Koala Genome Survey: An Open Data Resource to Improve Conservation Planning
Abstract
:1. Introduction
2. Systematic Review
3. Genomics and Conservation Planning
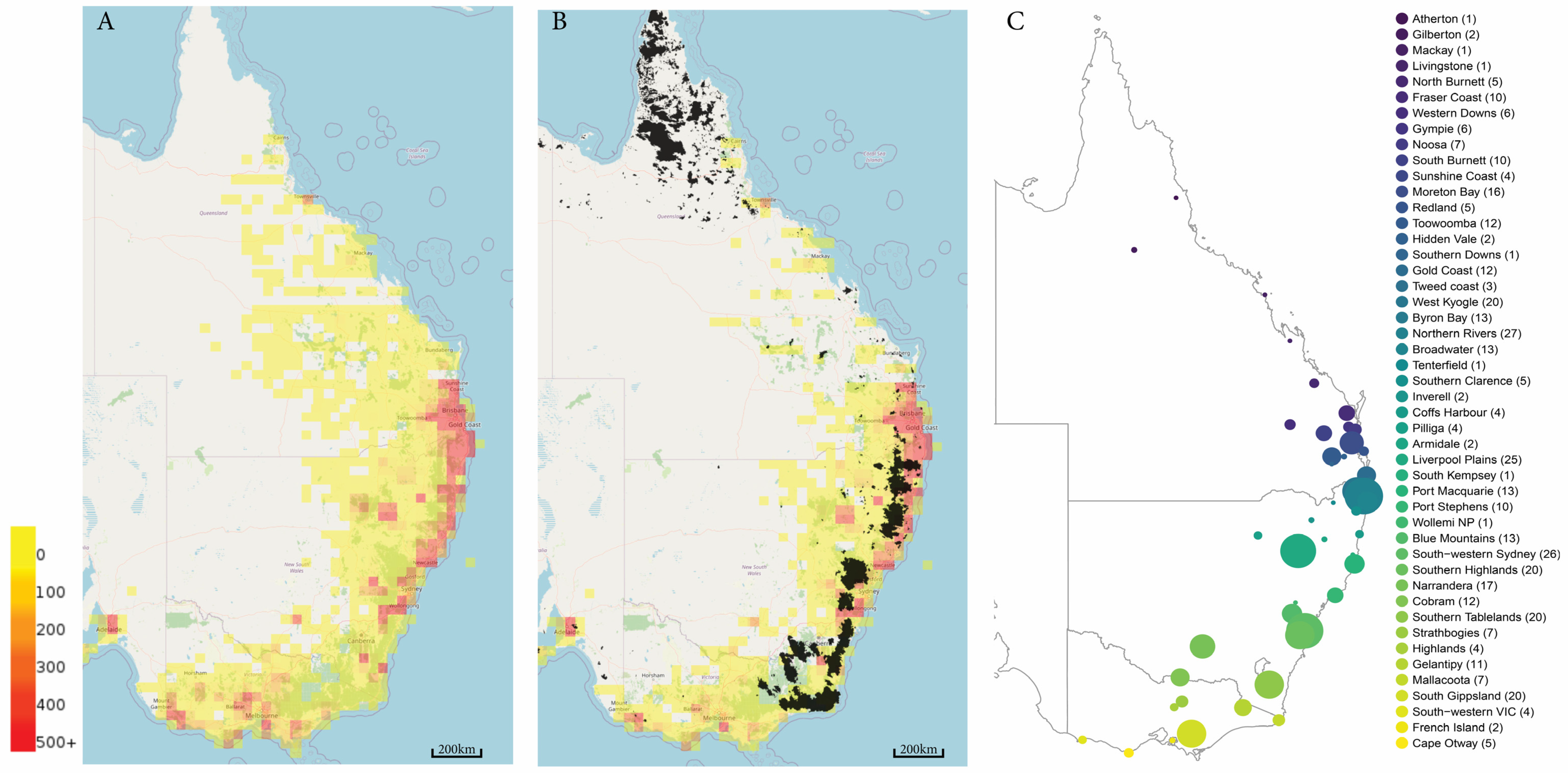
4. Koala Genome Survey
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lewin, H.A.; Robinson, G.E.; Kress, W.J.; Baker, W.J.; Coddington, J.; Crandall, K.A.; Durbin, R.; Edwards, S.V.; Forest, F.; Gilbert, M.T.P. Earth BioGenome Project: Sequencing life for the future of life. Proc. Natl. Acad. Sci. USA 2018, 115, 4325–4333. [Google Scholar] [CrossRef] [Green Version]
- Lewin, H.A.; Richards, S.; Lieberman Aiden, E.; Allende, M.L.; Archibald, J.M.; Bálint, M.; Barker, K.B.; Baumgartner, B.; Belov, K.; Bertorelle, G. The earth BioGenome project 2020: Starting the clock. Proc. Natl. Acad. Sci. USA 2022, 119, e2115635118. [Google Scholar] [CrossRef]
- Supple, M.A.; Shapiro, B. Conservation of biodiversity in the genomics era. Genome Biol. 2018, 19, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Hohenlohe, P.A.; Funk, W.C.; Rajora, O.P. Population genomics for wildlife conservation and management. Mol. Ecol. 2020, 30, 62–82. [Google Scholar] [CrossRef]
- Duntsch, L.; Whibley, A.; Brekke, P.; Ewen, J.G.; Santure, A.W. Genomic data of different resolutions reveal consistent inbreeding estimates but contrasting homozygosity landscapes for the threatened Aotearoa New Zealand hihi. Mol. Ecol. 2021, 30, 6006–6020. [Google Scholar] [CrossRef] [PubMed]
- Dussex, N.; Van Der Valk, T.; Morales, H.E.; Wheat, C.W.; Díez-del-Molino, D.; Von Seth, J.; Foster, Y.; Kutschera, V.E.; Guschanski, K.; Rhie, A. Population genomics of the critically endangered kākāpō. Cell Genom. 2021, 1, 100002. [Google Scholar] [CrossRef]
- Batley, K.C.; Sandoval-Castillo, J.; Kemper, C.M.; Zanardo, N.; Tomo, I.; Beheregaray, L.B.; Möller, L.M. Whole genomes reveal multiple candidate genes and pathways involved in the immune response of dolphins to a highly infectious virus. Mol. Ecol. 2021, 30, 6434–6448. [Google Scholar] [CrossRef]
- Wintle, B.A.; Legge, S.; Woinarski, J.C. After the megafires: What next for Australian wildlife? Trends Ecol. Evol. 2020, 35, 753–757. [Google Scholar] [CrossRef]
- Dickman, C.; McDonald, T. Some personal reflections on the present and future of Australia’s fauna in an increasingly fire-prone continent. Ecol. Manag. Restor. 2020, 21, 86–96. [Google Scholar] [CrossRef]
- Dong, E.; Du, H.; Gardner, L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020, 20, 533–534. [Google Scholar] [CrossRef]
- Wu, X.B.; Wang, Y.Q.; Zhou, K.Y.; Zhu, W.Q.; Nie, J.S.; Wang, C.L.; Xie, W.S. Genetic variation in captive population of chinese alligator, Alligator sinensis, revealed by random amplified polymorphic DNA (RAPD). Biol. Conserv. 2002, 106, 435–441. [Google Scholar] [CrossRef]
- Hogg, C.J.; Wright, B.; Morris, K.M.; Lee, A.V.; Ivy, J.A.; Grueber, C.E.; Belov, K. Founder relationships and conservation management: Empirical kinships reveal the effect on breeding programs when founders are assumed to be unrelated. Anim. Conserv. 2018, 22, 348–361. [Google Scholar] [CrossRef] [Green Version]
