The Developmental Origins of Cancer: A Review of the Genes Expressed in Embryonic Cells with Implications for Tumorigenesis
Abstract
1. Introduction
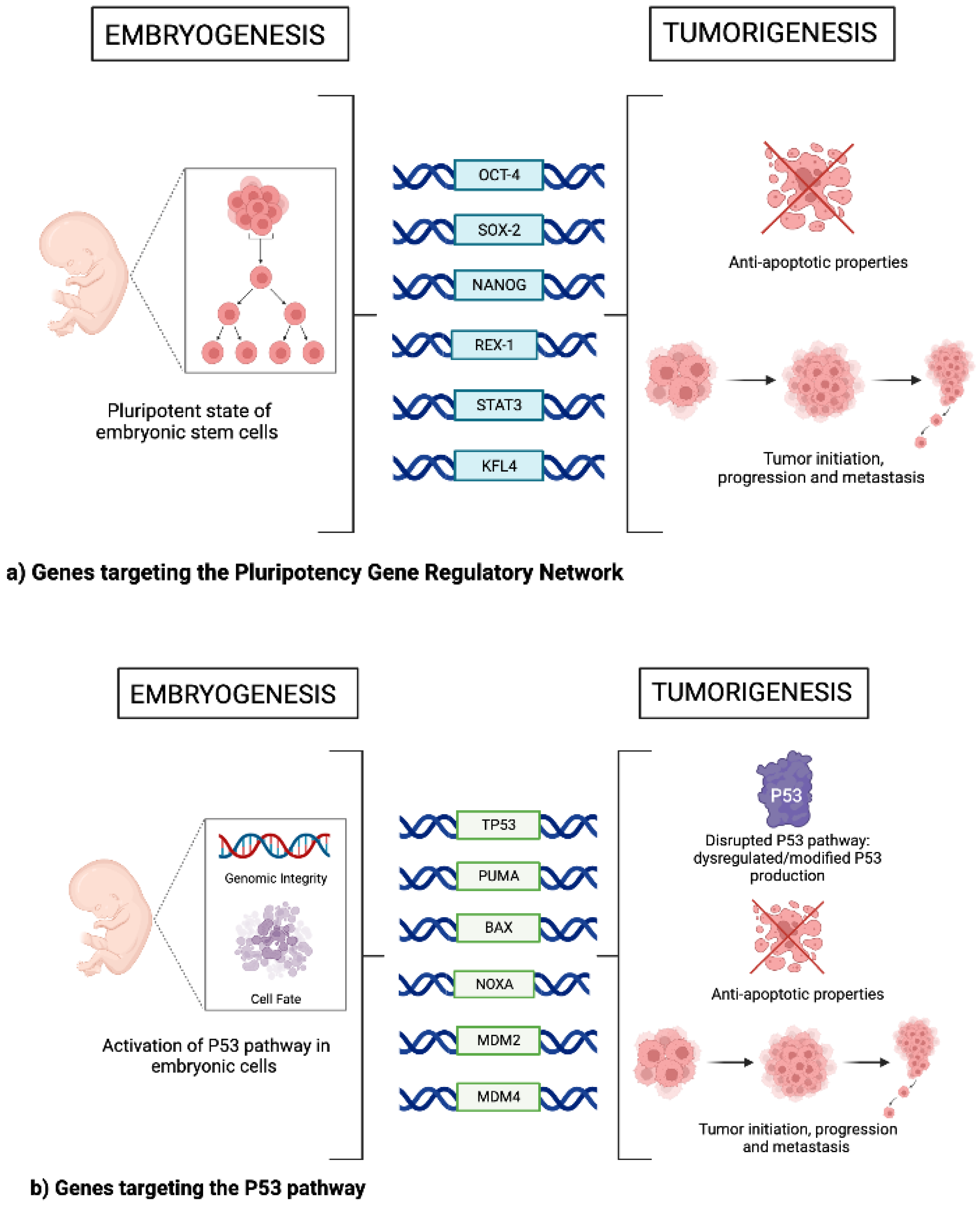
2. Pluripotency Gene Regulatory Network
3. The p53 Pathway
4. Epithelial Mesenchymal Transition (EMT)
5. Non-Coding RNAs
6. Clinical Relevance
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Biswas, A.; Hutchins, R. Embryonic Stem Cells. Stem Cells Dev. 2007, 16, 213–222. [Google Scholar] [CrossRef]
- Kim, J.; Orkin, S.H. Embryonic stem cell-specific signatures in cancer: Insights into genomic regulatory networks and implications for medicine. Genome Med. 2011, 3, 75–78. [Google Scholar] [CrossRef]
- Manzo, G. Similarities Between Embryo Development and Cancer Process Suggest New Strategies for Research and Therapy of Tumors: A New Point of View. Front. Cell Dev. Biol. 2019, 7, 20. [Google Scholar] [CrossRef]
- Kar, S.; Patra, S.K. Overexpression of OCT4 induced by modulation of histone marks plays crucial role in breast cancer progression. Gene 2018, 643, 35–45. [Google Scholar] [CrossRef]
- Lu, C.-S.; Shieh, G.-S.; Wang, C.-T.; Su, B.-H.; Su, Y.-C.; Chen, Y.-C.; Su, W.-C.; Wu, P.; Yang, W.-H.; Shiau, A.-L.; et al. Chemotherapeutics-induced Oct4 expression contributes to drug resistance and tumor recurrence in bladder cancer. Oncotarget 2016, 8, 30844–30858. [Google Scholar] [CrossRef]
- Villodre, E.S.; Kipper, F.C.; Pereira, M.B.; Lenz, G. Roles of OCT4 in tumorigenesis, cancer therapy resistance and prognosis. Cancer Treat. Rev. 2016, 51, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Rizzino, A.; Wuebben, E.L. Sox2/Oct4: A delicately balanced partnership in pluripotent stem cells and embryogenesis. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2016, 1859, 780–791. [Google Scholar] [CrossRef] [PubMed]
- de Wet, L.; Williams, A.; Gillard, M.; Kregel, S.; Lamperis, S.; Gutgesell, L.C.; Vellky, J.E.; Brown, R.; Conger, K.; Paner, G.P.; et al. SOX2 mediates metabolic reprogramming of prostate cancer cells. Oncogene 2022, 41, 1190–1202. [Google Scholar] [CrossRef]
- Xie, X.; Piao, L.; Cavey, G.S.; Old, M.; Teknos, T.N.; Mapp, A.K.; Pan, Q. Phosphorylation of Nanog is essential to regulate Bmi1 and promote tumorigenesis. Oncogene 2013, 33, 2040–2052. [Google Scholar] [CrossRef]
- Saga, K.; Park, J.; Nimura, K.; Kawamura, N.; Ishibashi, A.; Nonomura, N.; Kaneda, Y. NANOG helps cancer cells escape NK cell attack by downregulating ICAM1 during tumorigenesis. J. Exp. Clin. Cancer Res. 2019, 38, 416. [Google Scholar] [CrossRef] [PubMed]
