Genome Analysis Using Whole-Exome Sequencing of Non-Syndromic Cleft Lip and/or Palate from Malagasy Trios Identifies Variants Associated with Cilium-Related Pathways and Asian Genetic Ancestry
Abstract
1. Introduction
2. Results
2.1. Study Outline
2.2. Characterization of Madagascar Trios by Genetic Ancestry
2.3. Variant Analysis Identifies Genes That are Associated with Ciliopathies
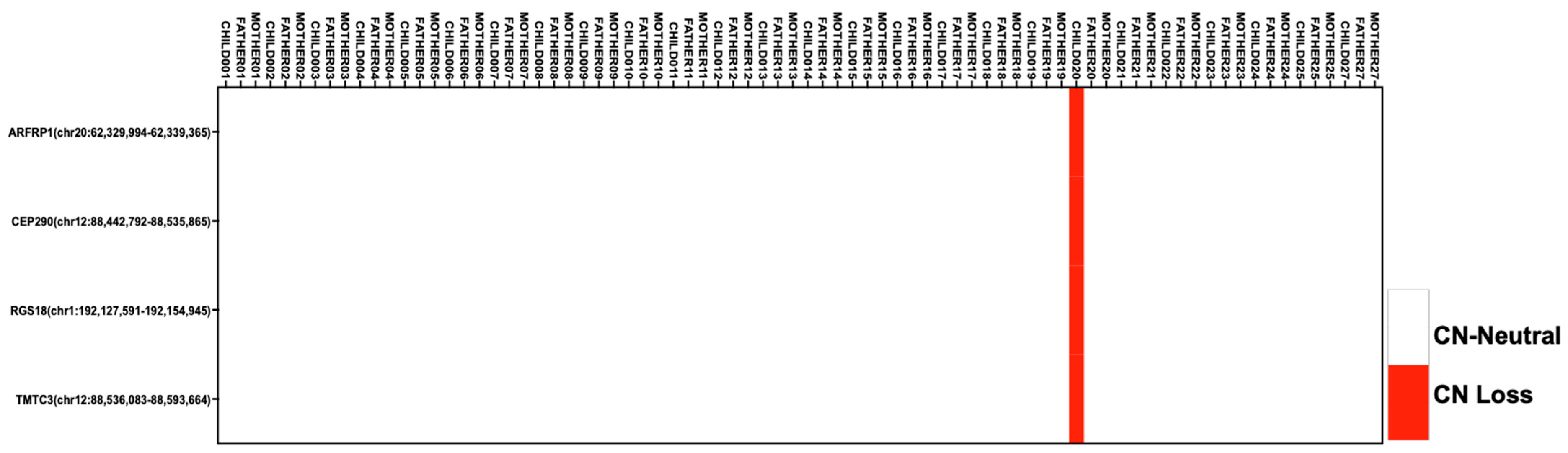
2.4. Copy-Number Analysis Reveals CNVs in Genes Associated with Metabolism and Ciliopathies
3. Discussion
4. Materials and Methods
4.1. Ethical Approval
4.2. Participants
4.3. Case Definition
4.4. Family Data Collection
4.5. Whole-Exome Sequencing
4.6. Primary Data Processing
4.7. Nucleotide-Variant Analysis
4.8. Copy-Number-Variant Analysis
4.9. Sample QC and Ancestry Analyses
4.10. Statistical Ancestry Analyses
5. Author Summary
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mossey, P.A.; Little, J.; Munger, R.G.; Dixon, M.J.; Shaw, W.C. Cleft lip and palate. Lancet 2009, 374, 1773–1785. [Google Scholar] [CrossRef]
- Mossey, P.A.; Modell, B. Epidemiology of oral clefts 2012: An international perspective. Front. Oral Biol. 2012, 16, 1–18. [Google Scholar] [PubMed]
- Cooper, M.E.; Ratay, J.S.; Marazita, M.L. Asian oral-facial cleft birth prevalence. Cleft Palate Craniofac. J. 2006, 43, 580–589. [Google Scholar] [CrossRef] [PubMed]
- Gundlach, K.K.; Maus, C. Epidemiological studies on the frequency of clefts in Europe and world-wide. J. Craniomaxillofac. Surg. 2006, 34 (Suppl. 2), 1–2. [Google Scholar] [CrossRef] [PubMed]
- Kadir, A.; Mossey, P.A.; Blencowe, H.; Moorthie, S.; Lawn, J.E.; Mastroiacovo, P.; Modell, B. Systematic Review and Meta-Analysis of the Birth Prevalence of Orofacial Clefts in Low- and Middle-Income Countries. Cleft Palate Craniofac. J. 2017, 54, 571–581. [Google Scholar] [CrossRef]
- Calzolari, E.; Pierini, A.; Astolfi, G.; Bianchi, F.; Neville, A.J.; Rivieri, F. Associated anomalies in multi-malformed infants with cleft lip and palate: An epidemiologic study of nearly 6 million births in 23 EUROCAT registries. Am. J. Med. Genet. A 2007, 143A, 528–537. [Google Scholar] [CrossRef]
- Rittler, M.; Lopez-Camelo, J.; Castilla, E.E. Sex ratio and associated risk factors for 50 congenital anomaly types: Clues for causal heterogeneity. Birth Defects Res. A Clin. Mol. Teratol. 2004, 70, 13–19. [Google Scholar] [CrossRef]
- Beaty, T.H.; Murray, J.C.; Marazita, M.L.; Munger, R.G.; Ruczinski, I.; Hetmanski, J.B.; Liang, K.Y.; Wu, T.; Murray, T.; Fallin, M.D.; et al. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat. Genet. 2010, 42, 525–529. [Google Scholar] [CrossRef]
- Beaty, T.H.; Taub, M.A.; Scott, A.F.; Murray, J.C.; Marazita, M.L.; Schwender, H.; Parker, M.M.; Hetmanski, J.B.; Balakrishnan, P.; Mansilla, M.A.; et al. Confirming genes influencing risk to cleft lip with/without cleft palate in a case-parent trio study. Hum. Genet. 2013, 132, 771–781. [Google Scholar] [CrossRef]
- Birnbaum, S.; Ludwig, K.U.; Reutter, H.; Herms, S.; Steffens, M.; Rubini, M.; Baluardo, C.; Ferrian, M.; Almeida de Assis, N.; Alblas, M.A.; et al. Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat. Genet. 2009, 41, 473–477. [Google Scholar] [CrossRef]
- Grant, S.F.; Wang, K.; Zhang, H.; Glaberson, W.; Annaiah, K.; Kim, C.E.; Bradfield, J.P.; Glessner, J.T.; Thomas, K.A.; Garris, M.; et al. A genome-wide association study identifies a locus for nonsyndromic cleft lip with or without cleft palate on 8q24. J. Pediatr. 2009, 155, 909–913. [Google Scholar] [CrossRef]