- Ogden, R.; Chuven, J.; Gilbert, T.; Hosking, C.; Gharbi, K.; Craig, M.; Al Dhaheri, S.S.; Senn, H. Benefits and pitfalls of captive conservation genetic management: Evaluating diversity in scimitar-horned oryx to support reintroduction planning. Biol. Conserv. 2020, 241, 108244. [Google Scholar] [CrossRef]
- Robert, A.; Couvet, D.; Sarrazin, F. Integration of demography and genetics in population restorations. Ecoscience 2007, 14, 463–471. [Google Scholar] [CrossRef]
- Pacioni, C.; Wayne, A.F.; Spencer, P.B.S. Genetic outcomes from the translocations of the critically endangered woylie. Curr. Zool. 2013, 59, 294–310. [Google Scholar] [CrossRef] [Green Version]
- Hogg, C.; McLennan, E.; Wise, P.; Lee, A.; Pemberton, D.; Fox, S.; Belov, K.; Grueber, C. Preserving the demographic and genetic integrity of a single source population during multiple translocations. Biol. Conserv. 2020, 241, 108318. [Google Scholar] [CrossRef]
- Farquharson, K.A.; McLennan, E.A.; Wayne, A.; Smith, M.; Peel, E.; Belov, K.; Hogg, C.J. Metapopulation management of a critically endangered marsupial in the age of genomics. Glob. Ecol. Conserv. 2021, 31, e01869. [Google Scholar] [CrossRef]
- Wallis, G.P. Thirty years of conservation genetics in New Zealand: What have we learnt? J. R. Soc. N. Z. 2019, 49, 320–346. [Google Scholar] [CrossRef]
- Pacioni, C.; Trocini, S.; Wayne, A.F.; Rafferty, C.; Page, M. Integrating population genetics in an adaptive management framework to inform management strategies. Biodivers. Conserv. 2020, 29, 947–966. [Google Scholar] [CrossRef]
- Kjeldsen, S.R.; Zenger, K.R.; Leigh, K.; Ellis, W.; Tobey, J.; Phalen, D.; Melzer, A.; FitzGibbon, S.; Raadsma, H.W. Genome-wide SNP loci reveal novel insights into koala (Phascolarctos cinereus) population variability across its range. Conserv. Genet. 2016, 17, 337–353. [Google Scholar] [CrossRef]
- Farquharson, K.A.; McLennan, E.A.; Cheng, Y.; Alexander, L.; Fox, S.; Lee, A.V.; Belov, K.; Hogg, C.J. Restoring faith in conservation action: Maintaining wild genetic diversity through the Tasmanian devil insurance program. Iscience 2022, 25, 104474. [Google Scholar] [CrossRef] [PubMed]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature 2004, 431, 931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steiner, C.C.; Putnam, A.S.; Hoeck, P.E.; Ryder, O.A. Conservation genomics of threatened animal species. Annu. Rev. Anim. Biosci. 2013, 1, 261–281. [Google Scholar] [CrossRef] [Green Version]
- Sims, D.; Sudbery, I.; Ilott, N.E.; Heger, A.; Ponting, C.P. Sequencing depth and coverage: Key considerations in genomic analyses. Nat. Rev. Genet. 2014, 15, 121–132. [Google Scholar] [CrossRef]
- Sansaloni, C.; Petroli, C.; Jaccoud, D.; Carling, J.; Detering, F.; Grattapaglia, D.; Kilian, A. Diversity Arrays Technology (DArT) and next-generation sequencing combined: Genome-wide, high throughput, highly informative genotyping for molecular breeding of Eucalyptus. BMC Proc. 2011, 5, P54. [Google Scholar] [CrossRef] [Green Version]
