Genome-Wide Association Analysis Identified Variants Associated with Body Measurement and Reproduction Traits in Shaziling Pigs
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Phenotypes
2.2. Animals and Genotypes
2.3. Data Quality Control
2.4. SNP-Based Heritability
2.5. Population Structure and Kinship Analyses
2.6. GWAS
2.7. Candidate Gene Search and Functional Annotation
3. Results
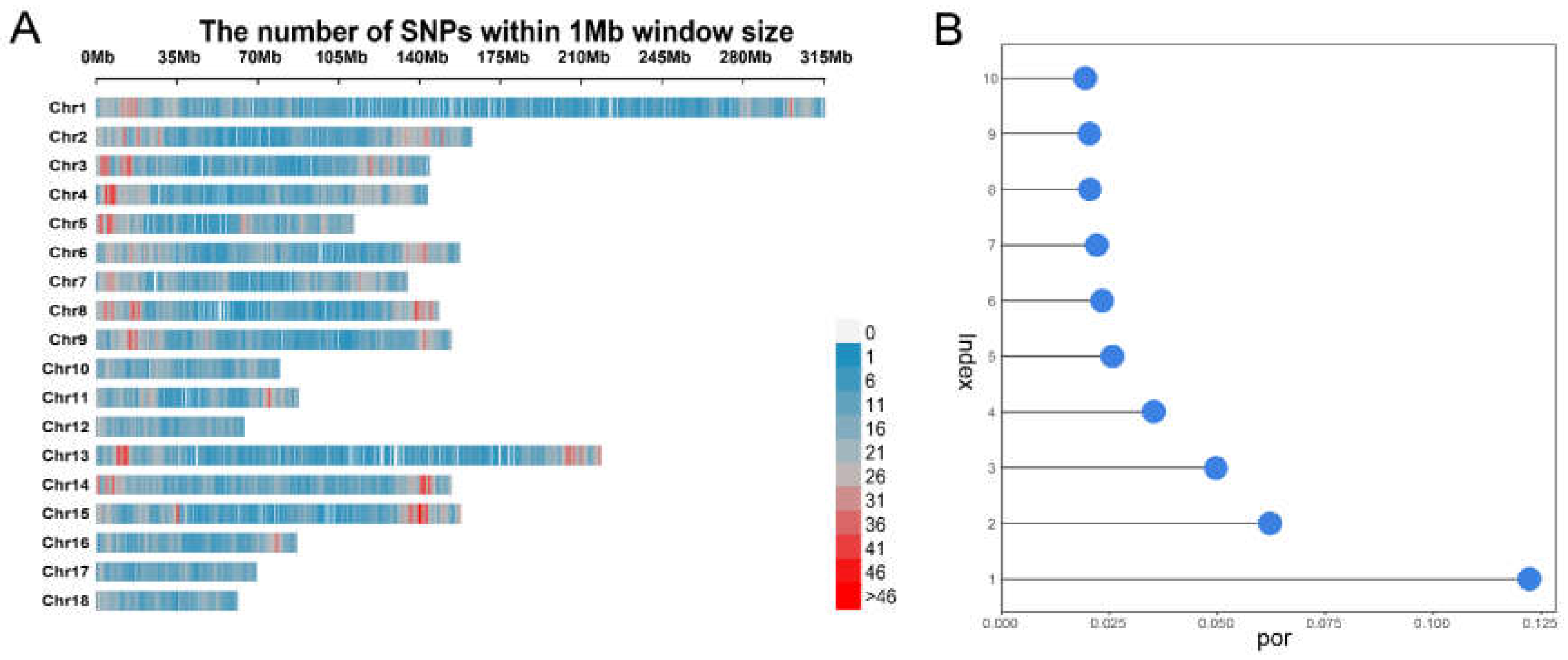
3.1. Description of Phenotypes and Genotypes
3.2. Results of Population Structure and Kinship Analyses
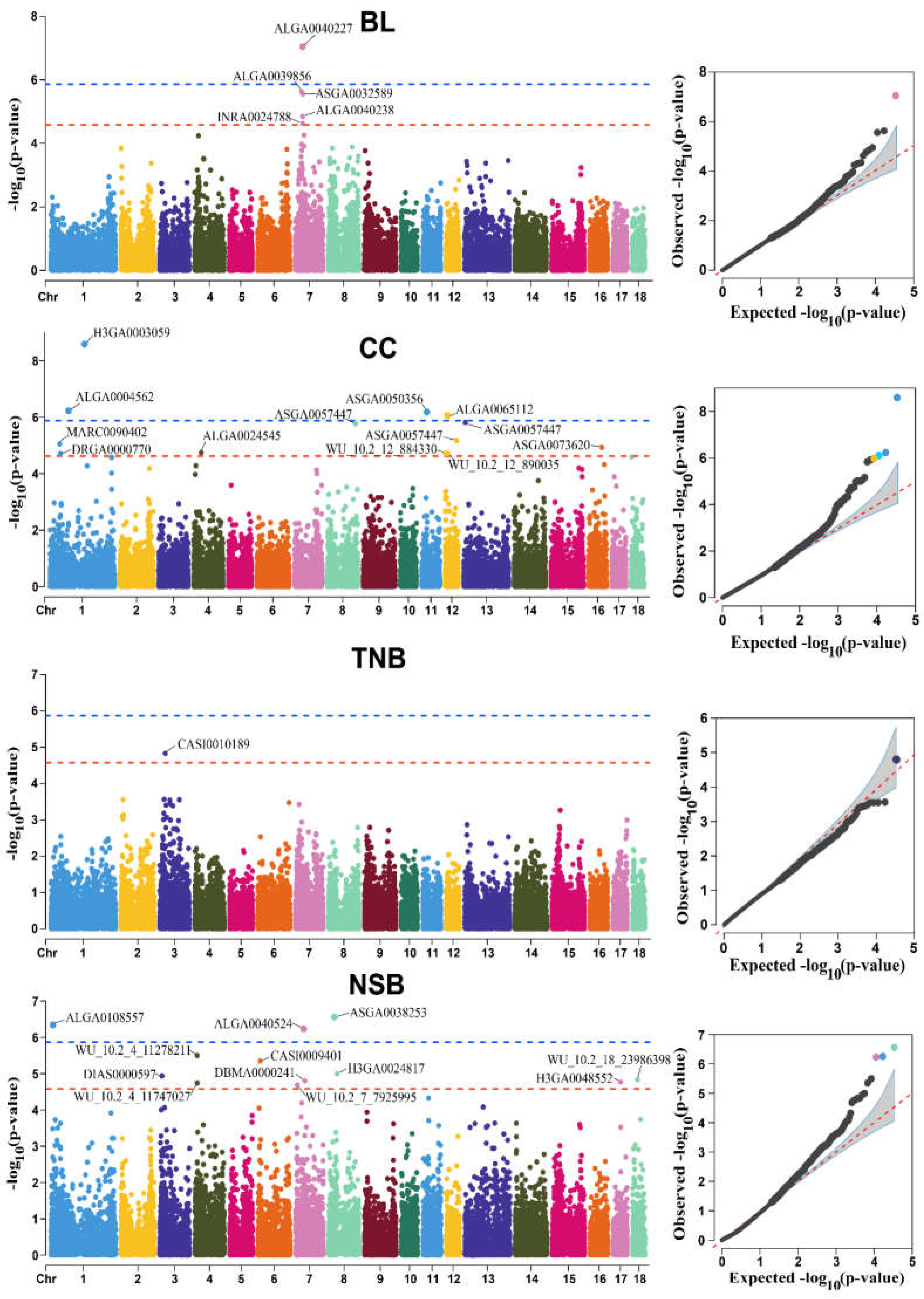
3.3. GWAS Results
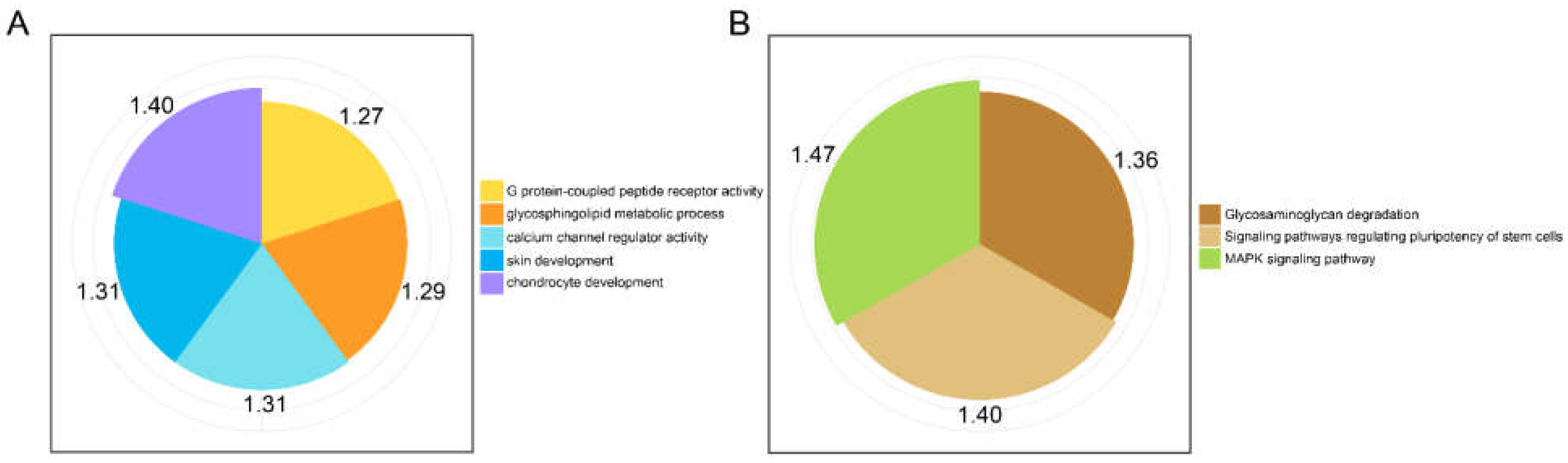
3.4. Enrichment Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lunney, J.K. Advances in swine biomedical model genomics. Int. J. Biol. Sci. 2007, 3, 179–184. [Google Scholar] [CrossRef]
- Ma, X.; Li, P.H.; Zhu, M.X.; He, L.C.; Sui, S.P.; Gao, S.; Su, G.S.; Ding, N.S.; Huang, Y.; Lu, Z.Q.; et al. Genome-wide association analysis reveals genomic regions on Chromosome 13 affecting litter size and candidate genes for uterine horn length in Erhualian pigs. Animal 2018, 12, 2453–2461. [Google Scholar] [CrossRef]
- Chen, P.; Baas, T.J.; Mabry, J.W.; Koehler, K.J.; Dekkers, J.C. Genetic parameters and trends for litter traits in U.S. Yorkshire, Duroc, Hampshire, and Landrace pigs. J. Anim. Sci. 2003, 81, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ober, U.; Erbe, M.; Zhang, H.; Gao, N.; He, J.; Li, J.; Simianer, H. Improving the accuracy of whole genome prediction for complex traits using the results of genome wide association studies. PLoS ONE 2014, 9, e93017. [Google Scholar] [CrossRef] [PubMed]
- Fukawa, K.; Sugiyama, T.; Kusuhara, S.; Kudoh, O.; Kameyama, K. Model selection and genetic parameter estimation for performance traits, body measurement traits and leg score traits in a closed population of Duroc pigs. Nihon Chikusan Gakkaiho 2001, 72, 97–106. [Google Scholar] [CrossRef]
- Ishida, T.; Kuroki, T.; Harada, H.; Fukuhara, R. Estimation of additive and dominance genetic variances in line breeding swine. Asian Australas. J. Anim. Sci. 2001, 14, 1–6. [Google Scholar] [CrossRef]
- Ogawa, S.; Yazaki, N.; Ohnishi, C.; Ishii, K.; Uemoto, Y.; Satoh, M. Maternal effect on body measurement and meat production traits in purebred Duroc pigs. J. Anim. Breed. Genet. 2021, 138, 237–245. [Google Scholar] [CrossRef]
- Ohnishi, C.; Satoh, M. Estimation of genetic parameters for performance and body measurement traits in Duroc pigs selected for average daily gain, loin muscle area, and backfat thickness. Livest. Sci. 2018, 214, 161–166. [Google Scholar] [CrossRef]
- Yazaki, N.; Ogawa, S.I.; Onishi, C.; Ishii, K.; Uemoto, Y.; Sato, M. Effectiveness of body measurement traits for improving production traits in Duroc pigs. Nihon Chikusan Gakkaiho 2020, 91, 9–16. [Google Scholar] [CrossRef]
- Tuomi, J. Mammalian reproductive strategies: A generalized relation of litter size to body size. Oecologia 1980, 45, 39–44. [Google Scholar] [CrossRef]