- Leslie, E.J.; Carlson, J.C.; Shaffer, J.R.; Butali, A.; Buxo, C.J.; Castilla, E.E.; Christensen, K.; Deleyiannis, F.W.; Leigh Field, L.; Hecht, J.T.; et al. Genome-wide meta-analyses of nonsyndromic orofacial clefts identify novel associations between FOXE1 and all orofacial clefts, and TP63 and cleft lip with or without cleft palate. Hum. Genet. 2017, 136, 275–286. [Google Scholar] [CrossRef]
- Leslie, E.J.; Carlson, J.C.; Shaffer, J.R.; Feingold, E.; Wehby, G.; Laurie, C.A.; Jain, D.; Laurie, C.C.; Doheny, K.F.; McHenry, T.; et al. A multi-ethnic genome-wide association study identifies novel loci for non-syndromic cleft lip with or without cleft palate on 2p24.2, 17q23 and 19q13. Hum. Mol. Genet. 2016, 25, 2862–2872. [Google Scholar] [CrossRef]
- Ludwig, K.U.; Ahmed, S.T.; Bohmer, A.C.; Sangani, N.B.; Varghese, S.; Klamt, J.; Schuenke, H.; Gultepe, P.; Hofmann, A.; Rubini, M.; et al. Meta-analysis Reveals Genome-Wide Significance at 15q13 for Nonsyndromic Clefting of Both the Lip and the Palate, and Functional Analyses Implicate GREM1 As a Plausible Causative Gene. PLoS Genet. 2016, 12, e1005914. [Google Scholar] [CrossRef]
- Ludwig, K.U.; Bohmer, A.C.; Bowes, J.; Nikolic, M.; Ishorst, N.; Wyatt, N.; Hammond, N.L.; Golz, L.; Thieme, F.; Barth, S.; et al. Imputation of orofacial clefting data identifies novel risk loci and sheds light on the genetic background of cleft lip +/− cleft palate and cleft palate only. Hum. Mol. Genet. 2017, 26, 829–842. [Google Scholar] [CrossRef]
- Ludwig, K.U.; Mangold, E.; Herms, S.; Nowak, S.; Reutter, H.; Paul, A.; Becker, J.; Herberz, R.; AlChawa, T.; Nasser, E.; et al. Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci. Nat. Genet. 2012, 44, 968–971. [Google Scholar] [CrossRef]
- Mangold, E.; Ludwig, K.U.; Birnbaum, S.; Baluardo, C.; Ferrian, M.; Herms, S.; Reutter, H.; de Assis, N.A.; Chawa, T.A.; Mattheisen, M.; et al. Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nat. Genet. 2010, 42, 24–26. [Google Scholar] [CrossRef]
- Sun, Y.; Huang, Y.; Yin, A.; Pan, Y.; Wang, Y.; Wang, C.; Du, Y.; Wang, M.; Lan, F.; Hu, Z.; et al. Genome-wide association study identifies a new susceptibility locus for cleft lip with or without a cleft palate. Nat. Commun. 2015, 6, 6414. [Google Scholar] [CrossRef]
- Yu, Y.; Zuo, X.; He, M.; Gao, J.; Fu, Y.; Qin, C.; Meng, L.; Wang, W.; Song, Y.; Cheng, Y.; et al. Genome-wide analyses of non-syndromic cleft lip with palate identify 14 novel loci and genetic heterogeneity. Nat. Commun. 2017, 8, 14364. [Google Scholar] [CrossRef]
- Wattanawong, K.; Rattanasiri, S.; McEvoy, M.; Attia, J.; Thakkinstian, A. Association between IRF6 and 8q24 polymorphisms and nonsyndromic cleft lip with or without cleft palate: Systematic review and meta-analysis. Birth Defects Res. A Clin. Mol. Teratol. 2016, 106, 773–788. [Google Scholar] [CrossRef]
- Butali, A.; Mossey, P.A.; Adeyemo, W.L.; Eshete, M.A.; Gowans, L.J.J.; Busch, T.D.; Jain, D.; Yu, W.; Huan, L.; Laurie, C.A.; et al. Genomic analyses in african populations identify novel risk loci for cleft palate. Hum. Mol. Genet. 2018, 28, 1038–1051. [Google Scholar] [CrossRef] [PubMed]
- Jezewski, P.A.; Vieira, A.R.; Nishimura, C.; Ludwig, B.; Johnson, M.; O’Brien, S.E.; Daack-Hirsch, S.; Schultz, R.E.; Weber, A.; Nepomucena, B.; et al. Complete sequencing shows a role for MSX1 in non-syndromic cleft lip and palate. J. Med. Genet. 2003, 40, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Vieira, A.R.; Avila, J.R.; Daack-Hirsch, S.; Dragan, E.; Felix, T.M.; Rahimov, F.; Harrington, J.; Schultz, R.R.; Watanabe, Y.; Johnson, M.; et al. Medical sequencing of candidate genes for nonsyndromic cleft lip and palate. PLoS Genet. 2005, 1, e64. [Google Scholar] [CrossRef] [PubMed]