- Peterson, B.K.; Weber, J.N.; Kay, E.H.; Fisher, H.S.; Hoekstra, H.E. Double digest RADseq: An inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 2012, 7, e37135. [Google Scholar] [CrossRef] [Green Version]
- Teixeira, J.C.; Huber, C.D. The inflated significance of neutral genetic diversity in conservation genetics. Proc. Natl. Acad. Sci. USA 2021, 118, e2015096118. [Google Scholar] [CrossRef]
- Gupta, P.; Balyan, H.; Sharma, P.; Ramesh, B. Microsatellites in plants: A new class of molecular markers. Curr. Sci. 1996, 70, 45–54. [Google Scholar]
- McLennan, E.A.; Wright, B.R.; Belov, K.; Hogg, C.J.; Grueber, C.E. Too much of a good thing? Finding the most informative genetic data set to answer conservation questions. Mol. Ecol. Resour. 2019, 19, 659–671. [Google Scholar] [CrossRef]
- Gao, R.; Liu, Y.; Gjesing, A.P.; Hollensted, M.; Wan, X.; He, S.; Pedersen, O.; Yi, X.; Wang, J.; Hansen, T. Evaluation of a target region capture sequencing platform using monogenic diabetes as a study-model. BMC Genet. 2014, 15, 13. [Google Scholar] [CrossRef] [Green Version]
- Reed, D.H.; Frankham, R. How closely correlated are molecular and quantitative measures of genetic variation? A meta-analysis. Evolution 2001, 55, 1095–1103. [Google Scholar]
- Marsh, K.J.; Moore, B.D.; Wallis, I.R.; Foley, W.J. Continuous monitoring of feeding by koalas highlights diurnal differences in tree preferences. Wildl. Res. 2014, 40, 639–646. [Google Scholar] [CrossRef]
- Reckless, H.J.; Murray, M.; Crowther, M.S. A review of climatic change as a determinant of the viability of koala populations. Wildl. Res. 2017, 44, 458–470. [Google Scholar] [CrossRef]
- Seabrook, L.; McAlpine, C.; Baxter, G.; Rhodes, J.; Bradley, A.; Lunney, D. Drought-driven change in wildlife distribution and numbers: A case study of koalas in south west Queensland. Wildl. Res. 2011, 38, 509–524. [Google Scholar] [CrossRef]
- Hindell, M.A.; Handasyde, K.A.; Lee, A.K. Tree species selection by free-ranging koala populations in Victoria. Wildl. Res. 1985, 12, 137–144. [Google Scholar] [CrossRef]
- Moore, B.D.; Foley, W.J.; Wallis, I.R.; Cowling, A.; Handasyde, K.A. Eucalyptus foliar chemistry explains selective feeding by koalas. Biol. Lett. 2005, 1, 64–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callaghan, J.; McAlpine, C.; Mitchell, D.; Thompson, J.; Bowen, M.; Rhodes, J.; de Jong, C.; Domalewski, R.; Scott, A. Ranking and mapping koala habitat quality for conservation planning on the basis of indirect evidence of tree-species use: A case study of Noosa Shire, south-eastern Queensland. Wildl. Res. 2011, 38, 89–102. [Google Scholar] [CrossRef]
- ANZECC. National Koala Conservation Strategy. Available online: https://www.environment.gov.au/system/files/resources/ec7db20d-0b2c-4b76-bc6d-5f61ec557561/files/koala-strategy-1998.pdf (accessed on 15 December 2020).
