The Role of PRMT5 in Immuno-Oncology
Abstract
1. Introduction
2. Immuno-Oncology
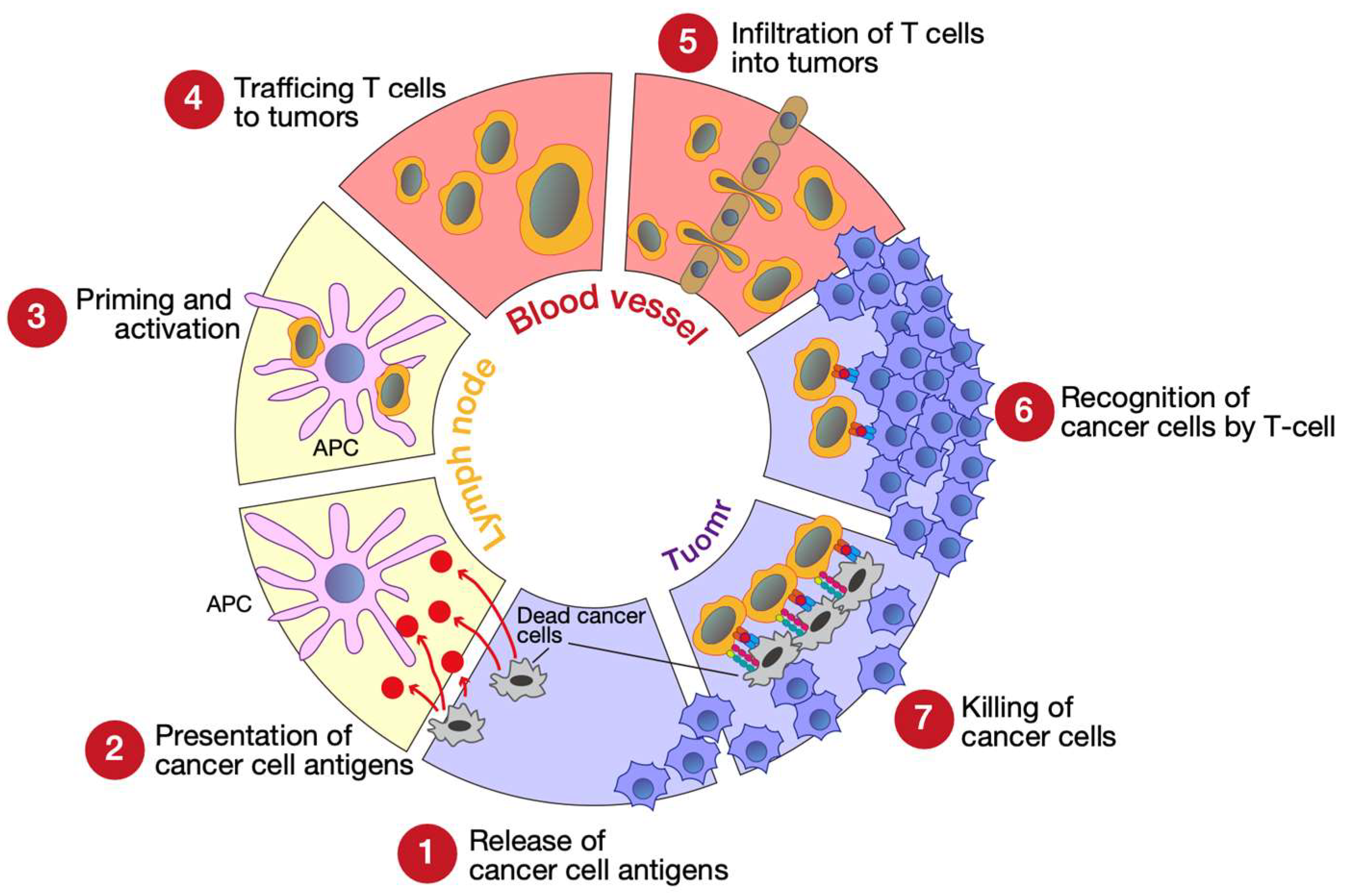
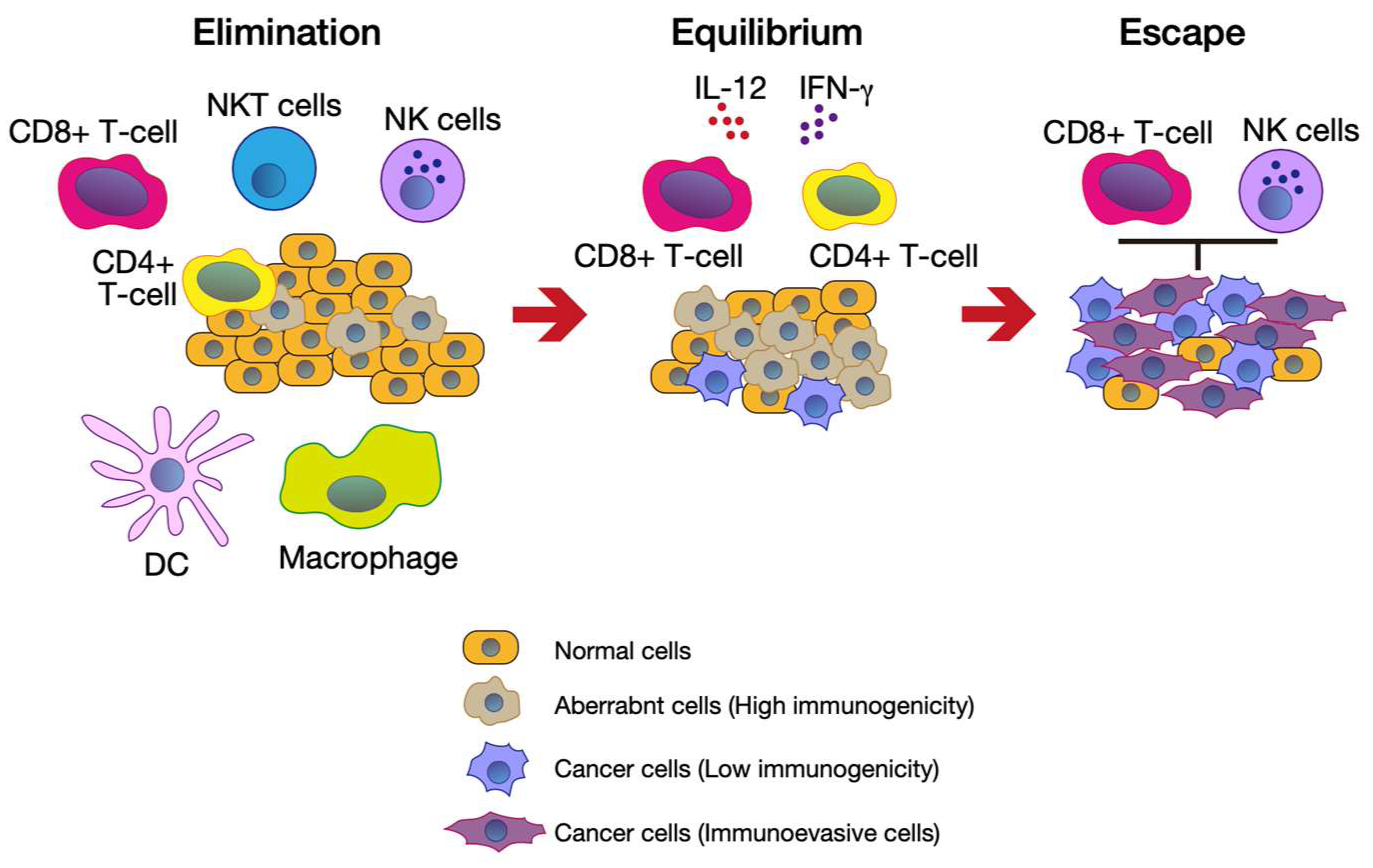
2.1. The Cancer-Immunity Cycle and Cancer Immunoediting
2.2. Immune Checkpoint Machinery–A Key Factor for Evading the Cancer-Immunity Cycle
3. Overview of PRMT5 Functions
3.1. PRMT Family
3.2. Overview of PRMT5
3.3. The Critical Role of PRMT5
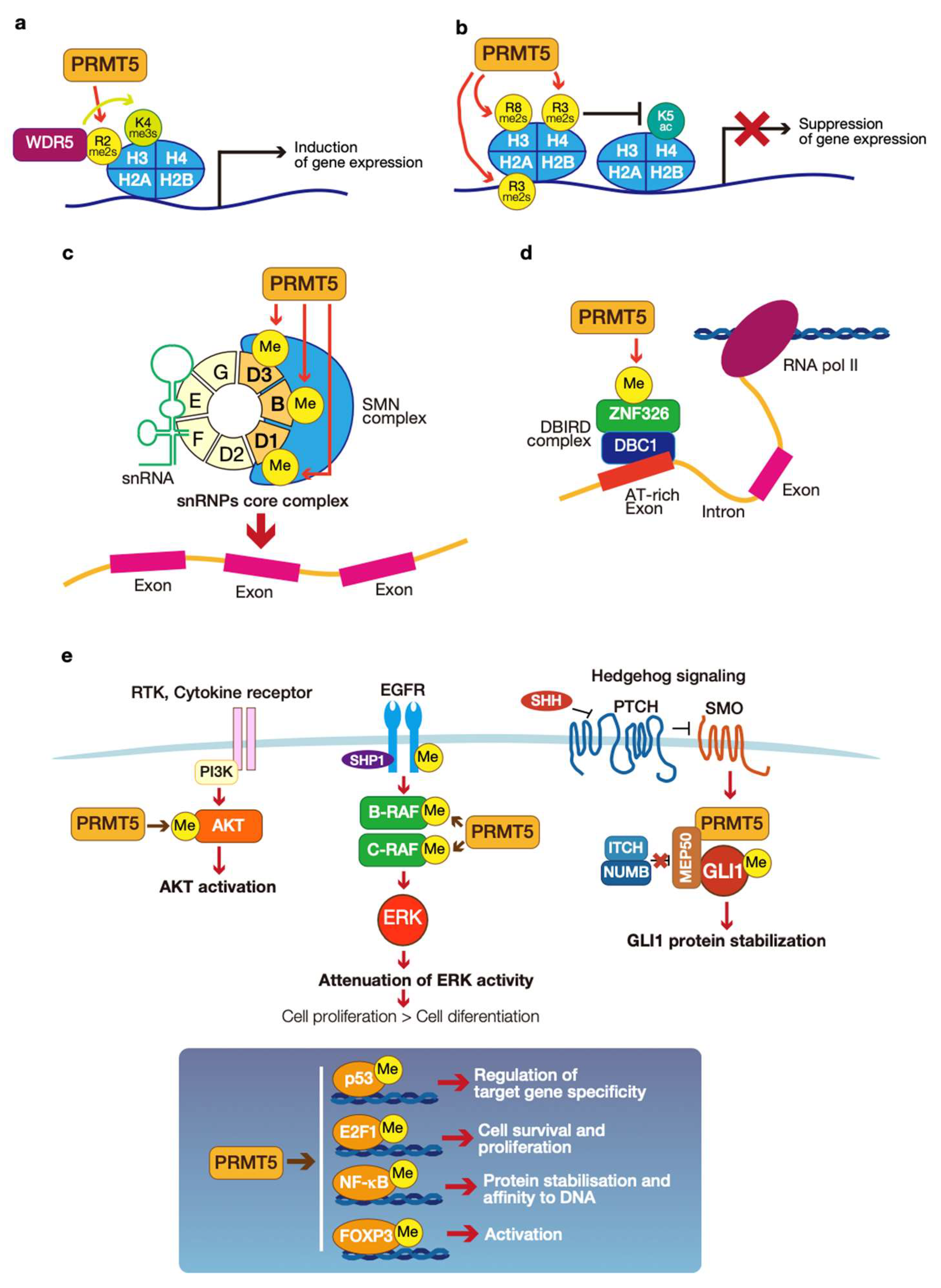
3.3.1. Epigenetic Regulation of Gene Expression
3.3.2. mRNA Splicing
3.3.3. Signal Transduction
4. The Role of PRMT5 in the Immune System
4.1. Roles of PRMT5 in T Cell Maintenance
4.2. Roles of PRMT5 in B Cell Development
5. The Roles of PRMT5 in Immuno-Oncology and Implication for Cancer Therapy Targeting PRMT5
5.1. PRMT5 as a Cancer Therapy Target
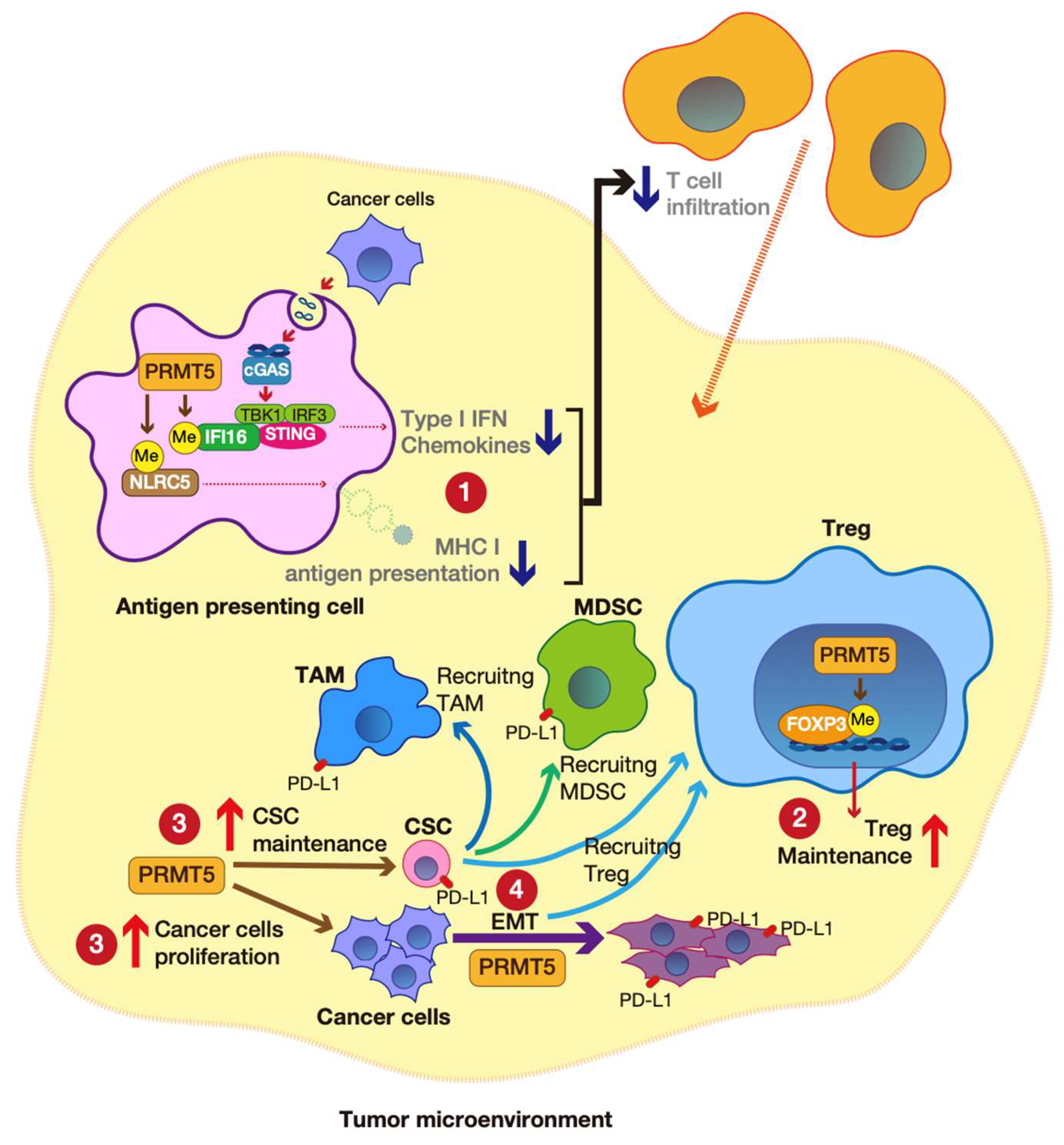
5.2. PRMT5-Mediated cGAS-STING Pathway
5.3. PRMT5-Mediated Treg Maintenance and Cancer
5.4. PRMT5-Mediated CSC Maintenance
5.5. PRMT5-Mediated EMT
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lengauer, C.; Kinzler, K.W.; Vogelstein, B. Genetic Instabilities in Human Cancers. Nature 1998, 396, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, H.; Lengauer, C. Aneuploidy and Cancer. Nature 2004, 432, 338–341. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.S.; Mellman, I. Oncology Meets Immunology: The Cancer-Immunity Cycle. Immunity 2013, 39, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, R.D.; Old, L.J.; Smyth, M.J. Cancer Immunoediting: Integrating Immunity’s Roles in Cancer Suppression and Promotion. Science 2011, 331, 1565–1570. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.; Koyasu, S. Cancer Immunoediting by Innate Lymphoid Cells. Trends Immunol. 2019, 40, 415–430. [Google Scholar] [CrossRef] [PubMed]
- Bayik, D.; Lathia, J.D. Cancer Stem Cell–Immune Cell Crosstalk in Tumour Progression. Nat. Rev. Cancer 2021, 21, 526–536. [Google Scholar] [CrossRef]
- Costa, A.; Kieffer, Y.; Scholer-Dahirel, A.; Pelon, F.; Bourachot, B.; Cardon, M.; Sirven, P.; Magagna, I.; Fuhrmann, L.; Bernard, C.; et al. Fibroblast Heterogeneity and Immunosuppressive Environment in Human Breast Cancer. Cancer Cell 2018, 33, 463–479.e10. [Google Scholar] [CrossRef]
- Sharma, P.; Siddiqui, B.A.; Anandhan, S.; Yadav, S.S.; Subudhi, S.K.; Gao, J.; Goswami, S.; Allison, J.P. The Next Decade of Immune Checkpoint Therapy. Cancer Discov. 2021, 11, 838–857. [Google Scholar] [CrossRef]
- Michot, J.M.; Bigenwald, C.; Champiat, S.; Collins, M.; Carbonnel, F.; Postel-Vinay, S.; Berdelou, A.; Varga, A.; Bahleda, R.; Hollebecque, A.; et al. Immune-Related Adverse Events with Immune Checkpoint Blockade: A Comprehensive Review. Eur. J. Cancer 2016, 54, 139–148. [Google Scholar] [CrossRef]
- Carbognin, L.; Pilotto, S.; Milella, M.; Vaccaro, V.; Brunelli, M.; Caliò, A.; Cuppone, F.; Sperduti, I.; Giannarelli, D.; Chilosi, M.; et al. Differential Activity of Nivolumab, Pembrolizumab and MPDL3280A According to the Tumor Expression of Programmed Death-Ligand-1 (PD-L1): Sensitivity Analysis of Trials in Melanoma, Lung and Genitourinary Cancers. PLoS ONE 2015, 10, e0130142. [Google Scholar] [CrossRef]