- Haldar, A.; Pal, P.; Datta, M.; Paul, R.; Pal, S.K.; Majumdar, D.; Biswas, C.K.; Pan, S. Prolificacy and Its Relationship with Age, Body Weight, Parity, Previous Litter Size and Body Linear Type Traits in Meat-type Goats. Asian Australas. J. Anim. Sci. 2014, 27, 628–634. [Google Scholar] [CrossRef]
- Song, B.; Zheng, C.; Zheng, J.; Zhang, S.; Zhong, Y.; Guo, Q.; Li, F.; Long, C.; Xu, K.; Duan, Y.; et al. Comparisons of carcass traits, meat quality, and serum metabolome between Shaziling and Yorkshire pigs. Anim. Nutr. 2022, 8, 125–134. [Google Scholar] [CrossRef]
- Ai, H.; Huang, L.; Ren, J. Genetic diversity, linkage disequilibrium and selection signatures in chinese and Western pigs revealed by genome-wide SNP markers. PLoS ONE 2013, 8, e56001. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Xu, X.L.; Ma, H.M.; Jiang, J. Integrative analysis of transcriptomics and proteomics of skeletal muscles of the Chinese indigenous Shaziling pig compared with the Yorkshire breed. BMC Genet. 2016, 17, 80. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xu, P.; Zhang, C.; Sun, C.; Li, X.; Han, X.; Li, M.; Qiao, R. Genome-wide association study identifies variants in the CAPN9 gene associated with umbilical hernia in pigs. Anim. Genet. 2019, 50, 162–165. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Gray, A.; Stewart, I.; Tenesa, A. Advanced complex trait analysis. Bioinformatics 2012, 28, 3134–3136. [Google Scholar] [CrossRef]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef]
- Xue, Y.; Liu, S.; Li, W.; Mao, R.; Zhuo, Y.; Xing, W.; Liu, J.; Wang, C.; Zhou, L.; Lei, M.; et al. Genome-Wide Association Study Reveals Additive and Non-Additive Effects on Growth Traits in Duroc Pigs. Genes 2022, 13, 1454. [Google Scholar] [CrossRef]
- Shi, L.; Wang, L.; Fang, L.; Li, M.; Tian, J.; Wang, L.; Zhao, F. Integrating genome-wide association studies and population genomics analysis reveals the genetic architecture of growth and backfat traits in pigs. Front. Genet. 2022, 13, 1078696. [Google Scholar] [CrossRef]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J.; et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef]
- Thiengpimol, P.; Koonawootrittriron, S.; Suwanasopee, T. Genetic and phenotypic correlations between backfat thickness and weight at 28 weeks of age, and reproductive performance in primiparous Landrace sows raised under tropical conditions. Trop. Anim. Health Prod. 2022, 54, 43. [Google Scholar] [CrossRef] [PubMed]
- Bohlouli, M.; Brandt, H.; Konig, S. Genetic parameters for linear conformation, stayability, performance and reproduction traits in German local Swabian-Hall landrace sows. J. Anim. Breed. Genet. 2023, 140, 144–157. [Google Scholar] [CrossRef]
- Nikkila, M.T.; Stalder, K.J.; Mote, B.E.; Rothschild, M.F.; Gunsett, F.C.; Johnson, A.K.; Karriker, L.A.; Boggess, M.V.; Serenius, T.V. Genetic associations for gilt growth, compositional, and structural soundness traits with sow longevity and lifetime reproductive performance. J. Anim. Sci. 2013, 91, 1570–1579. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Abdalla, I.M.; Nazar, M.; Fan, Y.; Zhang, Z.; Wu, X.; Xu, T.; Yang, Z. Genome-Wide Association Study on Reproduction-Related Body-Shape Traits of Chinese Holstein Cows. Animals 2021, 11, 1927. [Google Scholar] [CrossRef]
