |
|---|---|---|
| PBL | 625 | 11 |
| Breast tumour | 243 | 66 |
| Buccal swab | 193,704 | 15 |
| Urine | 51,904 | 23 |
| ∆ Type | ∆ Score a | Pre-mRNA Position b |
|---|---|---|
| Acceptor Loss | 0.70 | −147 bp |
| Donor Loss | 1.00 | −1 bp |
| Acceptor Gain | 0.00 | |
| Donor Gain | 0.01 | −21 bp |
| Case | Age of Diagnosis | Type of Cancer | Nucleotide Change | Amino Acid Change | Gene | VAF | Reference |
|---|---|---|---|---|---|---|---|
| 1 | 39 | Breast | DelEx16 | BRCA1 | ~33% PBL and saliva ~36% right breast tumor ~64% left breast tumor | [8] | |
| 2 | 43 | Breast | c.1953dupG | p.(Lys652Glufs*21) | BRCA1 | ~5% PBL, buccal swab and non-neoplastic breast ~47% in breast tumor | [2] |
| 3 | 41 | Breast | DelEx1-13 | BRCA1 | ~25% PBL | [9] | |
| 4 | 36 | Breast | c.9294C>G | p.(Tyr3098Ter) | BRCA2 | ~20 PBL | [3] |
| ~36% normal breast | |||||||
| ~21–25% ovary | |||||||
| ~57% in breast tumor | |||||||
| 5 | Mids 50 | Ovarian | c.7795G>T | p.(Glu2599Ter) | BRCA2 | ~26% PBL | [10] |
| ~21–23% buccal swab | |||||||
| ~77–78% ovarian tumor | |||||||
| 6 | 42 | Ovarian | c.5074G>A | p.(Asp1692Asn) | BRCA1 | ~10% PBL | [11] |
| ~52% ovarian tumor | |||||||
| 7 | 53 | Ovarian | c.2755_2758dupCCTG | p.(Val920Alafs*6) | BRCA1 | ~14% PBL | [11] |
| ~59% ovarian tumor | |||||||
| 8 | 25 | Breast | c.9648+1G>A | p.(Asn3168_Leu3216del) | BRCA2 | ~11% PBL | This case |
| ~15% buccal swab | |||||||
| ~23% urine | |||||||
| ~66% breast tumor |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hidalgo Mayoral, I.; Almeida Santiago, A.; Sánchez-Zapardiel, J.M.; Hidalgo Calero, B.; de la Hoya, M.; Gómez-Sanz, A.; de Miguel Reyes, M.; Robles, L. Unexpected Findings in Hereditary Breast and Ovarian Cancer Syndrome: Low-Level Constitutional Mosaicism in BRCA2. Genes 2023, 14, 502. https://doi.org/10.3390/genes14020502
Hidalgo Mayoral I, Almeida Santiago A, Sánchez-Zapardiel JM, Hidalgo Calero B, de la Hoya M, Gómez-Sanz A, de Miguel Reyes M, Robles L. Unexpected Findings in Hereditary Breast and Ovarian Cancer Syndrome: Low-Level Constitutional Mosaicism in BRCA2. Genes. 2023; 14(2):502. https://doi.org/10.3390/genes14020502
Chicago/Turabian StyleHidalgo Mayoral, Irene, Ainhoa Almeida Santiago, Jose Manuel Sánchez-Zapardiel, Beatriz Hidalgo Calero, Miguel de la Hoya, Alicia Gómez-Sanz, Montserrat de Miguel Reyes, and Luis Robles. 2023. "Unexpected Findings in Hereditary Breast and Ovarian Cancer Syndrome: Low-Level Constitutional Mosaicism in BRCA2" Genes 14, no. 2: 502. https://doi.org/10.3390/genes14020502
APA StyleHidalgo Mayoral, I., Almeida Santiago, A., Sánchez-Zapardiel, J. M., Hidalgo Calero, B., de la Hoya, M., Gómez-Sanz, A., de Miguel Reyes, M., & Robles, L. (2023). Unexpected Findings in Hereditary Breast and Ovarian Cancer Syndrome: Low-Level Constitutional Mosaicism in BRCA2. Genes, 14(2), 502. https://doi.org/10.3390/genes14020502