- Miyai, Y.; Sugiyama, D.; Hase, T.; Asai, N.; Taki, T.; Nishida, K.; Fukui, T.; Chen-Yoshikawa, T.F.; Kobayashi, H.; Mii, S.; et al. Meflin-Positive Cancer-Associated Fibroblasts Enhance Tumor Response to Immune Checkpoint Blockade. Life Sci. Alliance 2022, 5, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Ebert, P.J.R.; Cheung, J.; Yang, Y.; McNamara, E.; Hong, R.; Moskalenko, M.; Gould, S.E.; Maecker, H.; Irving, B.A.; Kim, J.M.; et al. MAP Kinase Inhibition Promotes T Cell and Anti-Tumor Activity in Combination with PD-L1 Checkpoint Blockade. Immunity 2016, 44, 609–621. [Google Scholar] [CrossRef] [PubMed]
- Loi, S.; Dushyanthen, S.; Beavis, P.A.; Salgado, R.; Denkert, C.; Savas, P.; Combs, S.; Rimm, D.L.; Giltnane, J.M.; Estrada, M.V.; et al. RAS/MAPK Activation Is Associated with Reduced Tumor-Infiltrating Lymphocytes in Triple-Negative Breast Cancer: Therapeutic Cooperation Between MEK and PD-1/PD-L1 Immune Checkpoint Inhibitors. Clin. Cancer Res. 2016, 22, 1499–1509. [Google Scholar] [CrossRef]
- Bedford, M.T.; Richard, S. Arginine Methylation: An Emerging Regulator of Protein Function. Mol. Cell 2005, 18, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Inoue, M.; Okamoto, K.; Terashima, A.; Nitta, T.; Muro, R.; Negishi-Koga, T.; Kitamura, T.; Nakashima, T.; Takayanagi, H. Arginine Methylation Controls the Strength of Γc-Family Cytokine Signaling in T Cell Maintenance. Nat. Immunol. 2018, 19, 1265–1276. [Google Scholar] [CrossRef]
- Litzler, L.C.; Zahn, A.; Meli, A.P.; Hébert, S.; Patenaude, A.-M.; Methot, S.P.; Sprumont, A.; Bois, T.; Kitamura, D.; Costantino, S.; et al. PRMT5 Is Essential for B Cell Development and Germinal Center Dynamics. Nat. Commun. 2019, 10, 22. [Google Scholar] [CrossRef]
- Yang, Y.; Bedford, M.T. Protein Arginine Methyltransferases and Cancer. Nat. Rev. Cancer 2013, 13, 37–50. [Google Scholar] [CrossRef]
- Jarrold, J.; Davies, C.C. PRMTs and Arginine Methylation: Cancer’s Best-Kept Secret? Trends Mol. Med. 2019, 25, 993–1009. [Google Scholar] [CrossRef]
- Chen, L.; Flies, D.B. Molecular Mechanisms of T Cell Co-Stimulation and Co-Inhibition. Nat. Rev. Immunol. 2013, 13, 227–242. [Google Scholar] [CrossRef]
- Krummel, M.F.; Allison, J.P. CD28 and CTLA-4 Have Opposing Effects on the Response of T Cells to Stimulation. J. Exp. Med. 1995, 182, 459–465. [Google Scholar] [CrossRef]
- Lanitis, E.; Irving, M.; Coukos, G. Targeting the Tumor Vasculature to Enhance T Cell Activity. Curr. Opin. Immunol. 2015, 33, 55–63. [Google Scholar] [CrossRef]
- Chen, L. Co-Inhibitory Molecules of the B7-CD28 Family in the Control of T-Cell Immunity. Nat. Rev. Immunol. 2004, 4, 336–347. [Google Scholar] [CrossRef]
- Zou, W.; Chen, L. Inhibitory B7-Family Molecules in the Tumour Microenvironment. Nat. Rev. Immunol. 2008, 8, 467–477. [Google Scholar] [CrossRef]
- Maruhashi, T.; Sugiura, D.; Okazaki, I.-M.; Okazaki, T. LAG-3: From Molecular Functions to Clinical Applications. J. Immunother. Cancer 2020, 8, e001014. [Google Scholar] [CrossRef]
- Zhao, L.; Cheng, S.; Fan, L.; Zhang, B.; Xu, S. TIM-3: An Update on Immunotherapy. Int. Immunopharmacol. 2021, 99, 107933. [Google Scholar] [CrossRef]