- Nielsen, S.; Kristensen, A.; Moustsen, V.A. Litter size of Danish crossbred sows increased without changes in sow body di mensions over a thirteen year period. Livest. Sci. 2018, 209, 73–76. [Google Scholar] [CrossRef]
- Ogawa, S.; Ohnishi, C.; Ishii, K.; Uemoto, Y.; Satoh, M. Genetic relationship between litter size traits at birth and body measurement and production traits in purebred Duroc pigs. Anim. Sci. J. 2020, 91, e13497. [Google Scholar] [CrossRef]
- Zhou, S.; Ding, R.; Zhuang, Z.; Zeng, H.; Wen, S.; Ruan, D.; Wu, J.; Qiu, Y.; Zheng, E.; Cai, G.; et al. Genome-Wide Association Analysis Reveals Genetic Loci and Candidate Genes for Chest, Abdominal, and Waist Circumferences in Two Duroc Pig Populations. Front. Vet. Sci. 2021, 8, 807003. [Google Scholar] [CrossRef]
- Li, C.; Duan, D.; Xue, Y.; Han, X.; Wang, K.; Qiao, R.; Li, X.L.; Li, X.J. An association study on imputed whole-genome resequencing from high-throughput sequencing data for body traits in crossbred pigs. Anim. Genet. 2022, 53, 212–219. [Google Scholar] [CrossRef]
- Manolio, T.A.; Collins, F.S.; Cox, N.J.; Goldstein, D.B.; Hindorff, L.A.; Hunter, D.J.; McCarthy, M.I.; Ramos, E.M.; Cardon, L.R.; Chakravarti, A.; et al. Finding the missing heritability of complex diseases. Nature 2009, 461, 747–753. [Google Scholar] [CrossRef]
- Jiang, N.; Liu, C.; Lan, T.; Zhang, Q.; Cao, Y.; Pu, G.; Niu, P.; Zhang, Z.; Li, Q.; Zhou, J.; et al. Polymorphism of VRTN Gene g.20311_20312ins291 Was Associated with the Number of Ribs, Carcass Diagonal Length and Cannon Bone Circumference in Suhuai Pigs. Animals 2020, 10, 484. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Gao, Q.; Wang, T.; Chai, W.; Zhan, Y.; Akhtar, F.; Zhang, Z.; Li, Y.; Shi, X.; Wang, C. Multi-Thoracolumbar Variations and NR6A1 Gene Polymorphisms Potentially Associated with Body Size and Carcass Traits of Dezhou Donkey. Animals 2022, 12, 1349. [Google Scholar] [CrossRef] [PubMed]
- Stewart, T.; Schinckel, A. Genetic Parameters for Swine Growth and Carcass Traits; FAO: Rome, Italy, 1989. [Google Scholar]
- Pereira, M.; Jeyabalan, J.; Jorgensen, C.S.; Hopkinson, M.; Al-Jazzar, A.; Roux, J.P.; Chavassieux, P.; Orriss, I.R.; Cleasby, M.E.; Chenu, C. Chronic administration of Glucagon-like peptide-1 receptor agonists improves trabecular bone mass and architecture in ovariectomised mice. Bone 2015, 81, 459–467. [Google Scholar] [CrossRef]
- Mabilleau, G.; Mieczkowska, A.; Irwin, N.; Flatt, P.R.; Chappard, D. Optimal bone mechanical and material properties require a functional glucagon-like peptide-1 receptor. J. Endocrinol. 2013, 219, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Philipp, M.; Berger, I.M.; Just, S.; Caron, M.G. Overlapping and opposing functions of G protein-coupled receptor kinase 2 (GRK2) and GRK5 during heart development. J. Biol. Chem. 2014, 289, 26119–26130. [Google Scholar] [CrossRef]
- Hwang, S.H.; White, K.A.; Somatilaka, B.N.; Shelton, J.M.; Richardson, J.A.; Mukhopadhyay, S. The G protein-coupled receptor Gpr161 regulates forelimb formation, limb patterning and skeletal morphogenesis in a primary cilium-dependent manner. Development 2018, 145, dev154054. [Google Scholar] [CrossRef]