- Luk, S.T.; Ng, K.; Zhou, L.; Tong, M.; Wong, T.; Yu, H.; Lo, C.; Man, K.; Guan, X.; Lee, T.K.; et al. Deficiency in embryonic stem cell marker reduced expression 1 activates mitogen-activated protein kinase kinase 6–dependent p38 mitogen-activated protein kinase signaling to drive hepatocarcinogenesis. Hepatology 2020, 72, 183–197. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, D.; Xu, J.; Wang, Y.; Ma, F.; Li, Z.; Liu, N. Stat3 activation is critical for pluripotency maintenance. J. Cell. Physiol. 2018, 234, 1044–1051. [Google Scholar] [CrossRef]
- Yang, J.; van Oosten, A.L.; Theunissen, T.W.; Guo, G.; Silva, J.C.; Smith, A. Stat3 Activation Is Limiting for Reprogramming to Ground State Pluripotency. Cell Stem Cell 2010, 7, 319–328. [Google Scholar] [CrossRef] [PubMed]
- van Oosten, A.L.; Costa, Y.; Smith, A.; Silva, J.C. JAK/STAT3 signalling is sufficient and dominant over antagonistic cues for the establishment of naive pluripotency. Nat. Commun. 2012, 3, 817. [Google Scholar] [CrossRef] [PubMed]
- Raz, R.; Lee, C.-K.; Cannizzaro, L.A.; D’Eustachio, P.; Levy, D.E. Essential role of STAT3 for embryonic stem cell pluripotency. Proc. Natl. Acad. Sci. USA 1999, 96, 2846–2851. [Google Scholar] [CrossRef]
- Humphrey, R.K.; Beattie, G.M.; Lopez, A.D.; Bucay, N.; King, C.C.; Firpo, M.T.; Rose-John, S.; Hayek, A. Maintenance of Pluripotency in Human Embryonic Stem Cells Is STAT3 Independent. Stem Cells 2004, 22, 522–530. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Torigoe, S.E.; Xiao, J.; Mai, D.H.; Li, L.; Davis, F.P.; Dong, P.; Marie-Nelly, H.; Grimm, J.; Lavis, L.; et al. A dynamic interplay of enhancer elements regulates Klf4 expression in naïve pluripotency. Genes Dev. 2017, 31, 1795–1808. [Google Scholar] [CrossRef] [PubMed]
- Timmerman, D.; Remmers, T.; Hillenius, S.; Looijenga, L. Mechanisms of TP53 pathway inactivation in embryonic and somatic cells—Relevance for understanding (germ cell) tumorigenesis. Int. J. Mol. Sci. 2021, 22, 5377. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Murphy, M.E. Genetic modifiers of the p53 pathway. Cold Spring Harb. Perspect. Med. 2016, 6, a026302. [Google Scholar] [CrossRef]
- Mogi, A.; Kuwano, H. TP53 mutations in nonsmall cell lung cancer. J. Biomed. Biotechnol. 2011, 2011, 583929. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Liu, N.; Liu, J.; Liu, Y.; Zhang, C.; Long, S.; Luo, G.; Zhang, L.; Zhang, Y. Mutant p53 drives cancer chemotherapy resistance due to loss of function on activating transcription of PUMA. Cell Cycle 2019, 18, 3442–3455. [Google Scholar] [CrossRef] [PubMed]
- Subhasree, N.; Jiangjiang, Q.; Kalkunte, S.; Minghai, W.; Ruiwen, Z. The MDM2-p53 pathway revisited. J. Biomed. Res. 2013, 27, 254–271. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef]
- Ramos, F.S.; Wons, L.; Cavalli, I.J.; Ribeiro, E.M. Epithelial-mesenchymal transition in cancer: An overview. Integr. Cancer Sci. Ther. 2017, 4, 1–5. [Google Scholar] [CrossRef]