- Blanc, R.S.; Richard, S. Arginine Methylation: The Coming of Age. Mol. Cell 2017, 65, 8–24. [Google Scholar] [CrossRef]
- Guccione, E.; Richard, S. The Regulation, Functions and Clinical Relevance of Arginine Methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 642–657. [Google Scholar] [CrossRef]
- Jain, K.; Clarke, S.G. PRMT7 as a Unique Member of the Protein Arginine Methyltransferase Family: A Review. Arch. Biochem. Biophys. 2019, 665, 36–45. [Google Scholar] [CrossRef]
- Kumar, S.; Zeng, Z.; Bagati, A.; Tay, R.E.; Sanz, L.A.; Hartono, S.R.; Ito, Y.; Abderazzaq, F.; Hatchi, E.; Jiang, P.; et al. Carm1 Inhibition Enables Immunotherapy of Resistant Tumors by Dual Action on Tumor Cells and t Cells. Cancer Discov. 2021, 11, 2050–2071. [Google Scholar] [CrossRef]
- Srour, N.; Villarreal, O.D.; Hardikar, S.; Yu, Z.; Preston, S.; Miller, W.H.; Szewczyk, M.M.; Barsyte-Lovejoy, D.; Xu, H.; Chen, T.; et al. PRMT7 Ablation Stimulates Anti-Tumor Immunity and Sensitizes Melanoma to Immune Checkpoint Blockade. Cell Rep. 2022, 38, 110582. [Google Scholar] [CrossRef]
- Abe, Y.; Tanaka, N. Fine-Tuning of GLI Activity through Arginine Methylation: Its Mechanisms and Function. Cells 2020, 9, 1973. [Google Scholar] [CrossRef] [PubMed]
- Pollack, B.P.; Kotenko, S.V.; He, W.; Izotova, L.S.; Barnoski, B.L.; Pestka, S. The Human Homologue of the Yeast Proteins Skb1 and Hsl7p Interacts with Jak Kinases and Contains Protein Methyltransferase Activity. J. Biol. Chem. 1999, 274, 31531–31542. [Google Scholar] [CrossRef]
- Friesen, W.J.; Wyce, A.; Paushkin, S.; Abel, L.; Rappsilber, J.; Mann, M.; Dreyfuss, G. A Novel WD Repeat Protein Component of the Methylosome Binds Sm Proteins. J. Biol. Chem. 2002, 277, 8243–8247. [Google Scholar] [CrossRef]
- Hosohata, K.; Li, P.; Hosohata, Y.; Qin, J.; Roeder, R.G.; Wang, Z. Purification and Identification of a Novel Complex Which Is Involved in Androgen Receptor-Dependent Transcription. Mol. Cell Biol. 2003, 23, 7019–7029. [Google Scholar] [CrossRef]
- Abe, Y.; Suzuki, Y.; Kawamura, K.; Tanaka, N. MEP50/PRMT5-Mediated Methylation Activates GLI1 in Hedgehog Signalling through Inhibition of Ubiquitination by the ITCH/NUMB Complex. Commun. Biol. 2019, 2, 23. [Google Scholar] [CrossRef] [PubMed]
- Guderian, G.; Peter, C.; Wiesner, J.; Sickmann, A.; Schulze-Osthoff, K.; Fischer, U.; Grimmler, M. RioK1, a New Interactor of Protein Arginine Methyltransferase 5 (PRMT5), Competes with PICln for Binding and Modulates PRMT5 Complex Composition and Substrate Specificity. J. Biol. Chem. 2011, 286, 1976–1986. [Google Scholar] [CrossRef]
- Meister, G.; Eggert, C.; Bühler, D.; Brahms, H.; Kambach, C.; Fischer, U. Methylation of Sm Proteins by a Complex Containing PRMT5 and the Putative U SnRNP Assembly Factor PICln. Curr. Biol. 2001, 11, 1990–1994. [Google Scholar] [CrossRef]
- Scaglione, A.; Patzig, J.; Liang, J.; Frawley, R.; Bok, J.; Mela, A.; Yattah, C.; Zhang, J.; Teo, S.X.; Zhou, T.; et al. PRMT5-Mediated Regulation of Developmental Myelination. Nat. Commun. 2018, 9, 2840. [Google Scholar] [CrossRef]