- Chen, Y.H.; Lin, Y.T.; Lee, G.H. Novel and unexpected functions of zebrafish CCAAT box binding transcription factor (NF-Y) B subunit during cartilages development. Bone 2009, 44, 777–784. [Google Scholar] [CrossRef]
- Basile, V.; Baruffaldi, F.; Dolfini, D.; Belluti, S.; Benatti, P.; Ricci, L.; Artusi, V.; Tagliafico, E.; Mantovani, R.; Molinari, S.; et al. NF-YA splice variants have different roles on muscle differentiation. Biochim. Biophys. Acta 2016, 1859, 627–638. [Google Scholar] [CrossRef]
- Go, M.J.; Takenaka, C.; Ohgushi, H. Forced expression of Sox2 or Nanog in human bone marrow derived mesenchymal stem cells maintains their expansion and differentiation capabilities. Exp. Cell Res. 2008, 314, 1147–1154. [Google Scholar] [CrossRef]
- arcOGEN Consortium; arcOGEN Collaborators. Identification of new susceptibility loci for osteoarthritis (arcOGEN): A genome-wide association study. Lancet 2012, 380, 815–823. [Google Scholar] [CrossRef]
- Zhu, M.; Yin, P.; Hu, F.; Jiang, J.; Yin, L.; Li, Y.; Wang, S. Integrating genome-wide association and transcriptome prediction model identifies novel target genes for osteoporosis. Osteoporos. Int. 2021, 32, 2493–2503. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.; Reynard, L.N.; Loughlin, J. Functional characterisation of the osteoarthritis susceptibility locus at chromosome 6q14.1 marked by the polymorphism rs9350591. BMC Med. Genet. 2015, 16, 81. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.R.; Ko, N.Y.; Chen, K.H. Medical Treatment for Osteoporosis: From Molecular to Clinical Opinions. Int. J. Mol. Sci. 2019, 20, 2213. [Google Scholar] [CrossRef]
- Ma, X.; Su, P.; Yin, C.; Lin, X.; Wang, X.; Gao, Y.; Patil, S.; War, A.R.; Qadir, A.; Tian, Y.; et al. The Roles of FoxO Transcription Factors in Regulation of Bone Cells Function. Int. J. Mol. Sci. 2020, 21, 692. [Google Scholar] [CrossRef]
- Shen, W.; Sun, B.; Zhou, C.; Ming, W.; Zhang, S.; Wu, X. CircFOXP1/FOXP1 promotes osteogenic differentiation in adipose-derived mesenchymal stem cells and bone regeneration in osteoporosis via miR-33a-5p. J. Cell. Mol. Med. 2020, 24, 12513–12524. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, P.; Xu, S.; Li, Y.; Dekker, J.D.; Li, B.; Fan, Y.; Zhang, Z.; Hong, Y.; Yang, G.; et al. FOXP1 controls mesenchymal stem cell commitment and senescence during skeletal aging. J. Clin. Investig. 2017, 127, 1241–1253. [Google Scholar] [CrossRef]
- Liu, P.; Huang, S.; Ling, S.; Xu, S.; Wang, F.; Zhang, W.; Zhou, R.; He, L.; Xia, X.; Yao, Z.; et al. Foxp1 controls brown/beige adipocyte differentiation and thermogenesis through regulating beta3-AR desensitization. Nat. Commun. 2019, 10, 5070. [Google Scholar] [CrossRef] [PubMed]
- Bolormaa, S.; Hayes, B.J.; van der Werf, J.H.; Pethick, D.; Goddard, M.E.; Daetwyler, H.D. Detailed phenotyping identifies genes with pleiotropic effects on body composition. BMC Genomics 2016, 17, 224. [Google Scholar] [CrossRef]
- Kausar, T.; Nayeem, S.M. Correlating interfacial water dynamics with protein-protein interaction in complex of GDF-5 and BMPRI receptors. Biophys. Chem. 2018, 240, 50–62. [Google Scholar] [CrossRef]