- Knott, T.; Lunney, D.; Coburn, D.; Callaghan, J. An ecological history of koala habitat in Port Stephens Shire and the Lower Hunter on the central coast of New South Wales, 1801–1998. Pac. Conserv. Biol. 1998, 4, 354–368. [Google Scholar] [CrossRef]
- Lunney, D.; Matthews, A.; Moon, C.; Ferrier, S. Incorporating habitat mapping into practical koala conservation on private lands. Conserv. Biol. 2000, 14, 669–680. [Google Scholar] [CrossRef]
- Commonwealth of Australia, List of Threatened Species Amendment (Phascolarctos cinereus (combined populations of Queensland, New South Wales and the Australian Capital Territory) (280)) Instrument 2022. F2022L00131. C. 2022; Federal Register of Legislation. Available online: https://www.legislation.gov.au/Details/F2022L00131 (accessed on 1 November 2022).
- Johnson, R.N.; O’Meally, D.; Chen, Z.; Etherington, G.J.; Ho, S.Y.W.; Nash, W.J.; Grueber, C.E.; Cheng, Y.; Whittington, C.M.; Dennison, S.; et al. Adaptation and conservation insights from the koala genome. Nat. Genet. 2018, 50, 1102–1111. [Google Scholar] [CrossRef]
- Adams-Hosking, C.; McBride, M.F.; Baxter, G.; Burgman, M.; De Villiers, D.; Kavanagh, R.; Lawler, I.; Lunney, D.; Melzer, A.; Menkhorst, P. Use of expert knowledge to elicit population trends for the koala (Phascolarctos cinereus). Divers. Distrib. 2016, 22, 249–262. [Google Scholar] [CrossRef] [Green Version]
- McAlpine, C.; Lunney, D.; Melzer, A.; Menkhorst, P.; Phillips, S.; Phalen, D.; Ellis, W.; Foley, W.; Baxter, G.; De Villiers, D. Conserving koalas: A review of the contrasting regional trends, outlooks and policy challenges. Biol. Conserv. 2015, 192, 226–236. [Google Scholar] [CrossRef]
- Cresswell, I.; Murphy, H. Australia state of the environment 2016: Biodiversity, independent report to the Australian Government Minister for the Environment and Energy. In Australian Government Department of the Environment and Energy: Canberra, ACT; Department of the Environment and Energy: Canberra, Australia, 2017. [Google Scholar]
- WWF-Australia. Briefing Koalas Face Extinction in Eastern Australia, a Deforestation Hotspot, November 2019; WWF-Australia: Sydney, Australia, 2019; Available online: File:///C:/Users/chog4569/Downloads/Briefing%20-%20koala%20extinction%20risk%20in%20Eastern%20Australia%20WWF-Aus%20Nov%202019.pdf (accessed on 15 July 2021).
- Lott, M.J.; Wright, B.R.; Neaves, L.E.; Frankham, G.J.; Dennison, S.; Eldridge, M.D.; Potter, S.; Alquezar-Planas, D.E.; Hogg, C.J.; Belov, K. Future-proofing the koala: Synergising genomic and environmental data for effective species management. Mol. Ecol. 2022, 31, 3035–3055. [Google Scholar] [CrossRef]
- Martin, W. Anatomy of Koala, Phascolarctos fuscus. Proc. Zool. Soc. Lond. 1836, 4, 109–113. [Google Scholar]
- Forbes, W.A. On some points in the anatomy of the Koala Phascolarctos cinereus. Proc. Zool. Soc. Lond. 1881, 49, 180–195. [Google Scholar] [CrossRef]
- Houlden, B.A.; England, P.; Sherwin, W.B. Paternity exclusion in koalas using hypervariable microsatellites. J. Hered. 1996, 87, 149–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houlden, B.A.; England, P.R.; Taylor, A.C.; Greville, W.D.; Sherwin, W.B. Low genetic variability of the koala Phascolarctos cinereus in south-eastern Australia following a severe population bottleneck. Mol. Ecol. 1996, 5, 269–281. [Google Scholar] [CrossRef]
- Taylor, A.C.; Graves, J.M.; Murray, N.D.; Obrien, S.J.; Yuhki, N.; Sherwin, B. Conservation genetics of the koala (Phascolarctos cinereus), low mitochondrial DNA variation amongst southern Australian populations. Genet. Res. 1997, 69, 25–33. [Google Scholar] [CrossRef] [Green Version]