- Pal, S.; Vishwanath, S.N.; Erdjument-Bromage, H.; Tempst, P.; Sif, S. Human SWI/SNF-Associated PRMT5 Methylates Histone H3 Arginine 8 and Negatively Regulates Expression of ST7 and NM23 Tumor Suppressor Genes. Mol. Cell Biol. 2004, 24, 9630–9645. [Google Scholar] [CrossRef]
- Migliori, V.; Müller, J.; Phalke, S.; Low, D.; Bezzi, M.; Mok, W.C.; Sahu, S.K.; Gunaratne, J.; Capasso, P.; Bassi, C.; et al. Symmetric Dimethylation of H3R2 Is a Newly Identified Histone Mark That Supports Euchromatin Maintenance. Nat. Struct. Mol. Biol. 2012, 19, 136–144. [Google Scholar] [CrossRef]
- Meister, G.; Fischer, U. Assisted RNP Assembly: SMN and PRMT5 Complexes Cooperate in the Formation of Spliceosomal UsnRNPs. EMBO J. 2002, 21, 5853–5863. [Google Scholar] [CrossRef]
- Matera, A.G.; Terns, R.M.; Terns, M.P. Non-Coding RNAs: Lessons from the Small Nuclear and Small Nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007, 8, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Close, P.; East, P.; Dirac-Svejstrup, A.B.; Hartmann, H.; Heron, M.; Maslen, S.; Chariot, A.; Söding, J.; Skehel, M.; Svejstrup, J.Q. DBIRD Complex Integrates Alternative MRNA Splicing with RNA Polymerase II Transcript Elongation. Nature 2012, 484, 386–389. [Google Scholar] [CrossRef] [PubMed]
- Rengasamy, M.; Zhang, F.; Vashisht, A.; Song, W.-M.; Aguilo, F.; Sun, Y.; Li, S.; Zhang, W.; Zhang, B.; Wohlschlegel, J.A.; et al. The PRMT5/WDR77 Complex Regulates Alternative Splicing through ZNF326 in Breast Cancer. Nucleic Acids Res. 2017, 45, 11106–11120. [Google Scholar] [CrossRef] [PubMed]
- Hsu, J.-M.; Chen, C.-T.; Chou, C.-K.; Kuo, H.-P.; Li, L.-Y.; Lin, C.-Y.; Lee, H.-J.; Wang, Y.-N.; Liu, M.; Liao, H.-W.; et al. Crosstalk between Arg 1175 Methylation and Tyr 1173 Phosphorylation Negatively Modulates EGFR-Mediated ERK Activation. Nat. Cell Biol. 2011, 13, 174–181. [Google Scholar] [CrossRef]
- Sapir, T.; Shifteh, D.; Pahmer, M.; Goel, S.; Maitra, R. Protein Arginine Methyltransferase 5 (PRMT5) and the ERK1/2 & PI3K Pathways: A Case for PRMT5 Inhibition and Combination Therapies in Cancer. Mol. Cancer Res. 2021, 19, 388–394. [Google Scholar] [CrossRef] [PubMed]
- Yin, S.; Liu, L.; Brobbey, C.; Palanisamy, V.; Ball, L.E.; Olsen, S.K.; Ostrowski, M.C.; Gan, W. PRMT5-Mediated Arginine Methylation Activates AKT Kinase to Govern Tumorigenesis. Nat. Commun. 2021, 12, 1–13. [Google Scholar] [CrossRef]
- Jansson, M.; Durant, S.T.; Cho, E.-C.; Sheahan, S.; Edelmann, M.; Kessler, B.; la Thangue, N.B. Arginine Methylation Regulates the P53 Response. Nat. Cell Biol. 2008, 10, 1431–1439. [Google Scholar] [CrossRef]
- Zheng, S.; Moehlenbrink, J.; Lu, Y.C.; Zalmas, L.P.; Sagum, C.A.; Carr, S.; McGouran, J.F.; Alexander, L.; Fedorov, O.; Munro, S.; et al. Arginine Methylation-Dependent Reader-Writer Interplay Governs Growth Control by E2F-1. Mol. Cell 2013, 52, 37–51. [Google Scholar] [CrossRef]
- Wei, H.; Wang, B.; Miyagi, M.; She, Y.; Gopalan, B.; Huang, D.-B.; Ghosh, G.; Stark, G.R.; Lu, T. PRMT5 Dimethylates R30 of the P65 Subunit to Activate NF-ΚB. Proc. Natl. Acad. Sci. USA 2013, 110, 13516–13521. [Google Scholar] [CrossRef]