- Lin, S.; Svoboda, K.K.; Feng, J.Q.; Jiang, X. The biological function of type I receptors of bone morphogenetic protein in bone. Bone Res. 2016, 4, 16005. [Google Scholar] [CrossRef]
- Lin, P.I.; Tai, Y.T.; Chan, W.P.; Lin, Y.L.; Liao, M.H.; Chen, R.M. Estrogen/ERalpha signaling axis participates in osteoblast maturation via upregulating chromosomal and mitochondrial complex gene expressions. Oncotarget 2018, 9, 1169–1186. [Google Scholar] [CrossRef] [PubMed]
- McIlwain, D.R.; Pan, Q.; Reilly, P.T.; Elia, A.J.; McCracken, S.; Wakeham, A.C.; Itie-Youten, A.; Blencowe, B.J.; Mak, T.W. Smg1 is required for embryogenesis and regulates diverse genes via alternative splicing coupled to nonsense-mediated mRNA decay. Proc. Natl. Acad. Sci. USA 2010, 107, 12186–12191. [Google Scholar] [CrossRef] [PubMed]
- Ali, T.E.; Burnside, E.B.; Schaeffer, L.R. Relationship between external body measurements and calving difficulties in Canadian Holstein-Friesian cattle. J. Dairy Sci. 1984, 67, 3034–3044. [Google Scholar] [CrossRef]
- Michalska, E.; Koppolu, A.; Dobrzanska, A.; Płoski, R.; Gruszfeld, D. A case of severe trichothiodystrophy 3 in a neonate due to mutation in the GTF2H5 gene: Clinical report. Eur. J. Med. Genet. 2019, 62, 103557. [Google Scholar] [CrossRef]
- Griffiths, M.; Beaumont, N.; Yao, S.Y.; Sundaram, M.; Boumah, C.E.; Davies, A.; Kwong, F.Y.; Coe, I.; Cass, C.E.; Young, J.D.; et al. Cloning of a human nucleoside transporter implicated in the cellular uptake of adenosine and chemotherapeutic drugs. Nat. Med. 1997, 3, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, E.; Parrott, J.; Lee, G.T.; Domalakes, E.; Zhou, H.; He, L.; Mason, C.W. Regulation of human placental drug transporters in HCV infection and their influence on direct acting antiviral medications. Placenta 2018, 69, 32–39. [Google Scholar] [CrossRef]
- Liu, X.; Pan, G. Drug Transporters in Drug Disposition, Effects and Toxicity; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- Zhang, W.; Zhou, M.; Lu, W.; Gong, J.; Gao, F.; Li, Y.; Xu, X.; Lin, Y.; Zhang, X.; Ding, L.; et al. CNTNAP4 deficiency in dopaminergic neurons initiates parkinsonian phenotypes. Theranostics 2020, 10, 3000–3021. [Google Scholar] [CrossRef]
- Yin, F.T.; Futagawa, T.; Li, D.; Ma, Y.X.; Lu, M.H.; Lu, L.; Li, S.; Chen, Y.; Cao, Y.J.; Yang, Z.Z.; et al. Caspr4 interaction with LNX2 modulates the proliferation and neuronal differentiation of mouse neural progenitor cells. Stem Cells Dev. 2015, 24, 640–652. [Google Scholar] [CrossRef]
- Forutan, M.; Engle, B.; Goddard, M.; Hayes, B. A Conditional Multi-Trait Sequence GWAS of Heifer Fertility in Tropically Adapted Beef Cattle. Available online: https://www.wageningenacademic.com/pb-assets/wagen/WCGALP2022/54_005.pdf (accessed on 7 December 2022).
- Kaiser, F.; Hartweg, J.; Jansky, S.; Pelusi, N.; Kubaczka, C.; Sharma, N.; Nitsche, D.; Langkabel, J.; Schorle, H. Persistent Human KIT Receptor Signaling Disposes Murine Placenta to Premature Differentiation Resulting in Severely Disrupted Placental Structure and Functionality. Int. J. Mol. Sci. 2020, 21, 5503. [Google Scholar] [CrossRef]