- Ribatti, D.; Tamma, R.; Annese, T. Epithelial-Mesenchymal Transition in Cancer: A Historical Overview. Transl. Oncol. 2020, 13, 100773. [Google Scholar] [CrossRef]
- Yang, J.; Mani, S.A.; Donaher, J.L.; Ramaswamy, S.; Itzykson, R.A.; Come, C.; Savagner, P.; Gitelman, I.; Richardson, A.; Weinberg, R.A. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 2004, 117, 927–939. [Google Scholar] [CrossRef]
- Zhao, Z.; Rahman, M.A.; Chen, Z.G.; Shin, D.M. Multiple biological functions of Twist1 in various cancers. Oncotarget 2017, 8, 20380–20393. [Google Scholar] [CrossRef]
- Zhou, H.; Li, G.; Huang, S.; Feng, Y.; Zhou, A. SOX9 promotes epithelial-mesenchymal transition via the Hippo-YAP signaling pathway in gastric carcinoma cells. Oncol. Lett. 2019, 18, 599–608. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, Z.; Yu, X.; Huang, X.; Liu, Z.; Chai, Y.; Yang, L.; Wang, Q.; Li, M.; Zhao, J.; et al. Unbalanced YAP–SOX9 circuit drives stemness and malignant progression in esophageal squamous cell carcinoma. Oncogene 2018, 38, 2042–2055. [Google Scholar] [CrossRef]
- Zeng, Y.-T.; Liu, X.-F.; Yang, W.-T.; Zheng, P.-S. REX1 promotes EMT-induced cell metastasis by activating the JAK2/STAT3-signaling pathway by targeting SOCS1 in cervical cancer. Oncogene 2019, 38, 6940–6957. [Google Scholar] [CrossRef] [PubMed]
- Maturi, V.; Morén, A.; Enroth, S.; Heldin, C.; Moustakas, A. Genomewide binding of transcription factor Snail1 in triple-negative breast cancer cells. Mol. Oncol. 2018, 12, 1153–1174. [Google Scholar] [CrossRef] [PubMed]
- Jiao, W.; Miyazaki, K.; Kitajima, Y. Inverse correlation between E-cadherin and Snail expression in hepatocellular carcinoma cell lines in vitro and in vivo. Br. J. Cancer 2002, 86, 98–101. [Google Scholar] [CrossRef] [PubMed]
- Elloul, S.; Elstrand, M.B.; Nesland, J.M.; Tropé, C.G.; Kvalheim, G.; Goldberg, I.; Reich, R.; Davidson, B. Snail, Slug, and Smad-interacting protein 1 as novel parameters of disease aggressiveness in metastatic ovarian and breast carcinoma. Cancer 2005, 103, 1631–1643. [Google Scholar] [CrossRef]
- Alves, C.L.; Elias, D.; Lyng, M.B.; Bak, M.; Ditzel, H.J. SNAI2 upregulation is associated with an aggressive phenotype in fulvestrant-resistant breast cancer cells and is an indicator of poor response to endocrine therapy in estrogen receptor-positive metastatic breast cancer. Breast Cancer Res. 2018, 20, 60. [Google Scholar] [CrossRef]
- Masuo, K.; Chen, R.; Yogo, A.; Sugiyama, A.; Fukuda, A.; Masui, T.; Uemoto, S.; Seno, H.; Takaishi, S. SNAIL2 contributes to tumorigenicity and chemotherapy resistance in pancreatic cancer by regulating IGFBP2. Cancer Sci. 2021, 112, 4987–4999. [Google Scholar] [CrossRef]