- Bureau, A.; Parker, M.M.; Ruczinski, I.; Taub, M.A.; Marazita, M.L.; Murray, J.C.; Mangold, E.; Noethen, M.M.; Ludwig, K.U.; Hetmanski, J.B.; et al. Whole exome sequencing of distant relatives in multiplex families implicates rare variants in candidate genes for oral clefts. Genetics 2014, 197, 1039–1044. [Google Scholar] [CrossRef]
- Liu, Y.; Cheng, L.; Weng, X.; Jin, X.; Yan, N.; Wang, H.; Pan, Q. Identification of a Tp63 gene variant in an abortus with Ectrodactyly, Ectodermal dysplasia, Cleft lip/palate syndrome by whole-exome sequencing. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2020, 37, 139–141. [Google Scholar]
- Liu, Y.-P.; Xu, L.-F.; Wang, Q.; Zhou, X.-L.; Zhou, J.-L.; Pan, C.; Zhang, J.-P.; Wu, Q.-R.; Li, Y.-Q.; Xia, Y.-J.; et al. Identification of susceptibility genes in non-syndromic cleft lip with or without cleft palate using whole-exome sequencing. Med. Oral Patol. Oral Cir. Bucal 2015, 20, e763–e770. [Google Scholar] [CrossRef]
- Meng, P.; Zhao, H.; Huang, W.; Zhang, Y.; Zhong, W.; Zhang, M.; Jia, P.; Zhou, Z.; Maimaitili, G.; Chen, F.; et al. Three GLI2 mutations combined potentially underlie non-syndromic cleft lip with or without cleft palate in a Chinese pedigree. Mol. Genet. Genom. Med. 2019, 7, e714. [Google Scholar]
- Sirugo, G.; Williams, S.M.; Tishkoff, S.A. The Missing Diversity in Human Genetic Studies. Cell 2019, 177, 1080. [Google Scholar] [CrossRef]
- Brucato, N.; Fernandes, V.; Kusuma, P.; Černý, V.; Mulligan, C.J.; Soares, P.; Rito, T.; Besse, C.; Boland, A.; Deleuze, J.-F.; et al. Evidence of Austronesian Genetic Lineages in East Africa and South Arabia: Complex Dispersal from Madagascar and Southeast Asia. Genome Biol. Evol. 2019, 11, 748–758. [Google Scholar] [CrossRef]
- Pierron, D.; Heiske, M.; Razafindrazaka, H.; Rakoto, I.; Rabetokotany, N.; Ravololomanga, B.; Rakotozafy, L.M.A.; Rakotomalala, M.M.; Razafiarivony, M.; Rasoarifetra, B.; et al. Genomic landscape of human diversity across Madagascar. Proc. Natl. Acad. Sci. USA 2017, 114, E6498–E6506. [Google Scholar] [CrossRef]
- Figueiredo, J.C.; Ly, S.; Raimondi, H.; Magee, K.; Baurley, J.W.; Sanchez-Lara, P.A.; Ihenacho, U.; Yao, C.; Edlund, C.K.; van den Berg, D.; et al. Genetic risk factors for orofacial clefts in Central Africans and Southeast Asians. Am. J. Med. Genet. A 2014, 164A, 2572–2580. [Google Scholar] [CrossRef]
- Bloch-Zupan, A.; Huckert, M.; Stoetzel, C.; Meyer, J.; Geoffroy, V.; Razafindrakoto, R.W.; Ralison, S.N.; Randrianaivo, J.-C.; Ralison, G.; Andriamasinoro, R.O.; et al. Detection of a Novel DSPP Mutation by NGS in a Population Isolate in Madagascar. Front. Physiol. 2016, 7, 70. [Google Scholar]
- Bessaud, M.; Razafindratsimandresy, R.; Nougairède, A.; Joffret, M.L.; Deshpande, J.M.; Dubot-Pérès, A.; Héraud, J.M.; de Lamballerie, X.; Delpeyroux, F.; Bailly, J.L. Molecular comparison and evolutionary analyses of VP1 nucleotide sequences of new African human enterovirus 71 isolates reveal a wide genetic diversity. PLoS ONE 2014, 9, e90624. [Google Scholar] [CrossRef]
- Capredon, M.; Brucato, N.; Tonasso, L.; Choesmel-Cadamuro, V.; Ricaut, F.X.; Razafindrazaka, H.; Rakotondrabe, A.B.; Ratolojanahary, M.A.; Randriamarolaza, L.P.; Champion, B.; et al. Tracing Arab-Islamic inheritance in Madagascar: Study of the Y-chromosome and mitochondrial DNA in the Antemoro. PLoS ONE 2013, 8, e80932. [Google Scholar] [CrossRef]