- Nardi, E.; Seravalli, V.; Serena, C.; Mecacci, F.; Massi, D.; Bertaccini, B.; Di Tommaso, M.; Castiglione, F. A study on the placenta in stillbirth: An evaluation of molecular alterations through next generation sequencing. Placenta 2022, 129, 7–11. [Google Scholar] [CrossRef]
- Itonori, S.; Shirai, T.; Kiso, Y.; Ohashi, Y.; Shiota, K.; Ogawa, T. Glycosphingolipid composition of rat placenta: Changes associated with stage of pregnancy. Biochem. J. 1995, 307 Pt 2, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Tzetis, M.; Konstantinidou, A.; Sofocleous, C.; Kosma, K.; Mitrakos, A.; Tzannatos, C.; Kitsiou-Tzeli, S. Compound heterozygosity of a paternal submicroscopic deletion and a maternal missense mutation in POR gene: Antley-bixler syndrome phenotype in three sibling fetuses. Birth Defects Res. A Clin. Mol. Teratol. 2016, 106, 536–541. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, G.S.; Miller, K.A.; Galileo, D.S.; Martin-DeLeon, P.A. Murine SPAM1 is secreted by the estrous uterus and oviduct in a form that can bind to sperm during capacitation: Acquisition enhances hyaluronic acid-binding ability and cumulus dispersal efficiency. Reproduction 2008, 135, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Martin-DeLeon, P.A. Epididymal SPAM1 and its impact on sperm function. Mol. Cell. Endocrinol. 2006, 250, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Yamada, S.; Sugahara, K.; Ozbek, S. Evolution of glycosaminoglycans: Comparative biochemical study. Commun. Integr. Biol. 2011, 4, 150–158. [Google Scholar] [CrossRef]
- Sugahara, K.; Kitagawa, H. Recent advances in the study of the biosynthesis and functions of sulfated glycosaminoglycans. Curr. Opin. Struct. Biol. 2000, 10, 518–527. [Google Scholar] [CrossRef]
- Schorle, H.; Meier, P.; Buchert, M.; Jaenisch, R.; Mitchell, P.J. Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature 1996, 381, 235–238. [Google Scholar] [CrossRef]
- Zhang, J.; Hagopian-Donaldson, S.; Serbedzija, G.; Elsemore, J.; Plehn-Dujowich, D.; McMahon, A.P.; Flavell, R.A.; Williams, T. Neural tube, skeletal and body wall defects in mice lacking transcription factor AP-2. Nature 1996, 381, 238–241. [Google Scholar] [CrossRef]
- Brewer, S.; Jiang, X.; Donaldson, S.; Williams, T.; Sucov, H.M. Requirement for AP-2alpha in cardiac outflow tract morphogenesis. Mech. Dev. 2002, 110, 139–149. [Google Scholar] [CrossRef]
- Moser, M.; Pscherer, A.; Roth, C.; Becker, J.; Mucher, G.; Zerres, K.; Dixkens, C.; Weis, J.; Guay-Woodford, L.; Buettner, R.; et al. Enhanced apoptotic cell death of renal epithelial cells in mice lacking transcription factor AP-2beta. Genes Dev. 1997, 11, 1938–1948. [Google Scholar] [CrossRef]