- Harris, D.P.; Bandyopadhyay, S.; Maxwell, T.J.; Willard, B.; DiCorleto, P.E. Tumor Necrosis Factor (TNF)-α Induction of CXCL10 in Endothelial Cells Requires Protein Arginine Methyltransferase 5 (PRMT5)-Mediated Nuclear Factor (NF)-ΚB P65 Methylation. J. Biol. Chem. 2014, 289, 15328–15339. [Google Scholar] [CrossRef] [PubMed]
- Nagai, Y.; Ji, M.Q.; Zhu, F.; Xiao, Y.; Tanaka, Y.; Kambayashi, T.; Fujimoto, S.; Goldberg, M.M.; Zhang, H.; Li, B.; et al. PRMT5 Associates with the FoXP3 Homomer and When Disabled Enhances Targeted P185ErbB2/Neu Tumor Immunotherapy. Front. Immunol. 2019, 10, 174. [Google Scholar] [CrossRef]
- Kim, H.; Kim, H.; Feng, Y.; Li, Y.; Tamiya, H.; Tocci, S.; Ronai, Z.A. PRMT5 Control of CGAS/STING and NLRC5 Pathways Defines Melanoma Response to Antitumor Immunity. Sci. Transl. Med. 2020, 12, eaaz5683. [Google Scholar] [CrossRef] [PubMed]
- Boyman, O.; Sprent, J. The Role of Interleukin-2 during Homeostasis and Activation of the Immune System. Nat. Rev. Immunol. 2012, 12, 180–190. [Google Scholar] [CrossRef] [PubMed]
- Richard, S.; Morel, M.; Cléroux, P. Arginine Methylation Regulates IL-2 Gene Expression: A Role for Protein Arginine Methyltransferase 5 (PRMT5). Biochem. J. 2005, 388, 379–386. [Google Scholar] [CrossRef]
- Leonard, W.J.; Lin, J.X.; O’Shea, J.J. The γ c Family of Cytokines: Basic Biology to Therapeutic Ramifications. Immunity 2019, 50, 832–850. [Google Scholar] [CrossRef]
- De Silva, N.S.; Klein, U. Dynamics of B Cells in Germinal Centres. Nat. Rev. Immunol. 2015, 15, 137–148. [Google Scholar] [CrossRef]
- Gerhart, S.V.; Kellner, W.A.; Thompson, C.; Pappalardi, M.B.; Zhang, X.-P.; Montes de Oca, R.; Penebre, E.; Duncan, K.; Boriack-Sjodin, A.; Le, B.; et al. Activation of the P53-MDM4 Regulatory Axis Defines the Anti-Tumour Response to PRMT5 Inhibition through Its Role in Regulating Cellular Splicing. Sci. Rep. 2018, 8, 9711. [Google Scholar] [CrossRef]
- Legube, G.; Linares, L.K.; Tyteca, S.; Caron, C.; Scheffner, M.; Chevillard-Briet, M.; Trouche, D. Role of the Histone Acetyl Transferase Tip60 in the P53 Pathway. J. Biol. Chem. 2004, 279, 44825–44833. [Google Scholar] [CrossRef]
- Ding, G.; Liu, H.-D.; Huang, Q.; Liang, H.-X.; Ding, Z.-H.; Liao, Z.-J.; Huang, G. HDAC6 Promotes Hepatocellular Carcinoma Progression by Inhibiting P53 Transcriptional Activity. FEBS Lett. 2013, 587, 880–886. [Google Scholar] [CrossRef]
- Shailesh, H.; Zakaria, Z.Z.; Baiocchi, R.; Sif, S. Protein Arginine Methyltransferase 5 (PRMT5) Dysregulation in Cancer. Oncotarget 2018, 9, 36705–36718. [Google Scholar] [CrossRef]
- Feustel, K.; Falchook, G.S. Protein Arginine Methyltransferase 5 (PRMT5) Inhibitors in Oncology Clinical Trials: A Review. J. Immunother. Precis. Oncol. 2022, 5, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Stopa, N.; Krebs, J.E.; Shechter, D. The PRMT5 Arginine Methyltransferase: Many Roles in Development, Cancer and Beyond. Cell. Mol. Life Sci. 2015, 72, 2041–2059. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Yang, J.; Adair, S.J.; Ozturk, H.; Kuscu, C.; Lee, K.Y.; Kane, W.J.; O’Hara, P.E.; Liu, D.; Demirlenk, Y.M.; et al. Targeted CRISPR Screening Identifies PRMT5 as Synthetic Lethality Combinatorial Target with Gemcitabine in Pancreatic Cancer Cells. Proc. Natl. Acad. Sci. USA 2020, 117, 28068–28079. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Yan, H.; Wu, D.; Zhao, Z.; Chen, X.; Long, Q.; Zhang, C.; Wang, X.; Deng, W.; Liu, X. PRMT5/Wnt4 Axis Promotes Lymph-Node Metastasis and Proliferation of Laryngeal Carcinoma. Cell Death Dis. 2020, 11, 864. [Google Scholar] [CrossRef]