- Fowler, E.V.; Hoeben, P.; Timms, P. Randomly amplified polymorphic DNA variation in populations of eastern Australian koalas, Phascolarctos cinereus. Biochem. Genet. 1998, 36, 381–393. [Google Scholar] [CrossRef] [PubMed]
- Sherwin, W.B.; Timms, P.; Wilcken, J.; Houlden, B. Analysis and conservation implications of koala genetics. Conserv. Biol. 2000, 14, 639–649. [Google Scholar] [CrossRef]
- Seymour, A.M.; Montgomery, M.E.; Costello, B.H.; Ihle, S.; Johnsson, G.; St John, B.; Taggart, D.; Houlden, B.A. High effective inbreeding coefficients correlate with morphological abnormalities in populations of South Australian koalas (Phascolarctos cinereus). Anim. Conserv. 2001, 4, 211–219. [Google Scholar] [CrossRef]
- Cristescu, R.; Cahill, V.; Sherwin, W.B.; Handasyde, K.; Carlyon, K.; Whisson, D.; Herbert, C.A.; Carlsson, B.L.J.; Wilton, A.N.; Cooper, D.W. Inbreeding and testicular abnormalities in a bottlenecked population of koalas (Phascolarctos cinereus). Wildl. Res. 2009, 36, 299–308. [Google Scholar] [CrossRef]
- Cristescu, R.; Sherwin, W.B.; Handasyde, K.; Cahill, V.; Cooper, D.W. Detecting bottlenecks using BOTTLENECK 1.2.02 in wild populations: The importance of the microsatellite structure. Conserv. Genet. 2010, 11, 1043–1049. [Google Scholar] [CrossRef]
- Lee, T.; Zenger, K.R.; Close, R.L.; Jones, M.; Phalen, D.N. Defining spatial genetic structure and management units for vulnerable koala (Phascolarctos cinereus) populations in the Sydney region, Australia. Wildl. Res. 2010, 37, 156–165. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.E.; Seddon, J.M.; Corley, S.W.; Ellis, W.A.H.; Johnston, S.D.; de Villiers, D.L.; Preece, H.J.; Carrick, F.N. Genetic variation and structuring in the threatened koala populations of Southeast Queensland. Conserv. Genet. 2010, 11, 2091–2103. [Google Scholar] [CrossRef]
- Lee, K.E.; Seddon, J.M.; Johnston, S.; FitzGibbon, S.I.; Carrick, F.; Melzer, A.; Bercovitch, F.; Ellis, W. Genetic diversity in natural and introduced island populations of koalas in Queensland. Aust. J. Zool. 2012, 60, 303–310. [Google Scholar] [CrossRef]
- Lee, T.; Zenger, K.R.; Close, R.L.; Phalen, D.N. Genetic analysis reveals a distinct and highly diverse koala (Phascolarctos cinereus) population in South Gippsland, Victoria, Australia. Aust. Mammal. 2012, 34, 68–74. [Google Scholar] [CrossRef]
- Dudaniec, R.Y.; Rhodes, J.R.; Wilmer, J.W.; Lyons, M.; Lee, K.E.; McAlpine, C.A.; Carrick, F.N. Using multilevel models to identify drivers of landscape-genetic structure among management areas. Mol. Ecol. 2013, 22, 3752–3765. [Google Scholar] [CrossRef]
- Lee, K.E.; Ellis, W.A.H.; Carrick, F.N.; Corley, S.W.; Johnston, S.D.; Baverstock, P.R.; Nock, C.J.; Rowe, K.C.; Seddon, J.M. Anthropogenic changes to the landscape resulted in colonization of koalas in north-east New South Wales, Australia. Austral Ecol. 2013, 38, 355–363. [Google Scholar] [CrossRef]
- Ruiz-Rodriguez, C.T.; Ishida, Y.; Greenwood, A.D.; Roca, A.L. Development of 14 microsatellite markers in the Queensland koala (Phascolarctos cinereus adustus) using next generation sequencing technology. Conserv. Genet. Resour. 2014, 6, 429–431. [Google Scholar] [CrossRef]
- Seddon, J.M.; Lee, K.E.; Johnston, S.D.; Nicolson, V.N.; Pyne, M.; Carrick, F.N.; Ellis, W.A.H. Testing the regional genetic representativeness of captive koala populations in South-East Queensland. Wildl. Res. 2014, 41, 277–286. [Google Scholar] [CrossRef]
- Dennison, S.; Frankham, G.J.; Neaves, L.E.; Flanagan, C.; FitzGibbon, S.; Eldridge, M.D.B.; Johnson, R.N. Population genetics of the koala (Phascolarctos cinereus) in north-eastern New South Wales and south-eastern Queensland. Aust. J. Zool. 2016, 64, 402–412. [Google Scholar] [CrossRef]