- Regueiro, M.; Mirabal, S.; Lacau, H.; Caeiro, J.L.; Garcia-Bertrand, R.L.; Herrera, R.J. Austronesian genetic signature in East African Madagascar and Polynesia. J. Hum. Genet. 2008, 53, 106–120. [Google Scholar] [CrossRef]
- Chow, R.A.; Caeiro, J.L.; Chen, S.J.; Garcia-Bertrand, R.L.; Herrera, R.J. Genetic characterization of four Austronesian-speaking populations. J. Hum. Genet. 2005, 50, 550–559. [Google Scholar] [CrossRef]
- Capredon, M.; Sanchez-Mazas, A.; Guitard, E.; Razafindrazaka, H.; Chiaroni, J.; Champion, B.; Dugoujon, J.M. The Arabo-Islamic migrations in Madagascar: First genetic study of the GM system in three Malagasy populations. Int. J. Immunogenet. 2012, 39, 161–169. [Google Scholar] [CrossRef]
- Razafindrazaka, H.; Monnereau, A.; Razafindrazaka, D.; Tonasso, L.; Schiavinato, S.; Rakotoarisoa, J.A.; Radimilahy, C.; Letellier, T.; Pierron, D. Genetic Admixture and Flavor Preferences: Androstenone Sensitivity in Malagasy Populations. Hum. Biol. 2015, 87, 59–70. [Google Scholar] [CrossRef]
- Howes, R.E.; Chan, E.R.; Rakotomanga, T.A.; Schulte, S.; Gibson, J.; Zikursh, M.; Franchard, T.; Ramiranirina, B.; Ratsimbasoa, A.; Zimmerman, P.A. Prevalence and genetic variants of G6PD deficiency among two Malagasy populations living in Plasmodium vivax-endemic areas. Malar. J. 2017, 16, 139. [Google Scholar] [CrossRef]
- Hodgson, J.A.; Pickrell, J.K.; Pearson, L.N.; Quillen, E.E.; Prista, A.; Rocha, J.; Soodyall, H.; Shriver, M.D.; Perry, G.H. Natural selection for the Duffy-null allele in the recently admixed people of Madagascar. Proc. Biol. Sci. 2014, 281, 20140930. [Google Scholar] [CrossRef]
- Andriantsoanirina, V.; Khim, N.; Ratsimbasoa, A.; Witkowski, B.; Benedet, C.; Canier, L.; Bouchier, C.; Tichit, M.; Durand, R.; Ménard, D. Plasmodium falciparum Na+/H+ exchanger (pfnhe-1) genetic polymorphism in Indian Ocean malaria-endemic areas. Am. J. Trop. Med. Hyg. 2013, 88, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Pierron, D.; Heiske, M.; Razafindrazaka, H.; Pereda-Loth, V.; Sanchez, J.; Alva, O.; Arachiche, A.; Boland, A.; Olaso, R.; Deleuze, J.F.; et al. Strong selection during the last millennium for African ancestry in the admixed population of Madagascar. Nat. Commun. 2018, 9, 932. [Google Scholar] [CrossRef] [PubMed]
- Grant, A.V.; El Baghdadi, J.; Sabri, A.; El Azbaoui, S.; Alaoui-Tahiri, K.; Abderrahmani Rhorfi, I.; Gharbaoui, Y.; Abid, A.; Benkirane, M.; Raharimanga, V.; et al. Age-dependent association between pulmonary tuberculosis and common TOX variants in the 8q12-13 linkage region. Am. J. Hum. Genet. 2013, 92, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Paganotti, G.M.; Gramolelli, S.; Tabacchi, F.; Russo, G.; Modiano, D.; Coluzzi, M.; Romano, R. Distribution of human CYP2C8*2 allele in three different African populations. Malar. J. 2012, 11, 125. [Google Scholar] [CrossRef] [PubMed]
- Tofanelli, S.; Bertoncini, S.; Castrì, L.; Luiselli, D.; Calafell, F.; Donati, G.; Paoli, G. On the origins and admixture of Malagasy: New evidence from high-resolution analyses of paternal and maternal lineages. Mol. Biol. Evol. 2009, 26, 2109–2124. [Google Scholar] [CrossRef]
- Rakotoarison, R.A.; Rakotoarivony, A.E.; Rabesandratana, N.; Razafindrabe, J.B.; Andriambololona, R.; Andriambololo-Nivo, R.; Feki, A. Cleft lip and palate in Madagascar 1998–2007. Br. J. Oral Maxillofac. Surg. 2012, 50, 430–434. [Google Scholar] [CrossRef]
- Moore, E.R. Primary Cilia: The New Face of Craniofacial Research. Biomolecules 2022, 12, 1724. [Google Scholar] [CrossRef]