- Chan-Penebre, E.; Kuplast, K.G.; Majer, C.R.; Boriack-Sjodin, P.A.; Wigle, T.J.; Johnston, L.D.; Rioux, N.; Munchhof, M.J.; Jin, L.; Jacques, S.L.; et al. A Selective Inhibitor of PRMT5 with in Vivo and in Vitro Potency in MCL Models. Nat. Chem. Biol. 2015, 11, 432–437. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, Z.-H.; Savarese, T.M. Codeletion of the Genes for P16INK4, Methylthioadenosine Phosphorylase, Interferon-A1, Interferon-Β1, and Other 9p21 Markers in Human Malignant Cell Lines. Cancer Genet. Cytogenet 1996, 86, 22–28. [Google Scholar] [CrossRef]
- Kryukov, G.V.; Kryukov, G.V.; Wilson, F.H.; Ruth, J.R.; Paulk, J.; Tsherniak, A.; Marlow, S.E.; Vazquez, F.; Weir, B.A.; Fitzgerald, M.E.; et al. MTAP Deletion Confers Enhanced Dependency on the PRMT5 Arginine Methyltransferase in Cancer Cells. Science 2016, 351, 1214–1218. [Google Scholar] [CrossRef]
- Marjon, K.; Cameron, M.J.; Quang, P.; Clasquin, M.F.; Mandley, E.; Kunii, K.; McVay, M.; Choe, S.; Kernytsky, A.; Gross, S.; et al. MTAP Deletions in Cancer Create Vulnerability to Targeting of the MAT2A/PRMT5/RIOK1 Axis. Cell Rep. 2016, 15, 574–587. [Google Scholar] [CrossRef]
- Cai, X.; Chiu, Y.-H.; Chen, Z.J. The CGAS-CGAMP-STING Pathway of Cytosolic DNA Sensing and Signaling. Mol. Cell 2014, 54, 289–296. [Google Scholar] [CrossRef]
- Almine, J.F.; O’Hare, C.A.J.; Dunphy, G.; Haga, I.R.; Naik, R.J.; Atrih, A.; Connolly, D.J.; Taylor, J.; Kelsall, I.R.; Bowie, A.G.; et al. IFI16 and CGAS Cooperate in the Activation of STING during DNA Sensing in Human Keratinocytes. Nat. Commun. 2017, 8, 14392. [Google Scholar] [CrossRef] [PubMed]
- Zou, W. Regulatory T Cells, Tumour Immunity and Immunotherapy. Nat. Rev. Immunol. 2006, 6, 295–307. [Google Scholar] [CrossRef] [PubMed]
- Hori, S.; Nomura, T.; Sakaguchi, S. Control of Regulatory T Cell Development by the Transcription Factor Foxp3. Science 2003, 299, 1057–1061. [Google Scholar] [CrossRef]
- Fontenot, J.D.; Gavin, M.A.; Rudensky, A.Y. Foxp3 Programs the Development and Function of CD4+ CD25+ Regulatory T Cells. Nat. Immunol. 2003, 4, 330–336. [Google Scholar] [CrossRef]
- Nishikawa, H.; Sakaguchi, S. Regulatory T Cells in Tumor Immunity. Int. J. Cancer 2010, 127, 759–767. [Google Scholar] [CrossRef]
- Smil, D.; Eram, M.S.; Li, F.; Kennedy, S.; Szewczyk, M.M.; Brown, P.J.; Barsyte-Lovejoy, D.; Arrowsmith, C.H.; Vedadi, M.; Schapira, M. Discovery of a Dual PRMT5–PRMT7 Inhibitor. ACS Med. Chem. Lett. 2015, 6, 408–412. [Google Scholar] [CrossRef]
- Batlle, E.; Clevers, H. Cancer Stem Cells Revisited. Nat. Med. 2017, 23, 1124–1134. [Google Scholar] [CrossRef]
- Miao, Y.; Yang, H.; Levorse, J.; Yuan, S.; Polak, L.; Sribour, M.; Singh, B.; Rosenblum, M.D.; Fuchs, E. Adaptive Immune Resistance Emerges from Tumor-Initiating Stem Cells. Cell 2019, 177, 1172–1186.e14. [Google Scholar] [CrossRef]