- Moser, M.; Dahmen, S.; Kluge, R.; Grone, H.; Dahmen, J.; Kunz, D.; Schorle, H.; Buettner, R. Terminal renal failure in mice lacking transcription factor AP-2β. Lab. Investig. 2003, 83, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Bosserhoff, A.K.; Buettner, R.; Moser, M. A heart-hand syndrome gene: Tfap2b plays a critical role in the development and remodeling of mouse ductus arteriosus and limb patterning. PLoS ONE 2011, 6, e22908. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ye, J.; Han, X.; Qiao, R.; Li, X.; Lv, G.; Wang, K. Whole-genome sequencing identifies potential candidate genes for reproductive traits in pigs. Genomics 2020, 112, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Ma, J.; Lin, J. Depletion of NEDD9, a target gene of TGF-β, inhibits the proliferation and invasion of ectopic endometriotic stromal cells. Int. J. Clin. Exp. Pathol. 2017, 10, 2619–2627. [Google Scholar]
- Oliviero, C.; Heinonen, M.; Valros, A.; Peltoniemi, O. Environmental and sow-related factors affecting the duration of farrowing. Anim. Reprod. Sci. 2010, 119, 85–91. [Google Scholar] [CrossRef]
- Soede, N.M.; Langendijk, P.; Kemp, B. Reproductive cycles in pigs. Anim. Reprod. Sci. 2011, 124, 251–258. [Google Scholar] [CrossRef]
- Fletcher, L.; Akhtar, N.; Zhan, X.; Jafarikia, M.; Sullivan, B.P.; Huber, L.A.; Li, J. Identification of Candidate Salivary, Urinary and Serum Metabolic Biomarkers for High Litter Size Potential in Sows (Sus scrofa). Metabolites 2022, 12, 1045. [Google Scholar] [CrossRef]
- Wen, W.; He, Z.; Gao, F.; Liu, J.; Jin, H.; Zhai, S.; Qu, Y.; Xia, X. A High-Density Consensus Map of Common Wheat Integrating Four Mapping Populations Scanned by the 90K SNP Array. Front. Plant Sci. 2017, 8, 1389. [Google Scholar] [CrossRef]
- Li, J.; Wang, Z.; Lubritz, D.; Arango, J.; Fulton, J.; Settar, P.; Rowland, K.; Cheng, H.; Wolc, A. Genome-wide association studies for egg quality traits in White Leghorn layers using low-pass sequencing and SNP chip data. J. Anim. Breed. Genet. 2022, 139, 380–397. [Google Scholar] [CrossRef]

| Population | Trait a | N | Mean | SD | Minimum | Maximum | h2 | CV (%) |
|---|---|---|---|---|---|---|---|---|
| Shaziling | BL (cm) | 190 | 133.2 | 10.6 | 110 | 158 | 0.15 | 7.96% |
| CC (cm) | 190 | 132.9 | 12.0 | 107 | 162 | 0.14 | 9.03% | |
| TNB | 190 | 9.88 | 2.90 | 2 | 20 | <0.01 | - | |
| NHB | 190 | 8.81 | 2.81 | 2 | 19 | <0.01 | - | |
| NSB | 190 | 0.14 | 0.48 | 0 | 2 | <0.01 | - | |
| MUMM | 190 | 0.93 | 1.60 | 0 | 8 | 0.02 | - |
| Trait | SSC 1 | SNP | Position | p-Value | MAF 2 | β 3 | Distance (bp) | Candidate Genes |
|---|---|---|---|---|---|---|---|---|
| BL | 7 | ALGA0040227 | 34835986 | 9.01 × 10−8 | 0.41 (A/G) | −6.49 | 301630 | GLP1R |
| BL | 7 | ALGA0039856 | 30719424 | 2.36 × 10−6 | 0.15 (A/G) | −7.27 | within | SNRPC |
| BL | 7 | ASGA0032589 | 36378825 | 2.76 × 10−6 | 0.22 (A/G) | −6.24 | 7890 | NFYA |
| BL | 7 | ALGA0040238 | 34856640 | 1.42 × 10−5 | 0.33 (G/A) | −5.48 | 322284 | GLP1R |
| BL | 7 | INRA0024788 | 34978383 | 2.38 × 10−5 | 0.35 (A/G) | −5.31 | 444027 | GLP1R |
| CC | 1 | H3GA0003059 | 168650104 | 2.56 × 10−9 | 0.02 (A/G) | −4.69 | 414030 | NANOG |
| CC | 1 | ALGA0004562 | 90654890 | 5.95 × 10−7 | 0.06 (G/A) | −2.54 | 41856 | COX7A2 |
| CC | 11 | ASGA0050356 | 24088736 | 7.95 × 10−7 | 0.02 (A/G) | −3.41 | 224869 | FAM216B |
| CC | 12 | ALGA0065112 | 13563645 | 1.05 × 10−6 | 0.01 (A/C) | −4.85 | 89321 | CACNG1 |
| CC | 13 | ASGA0057447 | 52893776 | 1.24 × 10−6 | 0.01 (T/A) | −5.19 | 16884 | FOXP1 |