- Hu, Y.; Dai, M.; Zheng, Y.; Wu, J.; Yu, B.; Zhang, H.; Kong, W.; Wu, H.; Yu, X. Epigenetic suppression of E-cadherin expression by Snail2 during the metastasis of colorectal cancer. Clin. Epigenetics 2018, 10, 154. [Google Scholar] [CrossRef]
- Taki, M.; Abiko, K.; Baba, T.; Hamanishi, J.; Yamaguchi, K.; Murakami, R.; Yamanoi, K.; Horikawa, N.; Hosoe, Y.; Nakamura, E.; et al. Snail promotes ovarian cancer progression by recruiting myeloid-derived suppressor cells via CXCR2 ligand upregulation. Nat. Commun. 2018, 9, 1685. [Google Scholar] [CrossRef]
- Slack, F.J.; Chinnaiyan, A.M. The Role of Non-coding RNAs in Oncology. Cell 2019, 179, 1033–1055. [Google Scholar] [CrossRef]
- Pauli, A.; Rinn, J.; Schier, A.F. Non-coding RNAs as regulators of embryogenesis. Nat. Rev. Genet. 2011, 12, 136–149. [Google Scholar] [CrossRef]
- Han, Y.-N.; Li, Y.; Xia, S.-Q.; Zhang, Y.-Y.; Zheng, J.-H.; Li, W. Piwi proteins and piwi-interacting RNA: Emerging roles in cancer. Cell. Physiol. Biochem. 2017, 44, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Navarro, A.; Tejero, R.; Viñolas, N.; Cordeiro, A.; Marrades, R.M.; Fuster, D.; Caritg, O.; Moises, J.; Muñoz, C.; Molins, L.; et al. The significance of PIWI family expression in human lung embryogenesis and non-small cell lung cancer. Oncotarget 2015, 6, 31544–31556. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Zhang, K.; Kong, J.; Wang, C.; Gu, Y.; Liang, C.; Jiang, T.; Qin, N.; Liu, J.; Guo, X.; et al. Cancer-testis gene PIWIL1 promotes cell proliferation, migration, and invasion in lung adenocarcinoma. Cancer Med. 2017, 7, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Erber, R.; Meyer, J.; Taubert, H.; Fasching, P.A.; Wach, S.; Häberle, L.; Gaß, P.; Schulz-Wendtland, R.; Landgraf, L.; Olbricht, S.; et al. PIWI-Like 1 and PIWI-Like 2 Expression in Breast Cancer. Cancers 2020, 12, 2742. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Yang, Z.-Z.; Liu, S.; Yang, F.; Lin, H. PIWIL1 promotes gastric cancer via a piRNA-independent mechanism. Proc. Natl. Acad. Sci. USA 2020, 117, 22390–22401. [Google Scholar] [CrossRef]
- Chu, H.; Xia, L.; Qiu, X.; Gu, D.; Zhu, L.; Jin, J.; Hui, G.; Hua, Q.; Du, M.; Tong, N.; et al. Genetic variants in noncoding PIWI-interacting RNA and colorectal cancer risk. Cancer 2015, 121, 2044–2052. [Google Scholar] [CrossRef]
- Chen, C.; Liu, J.; Xu, G. Overexpression of PIWI proteins in human stage III epithelial ovarian cancer with lymph node metastasis. Cancer Biomark. 2013, 13, 315–321. [Google Scholar] [CrossRef]
- Eckstein, M.; Jung, R.; Weigelt, K.; Sikic, D.; Stöhr, R.; Geppert, C.; Agaimy, A.; Lieb, V.; Hartmann, A.; Wullich, B.; et al. Piwi-like 1 and -2 protein expression levels are prognostic factors for muscle invasive urothelial bladder cancer patients. Sci. Rep. 2018, 8, 17693. [Google Scholar] [CrossRef]