- Badano, J.L.; Mitsuma, N.; Beales, P.L.; Katsanis, N. The ciliopathies: An emerging class of human genetic disorders. Annu. Rev. Genom. Hum. Genet. 2006, 7, 125–148. [Google Scholar] [CrossRef]
- Delous, M.; Baala, L.; Salomon, R.; Laclef, C.; Vierkotten, J.; Tory, K.; Golzio, C.; Lacoste, T.; Besse, L.; Ozilou, C.; et al. The ciliary gene RPGRIP1L is mutated in cerebello-oculo-renal syndrome (Joubert syndrome type B) and Meckel syndrome. Nat. Genet. 2007, 39, 875–881. [Google Scholar] [CrossRef]
- Manojlovic, Z.; Earwood, R.; Kato, A.; Perez, D.; Cabrera, O.A.; Didier, R.; Megraw, T.L.; Stefanovic, B.; Kato, Y. La-related protein 6 controls ciliated cell differentiation. Cilia 2017, 6, 4. [Google Scholar] [CrossRef]
- Zaghloul, N.A.; Brugmann, S.A. The emerging face of primary cilia. Genesis 2011, 49, 231–246. [Google Scholar] [CrossRef]
- Genomes Project, C.; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A global reference for human genetic variation. Nature 2015, 526, 68. [Google Scholar]
- Kramer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Panamonta, V.; Pradubwong, S.; Panamonta, M.; Chowchuen, B. Global Birth Prevalence of Orofacial Clefts: A Systematic Review. J. Med. Assoc. Thai 2015, 98 (Suppl. 7), S11–S21. [Google Scholar]
- Mangold, E.; Reutter, H.; Leon-Cachon, R.B.; Ludwig, K.U.; Herms, S.; Chacon-Camacho, O.; Ortiz-Lopez, R.; Paredes-Zenteno, M.; Arizpe-Cantu, A.; Munoz-Jimenez, S.G.; et al. Evaluating SKI as a candidate gene for non-syndromic cleft lip with or without cleft palate. Eur. J. Oral Sci. 2012, 120, 373–377. [Google Scholar] [CrossRef]
- Liu, X.; Sun, Y.; Weinberg, R.A.; Lodish, H.F. Ski/Sno and TGF-beta signaling. Cytokine Growth Factor Rev. 2001, 12, 1–8. [Google Scholar] [CrossRef]
- Chen, M.H.; Wilson, C.W.; Li, Y.J.; Law, K.K.; Lu, C.S.; Gacayan, R.; Zhang, X.; Hui, C.C.; Chuang, P.T. Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev. 2009, 23, 1910–1928. [Google Scholar] [CrossRef]
- Cheng, S.Y.; Bishop, J.M. Suppressor of Fused represses Gli-mediated transcription by recruiting the SAP18-mSin3 corepressor complex. Proc. Natl. Acad. Sci. USA 2002, 99, 5442–5447. [Google Scholar] [CrossRef]
- Kim, Y.J.; Osborn, D.P.; Lee, J.Y.; Araki, M.; Araki, K.; Mohun, T.; Kansakoski, J.; Brandstack, N.; Kim, H.T.; Miralles, F.; et al. WDR11-mediated Hedgehog signalling defects underlie a new ciliopathy related to Kallmann syndrome. EMBO Rep. 2018, 19, 269–289. [Google Scholar] [CrossRef]
- Van den Boogaard, M.J.; Dorland, M.; Beemer, F.A.; van Amstel, H.K. MSX1 mutation is associated with orofacial clefting and tooth agenesis in humans. Nat. Genet. 2000, 24, 342–343. [Google Scholar] [CrossRef]
- Suzuki, Y.; Jezewski, P.A.; Machida, J.; Watanabe, Y.; Shi, M.; Cooper, M.E.; Viet, L.T.; Nguyen, T.D.; Hai, H.; Natsume, N.; et al. In a Vietnamese population, MSX1 variants contribute to cleft lip and palate. Genet. Med. 2004, 6, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Tongkobpetch, S.; Siriwan, P.; Shotelersuk, V. MSX1 mutations contribute to nonsyndromic cleft lip in a Thai population. J. Hum. Genet. 2006, 51, 671. [Google Scholar] [CrossRef] [PubMed]
- Gowans, L.J.; Adeyemo, W.L.; Eshete, M.; Mossey, P.A.; Busch, T.; Aregbesola, B.; Donkor, P.; Arthur, F.K.; Bello, S.A.; Martinez, A.; et al. Association Studies and Direct DNA Sequencing Implicate Genetic Susceptibility Loci in the Etiology of Nonsyndromic Orofacial Clefts in Sub-Saharan African Populations. J. Dent. Res. 2016, 95, 1245–1256. [Google Scholar] [CrossRef] [PubMed]