| CC | 8 | ALGA0111294 | 124178084 | 1.47 × 10−6 | 0.04 (G/A) | −2.56 | 360686 | BMPR1B |
| CC | 12 | MARC0074335 | 56183179 | 6.87 × 10−6 | 0.02 (A/G) | −3.11 | within | DNAH9 |
| CC | 1 | MARC0090402 | 48528854 | 8.86 × 10−6 | 0.05 (A/G) | −2.66 | 175464 | ENSSSCG00000060802 |
| CC | 16 | ASGA0073620 | 60469163 | 1.08 × 10−5 | 0.01 (G/A) | −3.85 | 112426 | CCNG1 |
| CC | 4 | ALGA0024545 | 37210968 | 1.78 × 10−5 | 0.05 (G/A) | −2.58 | 121604 | COX6C |
| CC | 1 | DRGA0000770 | 51978066 | 1.90 × 10−5 | 0.03 (G/A) | −2.34 | within | RIMS1 |
| CC | 12 | WU_10.2_12_884330 | 884330 | 1.91 × 10−5 | 0.04 (G/A) | −2.22 | within | CCDC57 |
| CC | 12 | WU_10.2_12_890035 | 890035 | 1.91 × 10−5 | 0.04 (A/G) | −2.22 | within | CCDC57 |
| TNB | 3 | CASI0010189 | 26899862 | 1.60 × 10−5 | 0.03 (A/C) | 4.26 | 129540 | SMG1 |
| NSB | 8 | ASGA0038253 | 27689768 | 2.75 × 10−7 | 0.02 (G/A) | 0.88 | 81655 | ARAP2 |
| NSB | 1 | ALGA0108557 | 8855372 | 5.59 × 10−7 | 0.02 (A/C) | 1.08 | 61868 | GTF2H5 |
| NSB | 7 | ALGA0040524 | 39239703 | 5.87 × 10−7 | 0.01 (G/A) | 1.25 | 5018 | SLC29A1 |
| NSB | 4 | WU_10.2_4_11278211 | 11278211 | 3.18 × 10−6 | 0.01 (A/G) | 0.90 | 418666 | FAM49B |
| NSB | 6 | CASI0009401 | 10987986 | 4.47 × 10−6 | 0.03 (A/G) | 0.68 | 329863 | CNTNAP4 |
| NSB | 8 | H3GA0024817 | 41595542 | 1.01 × 10−5 | 0.07 (A/G) | 0.45 | 103236 | KIT |
| NSB | 3 | DIAS0000597 | 10273754 | 1.16 × 10−5 | 0.02 (G/A) | 0.88 | 2896 | POR |
| NSB | 18 | WU_10.2_18_23986398 | 23986398 | 1.46 × 10−5 | 0.08 (A/G) | 0.39 | 396451 | SPAM1 |
| NSB | 7 | DBMA0000241 | 45173179 | 1.58 × 10−5 | 0.09 (G/A) | 0.44 | 402885 | TFAP2B |
| NSB | 17 | H3GA0048552 | 36746539 | 1.70 × 10−5 | 0.15 (A/G) | 0.31 | within | BPIFA3 |
| NSB | 4 | WU_10.2_4_11747027 | 11747027 | 1.81 × 10−5 | 0.01 (C/A) | 0.96 | 45942 | ENSSSCG00000060440 |
| NSB | 7 | WU_10.2_7_7925995 | 7925995 | 2.07 × 10−5 | 0.12 (A/C) | 0.34 | 11934 | NEDD9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lan, Q.; Deng, Q.; Qi, S.; Zhang, Y.; Li, Z.; Yin, S.; Li, Y.; Tan, H.; Wu, M.; Yin, Y.; et al. Genome-Wide Association Analysis Identified Variants Associated with Body Measurement and Reproduction Traits in Shaziling Pigs. Genes 2023, 14, 522. https://doi.org/10.3390/genes14020522
Lan Q, Deng Q, Qi S, Zhang Y, Li Z, Yin S, Li Y, Tan H, Wu M, Yin Y, et al. Genome-Wide Association Analysis Identified Variants Associated with Body Measurement and Reproduction Traits in Shaziling Pigs. Genes. 2023; 14(2):522. https://doi.org/10.3390/genes14020522
Chicago/Turabian StyleLan, Qun, Qiuchun Deng, Shijin Qi, Yuebo Zhang, Zhi Li, Shishu Yin, Yulian Li, Hong Tan, Maisheng Wu, Yulong Yin, and et al. 2023. "Genome-Wide Association Analysis Identified Variants Associated with Body Measurement and Reproduction Traits in Shaziling Pigs" Genes 14, no. 2: 522. https://doi.org/10.3390/genes14020522
APA StyleLan, Q., Deng, Q., Qi, S., Zhang, Y., Li, Z., Yin, S., Li, Y., Tan, H., Wu, M., Yin, Y., He, J., & Liu, M. (2023). Genome-Wide Association Analysis Identified Variants Associated with Body Measurement and Reproduction Traits in Shaziling Pigs. Genes, 14(2), 522. https://doi.org/10.3390/genes14020522