- Zhuang, K.; Wu, Q.; Jin, C.-S.; Yuan, H.-J.; Cheng, J.-Z. Long non-coding RNA HNF1A-AS is upregulated and promotes cell proliferation and metastasis in nasopharyngeal carcinoma. Cancer Biomark. 2016, 16, 291–300. [Google Scholar] [CrossRef]
- Middelkamp, M.; Ruck, L.; Krisp, C.; Sumisławski, P.; Mohammadi, B.; Dottermusch, M.; Meister, V.; Küster, L.; Schlüter, H.; Windhorst, S.; et al. Overexpression of Lin28A in neural progenitor cells in vivo does not lead to brain tumor formation but results in reduced spine density. Acta Neuropathol. Commun. 2021, 9, 185. [Google Scholar] [CrossRef]
- Qiu, C.; Ma, Y.; Wang, J.; Peng, S.; Huang, Y. Lin28-mediated post-transcriptional regulation of Oct4 expression in human embryonic stem cells. Nucleic Acids Res. 2009, 38, 1240–1248. [Google Scholar] [CrossRef] [PubMed]
- Balzeau, J.; Menezes, M.R.; Cao, S.; Hagan, J.P. The LIN28/let-7 Pathway in Cancer. Front. Genet. 2017, 8, 31. [Google Scholar] [CrossRef]
- Pretzsch, E.; Max, N.; Kirchner, T.; Engel, J.; Werner, J.; Klauschen, F.; Angele, M.; Neumann, J. LIN28 promotes tumorigenesis in colorectal cancer but is not associated with metastatic spread. Pathol.—Res. Pract. 2021, 228, 153669. [Google Scholar] [CrossRef] [PubMed]
- Guilharducci, R.L.; Xavier, P.L.P.; da Silveira, J.C.; Cordeiro, Y.D.G.; Biondi, L.R.; Strefezzi, R.D.F.; Fukumasu, H. Expression of LIN28A/B and Let-7 miRNAs in canine mammary carcinomas. Cienc. Rural. 2022, 52, e20210171. [Google Scholar] [CrossRef]
- Skrzypek, K.; Majka, M. Interplay among snail transcription factor, MicroRNAs, long non-coding RNAS, and circular RNAS in the regulation of tumor growth and metastasis. Cancers 2020, 12, 209. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Sun, Q.; Zhang, J.; Yu, J.; Chen, W.; Zhang, Z. Downregulation of miR-153 contributes to epithelial-mesenchymal transition and tumor metastasis in human epithelial cancer. Carcinog. 2012, 34, 539–549. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Fu, T.; Zhang, L. MicroRNA-153 suppresses human laryngeal squamous cell carcinoma migration and invasion by targeting the SNAI1 gene. Oncol. Lett. 2018, 16, 5075–5083. [Google Scholar] [CrossRef]
- Luan, W.; Shi, Y.; Zhou, Z.; Xia, Y.; Wang, J. circRNA_0084043 promote malignant melanoma progression via miR-153-3p/Snail axis. Biochem. Biophys. Res. Commun. 2018, 502, 22–29. [Google Scholar] [CrossRef]
- Zhao, G.; Zhang, Y.; Zhao, Z.; Cai, H.; Zhao, X.; Yang, T.; Chen, W.; Yao, C.; Wang, Z.; Wang, Z.; et al. MiR-153 reduces stem cell-like phenotype and tumor growth of lung adenocarcinoma by targeting Jagged1. Stem Cell Res. Ther. 2020, 11, 170. [Google Scholar] [CrossRef]