- Ruiz-Rodriguez, C.T.; Ishida, Y.; Murray, N.D.; O’Brien, S.J.; Graves, J.A.M.; Greenwood, A.D.; Roca, A.L. Koalas (Phascolarctos cinereus) From Queensland Are Genetically Distinct From 2 Populations in Victoria. J. Hered. 2016, 107, 573–580. [Google Scholar] [CrossRef] [Green Version]
- Wedrowicz, F.; Mosse, J.; Wright, W.; Hogan, F.E. Genetic structure and diversity of the koala population in South Gippsland, Victoria: A remnant population of high conservation significance. Conserv. Genet. 2018, 19, 713–728. [Google Scholar] [CrossRef]
- Kjeldsen, S.R.; Raadsma, H.W.; Leigh, K.A.; Tobey, J.R.; Phalen, D.; Krockenberger, A.; Ellis, W.A.; Hynes, E.; Higgins, D.P.; Zenger, K.R. Genomic comparisons reveal biogeographic and anthropogenic impacts in the koala (Phascolarctos cinereus): A dietary-specialist species distributed across heterogeneous environments. Heredity 2019, 122, 525–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norman, J.A.; Phillips, S.S.; Blackmore, C.J.; Goldingay, R.; Christidis, L. Integrating measures of long-distance dispersal into vertebrate conservation planning: Scaling relationships and parentage-based dispersal analysis in the koala. Conserv. Genet. 2019, 20, 1163–1174. [Google Scholar] [CrossRef]
- Schultz, A.J.; Cristescu, R.H.; Hanger, J.; Loader, J.; de Villiers, D.; Frere, C.H. Inbreeding and disease avoidance in a free-ranging koala population. Mol. Ecol. 2020, 29, 2416–2430. [Google Scholar] [CrossRef]
- Seddon, J.M.; Schultz, B. Koala conservation in Queensland, Australia: A role for assisted gene flow for genetic rescue? In Conservation Genetics in Mammals: Integrative Research Using Novel Approaches; Springer: Berlin/Heidelberg, Germany, 2020; pp. 331–349. [Google Scholar] [CrossRef]
- Wedrowicz, F.; Karsa, M.; Mosse, J.; Hogan, F.E. Reliable genotyping of the koala (Phascolarctos cinereus) using DNA isolated from a single faecal pellet. Mol. Ecol. Resour. 2013, 13, 634–641. [Google Scholar] [CrossRef]
- Wedrowicz, F.; Mosse, J.; Wright, W.; Hogan, F. Validating the use of non-invasively sourced DNA for population genetic studies using pedigree data. Web Ecol. 2017, 17, 9–18. [Google Scholar] [CrossRef] [Green Version]
- Wedrowicz, F.; Mosse, J.; Wright, W.; Hogan, F.E. Isolating DNA sourced non-invasively from koala scats: A comparison of four commercial DNA stool kits. Conserv. Genet. Resour. 2019, 11, 219–229. [Google Scholar] [CrossRef]
- Schultz, A.J.; Cristescu, R.H.; Littleford-Colquhoun, B.L.; Jaccoud, D.; Frere, C.H. Fresh is best: Accurate SNP genotyping from koala scats. Ecol. Evol. 2018, 8, 3139–3151. [Google Scholar] [CrossRef] [PubMed]
- Cocciolone, R.A.; Timms, P. DNA profiling of queensland koalas reveals sufficient variability for individual identification and parentage determination. Wildl. Res. 1992, 19, 279–287. [Google Scholar] [CrossRef]
- Timms, P.; Kato, J.; Maugeri, M.; White, N. DNA fingerprint analysis of a free-range koala population. Biochem. Genet. 1993, 31, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Lambert, K.; Jones, M.; Shimmin, G.; Sofronidis, G.; Bowden, D.; Taggart, D.; TempleSmith, P. Informative enzyme/probe combinations for the multilocus DNA fingerprinting of marsupials. Electrophoresis 1997, 18, 1688–1692. [Google Scholar] [CrossRef]
- Wilmer, J.M.W.; Melzer, A.; Carrick, F.; Moritz, C. Low genetic diversity and inbreeding depression in Queensland koalas. Wildl. Res. 1993, 20, 177–188. [Google Scholar] [CrossRef]
- Takami, K.; Yoshida, M.; Yamamoto, Y.; Harada, M.; Furuyama, J. Genetic variation of mitochondrial cytochrome b genes among the subspecies of koala, Phascolarctos cinereus. J. Vet. Med. Sci. 1998, 60, 1161–1163. [Google Scholar] [CrossRef] [Green Version]