- Chen, M.; Sharma, A.; Lin, Y.; Wu, Y.; He, Q.; Gu, Y.; Xu, Z.P.; Monteiro, M.; Gu, W. Insluin and Epithelial Growth Factor (EGF) Promote Programmed Death Ligand 1(PD-L1) Production and Transport in Colon Cancer Stem Cells. BMC Cancer 2019, 19, 153. [Google Scholar] [CrossRef]
- Mansour, F.A.; Al-Mazrou, A.; Al-Mohanna, F.; Al-Alwan, M.; Ghebeh, H. PD-L1 Is Overexpressed on Breast Cancer Stem Cells through Notch3/MTOR Axis. Oncoimmunology 2020, 9, 1729299. [Google Scholar] [CrossRef]
- Chiang, K.; Zielinska, A.E.; Shaaban, A.M.; Sanchez-Bailon, M.P.; Jarrold, J.; Clarke, T.L.; Zhang, J.; Francis, A.; Jones, L.J.; Smith, S.; et al. PRMT5 Is a Critical Regulator of Breast Cancer Stem Cell Function via Histone Methylation and FOXP1 Expression. Cell Rep. 2017, 21, 3498–3513. [Google Scholar] [CrossRef]
- Jin, Y.; Zhou, J.; Xu, F.; Jin, B.; Cui, L.; Wang, Y.; Du, X.; Li, J.; Li, P.; Ren, R.; et al. Targeting Methyltransferase PRMT5 Eliminates Leukemia Stem Cells in Chronic Myelogenous Leukemia. J. Clin. Investig. 2016, 126, 3961–3980. [Google Scholar] [CrossRef]
- Gu, Z.; Gao, S.; Zhang, F.; Wang, Z.; Ma, W.; Davis, R.E.; Wang, Z. Protein Arginine Methyltransferase 5 Is Essential for Growth of Lung Cancer Cells. Biochem. J. 2012, 446, 235–241. [Google Scholar] [CrossRef]
- Iwatsuki, M.; Mimori, K.; Yokobori, T.; Ishi, H.; Beppu, T.; Nakamori, S.; Baba, H.; Mori, M. Epithelial-Mesenchymal Transition in Cancer Development and Its Clinical Significance. Cancer Sci. 2010, 101, 293–299. [Google Scholar] [CrossRef]
- Bailey, J.M.; Singh, P.K.; Hollingsworth, M. a Cancer Metastasis Facilitated by Developmental Pathways: Sonic Hedgehog, Notch, and Bone Morphogenic Proteins. J. Cell Biochem. 2007, 102, 829–839. [Google Scholar] [CrossRef]
- Ally, M.S.; Ransohoff, K.; Sarin, K.; Atwood, S.X.; Rezaee, M.; Bailey-Healy, I.; Kim, J.; Beachy, P.A.; Chang, A.L.S.; Oro, A.; et al. Effects of Combined Treatment With Arsenic Trioxide and Itraconazole in Patients With Refractory Metastatic Basal Cell Carcinoma. JAMA Dermatol. 2016, 152, 452. [Google Scholar] [CrossRef]
- Gentile, A.; Trusolino, L.; Comoglio, P.M. The Met Tyrosine Kinase Receptor in Development and Cancer. Cancer Metastasis Rev. 2008, 27, 85–94. [Google Scholar] [CrossRef]
- Sheng, W.; Shi, X.; Lin, Y.; Tang, J.; Jia, C.; Cao, R.; Sun, J.; Wang, G.; Zhou, L.; Dong, M. Musashi2 Promotes EGF-Induced EMT in Pancreatic Cancer via ZEB1-ERK/MAPK Signaling. J. Exp. Clin. Cancer Res. 2020, 39, 16. [Google Scholar] [CrossRef]
- Plaschka, M.; Benboubker, V.; Grimont, M.; Berthet, J.; Tonon, L.; Lopez, J.; Le-Bouar, M.; Balme, B.; Tondeur, G.; de la Fouchardière, A.; et al. ZEB1 Transcription Factor Promotes Immune Escape in Melanoma. J. Immunother. Cancer 2022, 10, e003484. [Google Scholar] [CrossRef]
- Canè, S.; van Snick, J.; Uyttenhove, C.; Pilotte, L.; van den Eynde, B.J. TGFβ1 Neutralization Displays Therapeutic Efficacy through Both an Immunomodulatory and a Non-Immune Tumor-Intrinsic Mechanism. J. Immunother. Cancer 2021, 9, e001798. [Google Scholar] [CrossRef]