- Zuo, Z.; Ye, F.; Liu, Z.; Huang, J.; Gong, Y. MicroRNA-153 inhibits cell proliferation, migration, invasion and epithelial-mesenchymal transition in breast cancer via direct targeting of RUNX2. Exp. Ther. Med. 2019, 17, 4693–4702. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, B.; Li, W.; Fu, L.; Zhu, Z.; Dong, J.-T. Epigenetic silencing of mir-203 upregulates snai2 and contributes to the invasiveness of malignant breast cancer cells. Genes Cancer 2011, 2, 782–791. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.H.; Robinton, D.A.; Seligson, M.T.; Wu, L.; Li, L.; Rakheja, D.; Comerford, S.A.; Ramezani, S.; Sun, X.; Parikh, M.S.; et al. LIN28B is sufficient to drive liver cancer and necessary for its maintenance in murine models. Cancer Cell 2014, 26, 248–261. [Google Scholar] [CrossRef] [PubMed]
- Penson, A.; Camacho, N.; Zheng, Y.; Varghese, A.M.; Al-Ahmadie, H.; Razavi, P.; Chandarlapaty, S.; Vallejo, C.E.; Vakiani, E.; Gilewski, T.; et al. Development of genome-derived tumor type prediction to inform clinical cancer care. JAMA Oncol. 2020, 6, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Barh, D.; Malhotra, R.; Ravi, B.; Sindhurani, P. Microrna let-7: An emerging next-generation cancer therapeutic. Curr. Oncol. 2010, 17, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Tavares, M.O.; Milan, T.M.; Bighetti-Trevisan, R.L.; Leopoldino, A.M.; de Almeida, L.O. Pharmacological inhibition of HDAC6 overcomes cisplatin chemoresistance by targeting cancer stem cells in oral squamous cell carcinoma. J. Oral Pathol. Med. 2022, 51, 529–537. [Google Scholar] [CrossRef]
- Fan, G.-T.; Ling, Z.-H.; He, Z.-W.; Wu, S.-J.; Zhou, G.-X. Suppressing CHD1L reduces the proliferation and chemoresistance in osteosarcoma. Biochem. Biophys. Res. Commun. 2021, 554, 214–221. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balachandran, S.; Narendran, A. The Developmental Origins of Cancer: A Review of the Genes Expressed in Embryonic Cells with Implications for Tumorigenesis. Genes 2023, 14, 604. https://doi.org/10.3390/genes14030604
Balachandran S, Narendran A. The Developmental Origins of Cancer: A Review of the Genes Expressed in Embryonic Cells with Implications for Tumorigenesis. Genes. 2023; 14(3):604. https://doi.org/10.3390/genes14030604
Chicago/Turabian StyleBalachandran, Savitha, and Aru Narendran. 2023. "The Developmental Origins of Cancer: A Review of the Genes Expressed in Embryonic Cells with Implications for Tumorigenesis" Genes 14, no. 3: 604. https://doi.org/10.3390/genes14030604
APA StyleBalachandran, S., & Narendran, A. (2023). The Developmental Origins of Cancer: A Review of the Genes Expressed in Embryonic Cells with Implications for Tumorigenesis. Genes, 14(3), 604. https://doi.org/10.3390/genes14030604