- Houlden, B.A.; Costello, B.H.; Sharkey, D.; Fowler, E.V.; Melzer, A.; Ellis, W.; Carrick, F.; Baverstock, P.R.; Elphinstone, M.S. Phylogeographic differentiation in the mitochondrial control region in the koala, Phascolarctos cinereus (Goldfuss 1817). Mol. Ecol. 1999, 8, 999–1011. [Google Scholar] [CrossRef]
- Fowler, E.V.; Houlden, B.A.; Hoeben, P.; Timms, P. Genetic diversity and gene flow among southeastern Queensland koalas (Phascolarctos cinereus). Mol. Ecol. 2000, 9, 155–164. [Google Scholar] [CrossRef]
- Munemasa, M.; Nikaido, M.; Donnellan, S.; Austin, C.C.; Okada, N.; Hasegawa, M. Phylogenetic analysis of diprotodontian marsupials based on complete mitochondrial genomes. Genes Genet. Syst. 2006, 81, 181–191. [Google Scholar] [CrossRef] [Green Version]
- Tsangaras, K.; Avila-Arcos, M.C.; Ishida, Y.; Helgen, K.M.; Roca, A.L.; Greenwood, A.D. Historically low mitochondrial DNA diversity in koalas (Phascolarctos cinereus). Bmc Genet. 2012, 13, 92. [Google Scholar] [CrossRef] [Green Version]
- Neaves, L.E.; Frankham, G.J.; Dennison, S.; FitzGibbon, S.; Flannagan, C.; Gillett, A.; Hynes, E.; Handasyde, K.; Helgen, K.M.; Tsangaras, K.; et al. Phylogeography of the Koala, (Phascolarctos cinereus), and Harmonising Data to Inform Conservation. PLoS ONE 2016, 11, e0162207. [Google Scholar] [CrossRef] [Green Version]
- Osborne, M.J.; Christidis, L.; Norman, J.A. Molecular phylogenetics of the Diprotodontia (kangaroos, wombats, koala, possums, and allies). Mol. Phylogenetics Evol. 2002, 25, 219–228. [Google Scholar] [CrossRef]
- Phillips, M.J.; Pratt, R.C. Family-level relationships among the Australasian marsupial “herbivores” (Diprotodontia: Koala, wombats, kangaroos and possums). Mol. Phylogenetics Evol. 2008, 46, 594–605. [Google Scholar] [CrossRef] [PubMed]
- Brandies, P.; Peel, E.; Hogg, C.J.; Belov, K. The Value of Reference Genomes in the Conservation of Threatened Species. Genes 2019, 10, 846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silver, L.W.; Cheng, Y.; Quigley, B.L.; Robbins, A.; Timms, P.; Hogg, C.J.; Belov, K. A targeted approach to investigating immune genes of an iconic Australian marsupial. Mol. Ecol. 2022, 31, 3286–3303. [Google Scholar] [CrossRef]
- Sutherland, W.J.; Pullin, A.S.; Dolman, P.M.; Knight, T.M. The need for evidence-based conservation. Trends Ecol. Evol. 2004, 19, 305–308. [Google Scholar] [CrossRef]
- Frankham, R.; Ballou, J.D.; Ralls, K.; Eldridge, M.; Dudash, M.R.; Fenster, C.B.; Lacy, R.C.; Sunnucks, P. Genetic Management of Fragmented Animal and Plant Populations; Oxford University Press: Oxford, UK, 2017. [Google Scholar]
- Lunney, D.; Predavec, M.; Sonawane, I.; Kavanagh, R.; Barrott-Brown, G.; Phillips, S.; Callaghan, J.; Mitchell, D.; Parnaby, H.; Paull, D.C. The remaining koalas (Phascolarctos cinereus) of the Pilliga forests, north-west New South Wales: Refugial persistence or a population on the road to extinction? Pac. Conserv. Biol. 2017, 23, 277–294. [Google Scholar] [CrossRef]
- Quinn, N.L.; McGowan, C.R.; Cooper, G.A.; Koop, B.F.; Davidson, W.S. Identification of genes associated with heat tolerance in Arctic charr exposed to acute thermal stress. Physiol. Genom. 2011, 43, 685–696. [Google Scholar] [CrossRef] [Green Version]
- Tedeschi, J.N.; Kennington, W.; Tomkins, J.L.; Berry, O.; Whiting, S.; Meekan, M.G.; Mitchell, N.J. Heritable variation in heat shock gene expression: A potential mechanism for adaptation to thermal stress in embryos of sea turtles. Proc. R. Soc. B Biol. Sci. 2016, 283, 20152320. [Google Scholar] [CrossRef] [Green Version]
- Lau, Q.; Griffith, J.E.; Higgins, D.P. Identification of MHCII variants associated with chlamydial disease in the koala (Phascolarctos cinereus). PeerJ 2014, 2, e443. [Google Scholar] [CrossRef] [Green Version]