- Butali, A.; Mossey, P.A.; Adeyemo, W.L.; Jezewski, P.A.; Onwuamah, C.K.; Ogunlewe, M.O.; Ugboko, V.I.; Adejuyigbe, O.; Adigun, A.I.; Abdur-Rahman, L.O.; et al. Genetic studies in the Nigerian population implicate an MSX1 mutation in complex oral facial clefting disorders. Cleft Palate Craniofac. J. 2011, 48, 646–653. [Google Scholar] [CrossRef] [PubMed]
- Fuchtbauer, A.; Lassen, L.B.; Jensen, A.B.; Howard, J.; Quiroga Ade, S.; Warming, S.; Sorensen, A.B.; Pedersen, F.S.; Fuchtbauer, E.M. Septin9 is involved in septin filament formation and cellular stability. Biol. Chem. 2011, 392, 769–777. [Google Scholar] [CrossRef] [PubMed]
- Amor, D.J.; Stephenson, S.E.M.; Mustapha, M.; Mensah, M.A.; Ockeloen, C.W.; Lee, W.S.; Tankard, R.M.; Phelan, D.G.; Shinawi, M.; de Brouwer, A.P.M.; et al. Pathogenic Variants in GPC4 Cause Keipert Syndrome. Am. J. Hum. Genet. 2019, 104, 914–924. [Google Scholar] [CrossRef]
- Rochard, L.; Monica, S.D.; Ling, I.T.; Kong, Y.; Roberson, S.; Harland, R.; Halpern, M.; Liao, E.C. Roles of Wnt pathway genes wls, wnt9a, wnt5b, frzb and gpc4 in regulating convergent-extension during zebrafish palate morphogenesis. Development 2016, 143, 2541–2547. [Google Scholar]
- Sakane, H.; Yamamoto, H.; Matsumoto, S.; Sato, A.; Kikuchi, A. Localization of glypican-4 in different membrane microdomains is involved in the regulation of Wnt signaling. J. Cell Sci. 2012, 125, 449–460. [Google Scholar] [CrossRef]
- D’Avola, A.; Legrave, N.; Tajan, M.; Chakravarty, P.; Shearer, R.L.; King, H.W.; Kluckova, K.; Cheung, E.C.; Clear, A.J.; Gunawan, A.S.; et al. PHGDH is required for germinal center formation and is a therapeutic target in MYC-driven lymphoma. J. Clin. Investig. 2022, 132, e153436. [Google Scholar] [CrossRef]
- Song, Z.; Feng, C.; Lu, Y.; Lin, Y.; Dong, C. PHGDH is an independent prognosis marker and contributes cell proliferation, migration and invasion in human pancreatic cancer. Gene 2018, 642, 43–50. [Google Scholar] [CrossRef]
- Drivas, T.G.; Bennett, J. CEP290 and the primary cilium. Adv. Exp. Med. Biol. 2014, 801, 519–525. [Google Scholar]
- Graham, J.B.; Sunryd, J.C.; Mathavan, K.; Weir, E.; Larsen, I.S.B.; Halim, A.; Clausen, H.; Cousin, H.; Alfandari, D.; Hebert, D.N. Endoplasmic reticulum transmembrane protein TMTC3 contributes to O-mannosylation of E-cadherin, cellular adherence, and embryonic gastrulation. Mol. Biol. Cell 2020, 31, 167–183. [Google Scholar] [CrossRef]
- Patmanathan, S.N.; Tong, B.T.; Teo, J.H.J.; Ting, Y.Z.J.; Tan, N.S.; Sim, S.H.K.; Ta, Y.C.; Woo, W.M. A PDZ Protein GIPC3 Positively Modulates Hedgehog Signaling and Melanoma Growth. J. Investig. Dermatol. 2022, 142, 179–188.e4. [Google Scholar] [CrossRef]
- Louwette, S.; Labarque, V.; Wittevrongel, C.; Thys, C.; Metz, J.; Gijsbers, R.; Debyser, Z.; Arnout, J.; Van Geet, C.; Freson, K. Regulator of G-protein signaling 18 controls megakaryopoiesis and the cilia-mediated vertebrate mechanosensory system. FASEB J. 2012, 26, 2125–2136. [Google Scholar] [CrossRef]
- Li, Y.; Ling, K.; Hu, J. The emerging role of Arf/Arl small GTPases in cilia and ciliopathies. J. Cell Biochem. 2012, 113, 2201–2207. [Google Scholar] [CrossRef]
- Figueiredo, J.C.; Ly, S.; Magee, K.S.; Ihenacho, U.; Baurley, J.W.; Sanchez-Lara, P.A.; Brindopke, F.; Nguyen, T.H.; Nguyen, V.; Tangco, M.I.; et al. Parental risk factors for oral clefts among Central Africans, Southeast Asians, and Central Americans. Birth Defects Res. A Clin. Mol. Teratol. 2015, 103, 863–879. [Google Scholar] [CrossRef]
- ICD-10 Classifications of Mental and Behavioural Disorder: Clinical Descriptions and Disgnostic Guidelines; World Health Organisation: Geneva, Switzerland, 1992.