- Taki, M.; Abiko, K.; Ukita, M.; Murakami, R.; Yamanoi, K.; Yamaguchi, K.; Hamanishi, J.; Baba, T.; Matsumura, N.; Mandai, M. Tumor Immune Microenvironment during Epithelial–Mesenchymal Transition. Clin. Cancer Res. 2021, 27, 4669–4679. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Massi, D.; Hemmings, B.A.; Mandalà, M.; Hu, Z.; Wicki, A.; Xue, G. AKT-Ions with a TWIST between EMT and MET. Oncotarget 2016, 7, 62767–62777. [Google Scholar] [CrossRef]
- Zhou, B.P.; Deng, J.; Xia, W.; Xu, J.; Li, Y.M.; Gunduz, M.; Hung, M.-C. Dual Regulation of Snail by GSK-3beta-Mediated Phosphorylation in Control of Epithelial-Mesenchymal Transition. Nat. Cell Biol. 2004, 6, 931–940. [Google Scholar] [CrossRef] [PubMed]
- Xue, G.; Restuccia, D.F.; Lan, Q.; Hynx, D.; Dirnhofer, S.; Hess, D.; Rüegg, C.; Hemmings, B.A. Akt/PKB-Mediated Phosphorylation of Twist1 Promotes Tumor Metastasis via Mediating Cross-Talk between PI3K/Akt and TGF-β Signaling Axes. Cancer Discov. 2012, 2, 248–259. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Zhang, X.-O.; Rozen, E.J.; Sun, X.; Sallis, B.; Verdejo-Torres, O.; Wigglesworth, K.; Moon, D.; Huang, T.; Cavaretta, J.P.; et al. PRMT5 Activates AKT via Methylation to Promote Tumor Metastasis. Nat. Commun. 2022, 13, 3955. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Liu, R.; Feng, D.; Huang, W.; Huo, M.; Zhang, J.; Leng, S.; Yang, Y.; Yang, T.; Yin, X.; et al. Snail/PRMT5/NuRD Complex Contributes to DNA Hypermethylation in Cervical Cancer by TET1 Inhibition. Cell Death Differ. 2021, 28, 2818–2836. [Google Scholar] [CrossRef]
- Zheng, Y.; Dai, M.; Dong, Y.; Yu, H.; Liu, T.; Feng, X.; Yu, B.; Zhang, H.; Wu, J.; Kong, W.; et al. ZEB2/TWIST1/PRMT5/NuRD Multicomplex Contributes to the Epigenetic Regulation of EMT and Metastasis in Colorectal Carcinoma. Cancers 2022, 14, 3426. [Google Scholar] [CrossRef]

| PRMT5 Overexpression | Effect of PRMT5 Overexpression |
| Lung cancer | Larger tumor size, advanced tumor grade, lymph node metastasis, worse survival |
| Breast cancer | Larger tumor size, advanced tumor grade, lymph node metastasis |
| Hepatocellular carcinoma | Larger tumor size, advanced tumor grade |
| Glioblastoma multiforme | Larger tumor size, advanced tumor grade |
| Pancreatic cancer | Worse survival |
| Laryngeal carcinoma | Worse survival |
| Gastric cancer | Lymph node metastasis |
| Head and neck cancer | Lymph node metastasis |
| Colorectal cancer | Lymph node metastasis |
| Ovarian caner | Lymph node metastasis |
| Bladder cancer | Lymph node metastasis |
| Melanoma | Metastasis |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abe, Y.; Sano, T.; Tanaka, N. The Role of PRMT5 in Immuno-Oncology. Genes 2023, 14, 678. https://doi.org/10.3390/genes14030678
Abe Y, Sano T, Tanaka N. The Role of PRMT5 in Immuno-Oncology. Genes. 2023; 14(3):678. https://doi.org/10.3390/genes14030678
Chicago/Turabian StyleAbe, Yoshinori, Takumi Sano, and Nobuyuki Tanaka. 2023. "The Role of PRMT5 in Immuno-Oncology" Genes 14, no. 3: 678. https://doi.org/10.3390/genes14030678
APA StyleAbe, Y., Sano, T., & Tanaka, N. (2023). The Role of PRMT5 in Immuno-Oncology. Genes, 14(3), 678. https://doi.org/10.3390/genes14030678