- Aljanabi, S.M.; Martinez, I. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 1997, 25, 4692–4693. [Google Scholar] [CrossRef] [PubMed]
- Foote, A.D.; Hooper, R.; Alexander, A.; Baird, R.W.; Baker, C.S.; Ballance, L.; Barlow, J.; Brownlow, A.; Collins, T.; Constantine, R. Runs of homozygosity in killer whale genomes provide a global record of demographic histories. Mol. Ecol. 2021, 30, 6162–6177. [Google Scholar] [CrossRef] [PubMed]
- Escoda, L.; Castresana, J. The genome of the Pyrenean desman and the effects of bottlenecks and inbreeding on the genomic landscape of an endangered species. Evol. Appl. 2021, 14, 1898–1913. [Google Scholar] [CrossRef] [PubMed]
| Publication Year | Reference | Location | Sample Collection | Data Type |
|---|---|---|---|---|
| 1996 | Houlden et al. [50] | NSW and SA | Unknown | 6 microsatellites |
| 1996 | Houlden et al. [51] | QLD, NSW, VIC, SA | Unknown | 6 microsatellites [50] |
| 1997 | Taylor et al. [52] | VIC | Unknown | 6 minisatellite probes |
| 1998 | Fowler et al. [53] | QLD, NSW, VIC, and SA | Unknown | 20 randomly amplified polymorphic DNA (RAPD) |
| 2000 | Sherwin et al. [54] | Review | ||
| 2001 | Seymour et al. [55] | NSW, VIC, and SA | Unknown | 6 microsatellites [50] |
| 2009 | Cristescu et al. [56] | VIC and SA | 2002–2006 | 15 microsatellites inc. [50] |
| 2010 | Cristescu et al. [57] | VIC and SA | 2002–2006 | 15 microsatellites inc. [50] |
| 2010 | Lee et al. [58] | NSW | 1998–2008 | 17 microsatellites inc. [50,56] |
| 2010 | Lee et al. [59] | QLD | Unknown | 6 microsatellites [50] |
| 2012 | Lee et al. [60] | QLD | Unknown | 6 microsatellites [50] |
| 2012 | Lee et al. [61] | VIC | 2008–2009 | 12 microsatellites inc. [50,56] |
| 2013 | Dudaniec et al. [62] | QLD | 2006–2009 | 6 microsatellites [50] |
| 2013 | Lee et al. [63] | NSW | Unknown | 6 microsatellites [50] and mitochondrial DNA |
| 2014 | Ruiz-Rodriguez et al. [64] | QLD | Unknown | 14 microsatellites (new ones developed) |
| 2014 | Seddon et al. [65] | QLD | Unknown | 6 microsatellites [50] |
| 2016 | Kjeldsen et al. [20] | QLD, NSW, VIC, and SA | Unknown | ddRAD (3060 SNPs after filtering) |
| 2016 | Dennison et al. [66] | NSW and QLD | Unknown | 14 microsatellites (new ones developed) |
| 2016 | Ruiz-Rodriguez et al. [67] | QLD and VIC | Unknown | 13 microsatellites inc. [64] and mitochondrial DNA |
| 2018 | Wedrowicz et al. [68] | QLD, NSW, VIC, and SA | 2013–2016 | 12 microsatellites inc. [50,56] |
| 2019 | Kjeldsen et al. [69] | QLD, NSW, VIC, and SA | Unknown | DArTseq (4606 SNPs after filtering) |
| 2019 | Norman et al. [70] | NSW | 2012–2015 | 17 microsatellites inc. [50,56,66] |
| 2020 | Schultz et al. [71] | QLD | 2013–2017 | DArTseq (427 SNPs filtering) |
| 2020 | Seddon and Schultz [72] | Review |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hogg, C.J.; Silver, L.; McLennan, E.A.; Belov, K. Koala Genome Survey: An Open Data Resource to Improve Conservation Planning. Genes 2023, 14, 546. https://doi.org/10.3390/genes14030546
Hogg CJ, Silver L, McLennan EA, Belov K. Koala Genome Survey: An Open Data Resource to Improve Conservation Planning. Genes. 2023; 14(3):546. https://doi.org/10.3390/genes14030546
Chicago/Turabian StyleHogg, Carolyn J., Luke Silver, Elspeth A. McLennan, and Katherine Belov. 2023. "Koala Genome Survey: An Open Data Resource to Improve Conservation Planning" Genes 14, no. 3: 546. https://doi.org/10.3390/genes14030546
APA StyleHogg, C. J., Silver, L., McLennan, E. A., & Belov, K. (2023). Koala Genome Survey: An Open Data Resource to Improve Conservation Planning. Genes, 14(3), 546. https://doi.org/10.3390/genes14030546