- Robinson, J.T.; Thorvaldsdottir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Landrum, M.J.; Chitipiralla, S.; Brown, G.R.; Chen, C.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; Kaur, K.; Liu, C.; et al. ClinVar: Improvements to accessing data. Nucleic Acids Res. 2020, 48, D835–D844. [Google Scholar] [CrossRef]
- Chun, S.; Fay, J.C. Identification of deleterious mutations within three human genomes. Genome Res. 2009, 19, 1553–1561. [Google Scholar] [CrossRef]
- Pollard, K.S.; Hubisz, M.J.; Rosenbloom, K.R.; Siepel, A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010, 20, 110–121. [Google Scholar] [CrossRef]
- Reva, B.; Antipin, Y.; Sander, C. Predicting the functional impact of protein mutations: Application to cancer genomics. Nucleic Acids Res. 2011, 39, e118. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, J.M.; Rodelsperger, C.; Schuelke, M.; Seelow, D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods 2010, 7, 575–576. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Chan, A.P. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015, 31, 2745–2747. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.C.; Henikoff, S. Predicting deleterious amino acid substitutions. Genome Res. 2001, 11, 863–874. [Google Scholar] [CrossRef] [PubMed]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M.; et al. UniProt: The Universal Protein knowledgebase. Nucleic Acids Res. 2004, 32, D115–D119. [Google Scholar] [CrossRef]
- Garber, M.; Guttman, M.; Clamp, M.; Zody, M.C.; Friedman, N.; Xie, X. Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics 2009, 25, i54–i62. [Google Scholar] [CrossRef]
- Kanehisa, M. The KEGG database. Novartis Found Symp. 2002, 247, 91–101, discussion 101–103, 119–128, 144–152. [Google Scholar]
- Kanehisa, M.; Sato, Y.; Kawashima, M. KEGG mapping tools for uncovering hidden features in biological data. Protein Sci. 2022, 31, 47–53. [Google Scholar] [CrossRef]
- Sudmant, P.H.; Mallick, S.; Nelson, B.J.; Hormozdiari, F.; Krumm, N.; Huddleston, J.; Coe, B.P.; Baker, C.; Nordenfelt, S.; Bamshad, M.; et al. Global diversity, population stratification, and selection of human copy-number variation. Science 2015, 349, aab3761. [Google Scholar] [CrossRef]
- Sudmant, P.H.; Rausch, T.; Gardner, E.J.; Handsaker, R.E.; Abyzov, A.; Huddleston, J.; Zhang, Y.; Ye, K.; Jun, G.; Fritz, M.H.; et al. An integrated map of structural variation in 2504 human genomes. Nature 2015, 526, 75–81. [Google Scholar] [CrossRef]
- Zhang, F.; Flickinger, M.; Taliun, S.A.G.; In, P.P.G.C.; Abecasis, G.R.; Scott, L.J.; McCaroll, S.A.; Pato, C.N.; Boehnke, M.; Kang, H.M. Ancestry-agnostic estimation of DNA sample contamination from sequence reads. Genome Res. 2020, 30, 185–194. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef]
- Lischer, H.E.; Excoffier, L. PGDSpider: An automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics 2012, 28, 298–299. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Pasaniuc, B.; Sankararaman, S.; Kimmel, G.; Halperin, E. Inference of locus-specific ancestry in closely related populations. Bioinformatics 2009, 25, i213–i221. [Google Scholar] [CrossRef]


| Trio Complete | Count | % |
|---|---|---|
| Yes | 26 | 100% |
| Proband Gender | ||
| Male | 13 | 50% |
| Female | 13 | 50% |
| Cleft type | ||
| Cleft lip and palate (unilateral) | 16 | 62% |
| Cleft lip and palate (bilateral) | 2 | 8% |
| Isolated cleft lip (unilateral) | 8 | 31% |
| Isolated cleft lip (bilateral) | 0 | 0% |
| Avg | Range | |
| Estimated coverage | 172.1 | 147–199 |
| Target bases 10× | 95% | 89–98% |
| FamilyID | Child015 | Child015 | Child005 | Child006 | Child009 | Child023 | Child005 | Child019 | Child004 | Child019 |
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | WNT5B | GPC4 | MSX1 | MSX1 | MSX1 | MSX1 | SEPTIN9 | WDR11 | PHGDH | SKI |
| Chromosome | 12 | X | 4 | 4 | 4 | 4 | 17 | 10 | 1 | 1 |
| Start | 1742006 | 132440095 | 4861877 | 4861877 | 4861877 | 4861877 | 75484846 | 122664299 | 120279876 | 2160660 |
| Stop | 1742006 | 132440095 | 4861877 | 4861877 | 4861877 | 4861877 | 75484846 | 122664299 | 120279876 | 2160660 |
| Read depth | 123 | 169 | 89 | 66 | 82 | 81 | 131 | 51 | 131 | 82 |
| Reference | G | G | A | A | A | A | G | A | C | G |
| Genotype (Proband) | G|T | C|G | T|A | T|A | T|A | T|A | A|G | G|A | T|T | G|A |
| Genotype (Mother) | G|G | G|C | A|T | A|T | A|T | A|T | G|A | A|G | C|T | G|G |
| Genotype (Father) | G|T | G|G | A|A | A|A | A|A | A|A | G|G | A|T | C|T | G|A |
| Inheritance mode | Heterozygous | Heterozygous | Heterozygous | Heterozygous | Heterozygous | Heterozygous | Heterozygous | Heterozygous | Homozygous | Heterozygous |
| Inherited from | Father | Mother | Mother | Mother | Mother | Mother | Mother | Mother | Both | Father |
| cDNA | c.263G > T | c.965C > G | c.251A > T | c.251A > T | c.251A > T | c.251A > T | c.1108G > A | c.3169A > G | c.932C > T | c.455G > A |
| HGVS | p.Arg88Leu | p.Ala322Gly | p.Glu84Val | p.Glu84Val | p.Glu84Val | p.Glu84Val | p.Glu370Lys | p.Met1057Val | p.Ser311Phe | p.Arg152His |
| Exon | 3 | 5 | 1 | 1 | 1 | 1 | 6 | 25 | 8 | 1 |
| dbSNP | rs200966877 | rs28928890 | rs28928890 | rs28928890 | rs28928890 | rs1297513860 | rs143340742 | |||
| OMIM(r) refs | 606361 | 604061 | ||||||||
| clinVar ClinicalSig | pathogenic | pathogenic | pathogenic | pathogenic | ||||||
| clinVar Gene Disease | Orofacial cleft 5 | Orofacial cleft 5 | Orofacial cleft 5 | Orofacial cleft 5 | ||||||
| ClinVar ID | 14883 | 14883 | 14883 | 14883 | ||||||
| LRT prediction | Deleterious | Deleterious | Neutral | Neutral | Neutral | Neutral | Deleterious | Deleterious | Neutral | Deliterious |
| Mutation Assessor Prediction | Medium | Medium | Low | Low | Low | Low | Medium | Medium | Low | Medium |
| Mutation Taster Prediction | Disease causing | Disease causing | Disease causing automatic | Disease causing automatic | Disease causing automatic | Disease causing automatic | Disease causing | Disease causing | Disease causing | Disease causing |
| phyloP | 0.917 | 0.917 | 1.039 | 1.039 | 1.039 | 1.039 | 0.913 | 1.062 | 0.871 | 0.917 |
| PPH2HumVar Prediction | Benign | Probably damaging | Benign | Benign | Benign | Benign | Probably damaging | Possibly damaging | Possibly damaging | Probably damaging |
| PROVEAN Prediction | Damaging | Damaging | Neutral | Neutral | Neutral | Neutral | Damaging | Neutral | Damaging | Damaging |
| SIFT score | 0.025 | 0.058 | 0.102 | 0.102 | 0.102 | 0.102 | 0.002 | 0.013 | 0.005 | 0.001 |
| SiPhyScore | 12.313 | 17.852 | 10.831 | 10.831 | 10.831 | 10.831 | 15.005 | 16.461 | 14.386 | 15.381 |
| gnomAD_AF All | 1/246220 = 0 | 1/102294 = 0 | 1/102294 = 0 | 1/102294 = 0 | 1/102294 = 0 | 1/246242 = 0 | 2/276962 = 0 | |||
| gnomAD_AF AMR | 0/33580 = 0 | 0/19614 = 0 | 0/19614 = 0 | 0/19614 = 0 | 0/19614 = 0 | 0/33578 = 0 | 0/34414 = 0 | |||
| gnomAD_AF AFR | 1/15304 = 0 | 1/2042 = 0 | 1/2042 = 0 | 1/2042 = 0 | 1/2042 = 0 | 0/15302 = 0 | 2/24024 = 0 | |||
| gnomAD_AF ASJ | 0/9848 = 0 | 0/6996 = 0 | 0/6996 = 0 | 0/6996 = 0 | 0/6996 = 0 | 0/9850 = 0 | 0/10142 = 0 | |||
| gnomAD_AF EAS | 0/17248 = 0 | 0/6594 = 0 | 0/6594 = 0 | 0/6594 = 0 | 0/6594 = 0 | 0/17248 = 0 | 0/18868 = 0 | |||
| gnomAD_AF FIN | 0/22284 = 0 | 0/6182 = 0 | 0/6182 = 0 | 0/6182 = 0 | 0/6182 = 0 | 0/22292 = 0 | 0/25718 = 0 | |||
| gnomAD_AF NFE | 0/111690 = 0 | 0/38366 = 0 | 0/38366 = 0 | 0/38366 = 0 | 0/38366 = 0 | 1/111704 = 0 | 0/126562 = 0 | |||
| gnomAD_AF OTH | 0/5484 = 0 | 0/2888 = 0 | 0/2888 = 0 | 0/2888 = 0 | 0/2888 = 0 | 0/5486 = 0 | 0/6456 = 0 | |||
| gnomAD_AF SAS | 0/30782 = 0 | 0/19612 = 0 | 0/19612 = 0 | 0/19612 = 0 | 0/19612 = 0 | 0/30782 = 0 | 0/30778 = 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manojlovic, Z.; Auslander, A.; Jin, Y.; Schmidt, R.J.; Xu, Y.; Chang, S.; Song, R.; Ingles, S.A.; Nunes, A.; Vavra, K.; et al. Genome Analysis Using Whole-Exome Sequencing of Non-Syndromic Cleft Lip and/or Palate from Malagasy Trios Identifies Variants Associated with Cilium-Related Pathways and Asian Genetic Ancestry. Genes 2023, 14, 665. https://doi.org/10.3390/genes14030665
Manojlovic Z, Auslander A, Jin Y, Schmidt RJ, Xu Y, Chang S, Song R, Ingles SA, Nunes A, Vavra K, et al. Genome Analysis Using Whole-Exome Sequencing of Non-Syndromic Cleft Lip and/or Palate from Malagasy Trios Identifies Variants Associated with Cilium-Related Pathways and Asian Genetic Ancestry. Genes. 2023; 14(3):665. https://doi.org/10.3390/genes14030665
Chicago/Turabian StyleManojlovic, Zarko, Allyn Auslander, Yuxin Jin, Ryan J. Schmidt, Yili Xu, Sharon Chang, Ruocen Song, Sue A. Ingles, Alana Nunes, KC Vavra, and et al. 2023. "Genome Analysis Using Whole-Exome Sequencing of Non-Syndromic Cleft Lip and/or Palate from Malagasy Trios Identifies Variants Associated with Cilium-Related Pathways and Asian Genetic Ancestry" Genes 14, no. 3: 665. https://doi.org/10.3390/genes14030665
APA StyleManojlovic, Z., Auslander, A., Jin, Y., Schmidt, R. J., Xu, Y., Chang, S., Song, R., Ingles, S. A., Nunes, A., Vavra, K., Feigelson, D., Rakotoarison, S., DiBona, M., Magee, K., Smile, O., Ramamonjisoa, A., & Magee III, W. (2023). Genome Analysis Using Whole-Exome Sequencing of Non-Syndromic Cleft Lip and/or Palate from Malagasy Trios Identifies Variants Associated with Cilium-Related Pathways and Asian Genetic Ancestry. Genes, 14(3), 665. https://doi.org/10.3390/genes14030665






