1. Introduction
Non-communicable diseases (NCDs) are becoming the leading cause of mortality, disability, decreased quality of life, and growing healthcare expenses throughout the world [
1]. The global mortality rate from these causes is double that of infectious illnesses, and nutritional deficiencies combined [
2]. The most frequent among them include cardiovascular diseases (CVDs) are diabetes, malignancies, and chronic respiratory disorders [
3]. They are the major cause of mortality in industrialized countries and the second largest cause in developing and underdeveloped nations, accounting for about 74.4% of all fatalities in 2019, which increased by 20.5% from 2009 to 2019 [
4].
Coronary Artery Disease (CAD) is a class of CVD which include Myocardial Infarction (MI), hypertension (HTN), and congenital heart disease. Pathology defines MI as myocardial cell loss caused by persistent ischemia [
5]. MI is well-known throughout the world for its high mortality and disability rates and is one of the top 10 causes of death in Pakistan [
2,
6]. There were 4 million MI-related fatalities between 1999 and 2019 [
7].
Lifestyle, environmental, and genetic factors have a key role in the development of CVD [
8]. Among the former, some major factors to mention are physical activity, diet, smoking habits, obesity, etc. About the latter, information from different studies has recommended a 40–80% genetic link [
9,
10]. A person with parental history of premature atherosclerosis has a 1.5- to 2-fold risk of developing the same [
11]. Risk can easily be predicted through a better understanding of the genetic components [
12,
13]. In the last decades, human genetics approaches have recognized genes that have possible contributions towards developing MI [
14]. Some of these include variants of different genes such as
Apolipoprotein (
APOE),
Paroxonase 1 (
PON1),
Cytochrome P4501A1 (
CYP1A1),
interleukin-6,
Cholesteryl Ester Transfer Protein (
CETP) and many others [
15,
16,
17,
18,
19].
One of the major cause of the development of CAD is Atherosclerosis which is a pathological procedure in which lipid is accumulated in the intima and media of the blood vessel and thus leads to the formation of plaques [
20]. Both youth and adolescents may develop coronary atherosclerosis [
21]. Abnormalities in two proteins, namely
APOE and human
PON1, play an important role in its development [
22,
23]. APOE is a serum glycoprotein that plays an important role in the transport and metabolism of lipids and is encoded by the
APOE gene, which is located on chromosome 19. Exon 4 of the gene has two common SNPs: rs429358 (388 T > C) and rs7412 (526 C > T). Moreover, three alleles (ɛ2(388 T–526 T), ɛ3(388 T-526C), ɛ4(388C-526C)) and six genotypes (ɛ2/ɛ2, ɛ2/ɛ3, ɛ2/ɛ4, ɛ3/ɛ3, ɛ3/ɛ4 and ɛ4/ɛ4) can be formed by the two SNPs [
24,
25]. Since allele 3 is the most prevalent in populations, it is referred to as “wild-type.”The alleles 2 and 4 are considered variants [
26]. Various studies have reported their association with MI in different ethnicities such as Chinese and Russian etc. [
27,
28]. Similarly, PON1 is a membrane-bound glycoprotein encoded by the
PON1 gene that is located on chromosome 7q21.3-q22.1 [
29,
30]. It is associated with highly-density lipoprotein (HDL) and is found in a variety of tissues but is predominantly synthesized in the liver [
31]. It inhibits the concentration of low-density lipoprotein cholesterol (LDL-C) by hydrolysis of lipid peroxides [
31]. PON1 has considerable anti-inflammatory and anti-oxidative actions through its enzymatic Paroxonase, lactonase, and esterase activities [
29]. There is some evidence of low serum PON1 activity in patients with lipid disorders such as diabetes mellitus (DM), MI, atherosclerosis, and familial hypercholesterolemia [
30].
PON1 gene has two common polymorphisms, namely L55M and Q192R, of which L55L and Q192Q are regarded as wild type, and Q192R, R192R, L55M, and M55M are considered variant genotypes [
8,
32,
33].
To the best of our knowledge, no such study hasreported the association of these variants in the Pashtun population of Khyber Pakhtunkhwa (KP), Pakistan, despite the reports of increasing incidence of CAD in recent years [
34,
35]. Owing to their unique cultural practices, social values, lifestyle, and behaviors make them suitable for such studies [
34,
36]. Considering the importance of the above-mentioned gene variants, it seems suitable to know their association with MI in the said population.
Therefore, this case-control study has been designed to investigate the possible association of APOE and PON1 variants with the risk of MI in the Pashtun ethnic population of KP, Pakistan.
2. Materials and Methods
2.1. Ethics Statement
Ethical approval was obtained from the Ethical Committee of the Department of Pharmacy, University of Peshawar (No: 906/Pham). Written informed consent was obtained from all the study subjects. The study was conducted in compliance with the ethical guidelines of the 1975 Declaration of Helsinki.
2.2. Study Population
A total of 300 age and gender-matched individuals (n = 200 MI cases and n = 100 healthy controls) of Pashtun ethnicity belonging from different districts such as Peshawar, Mardan, Swabi, Charsadda, Nowshehra, Swat, and others of Khyber Pakhtunkhwa were included in the study. The study period was from July 2018 to July 2019. The mean age of the control subjects was 58.43 ± 12.65 (140 males and 60 females), and the control was 56.63 ± 11.87(63 males and 37 females). The diagnosis of MI was based on the American College of Cardiology/American Heart Association (ACC/AHA) classification. A senior cardiologist diagnosed MI based on medical records that revealed medical indications, abnormal cardiac enzymes, ECG (Electrocardiogram) abnormalities, and angiography/echocardiography results. CAD was defined as stenosis ˃50% in at least one of the significant segments of the coronary artery. The control subjects had no lumen stenosis (˂50%) on coronary angiography or physical indications of cardiovascular disease. HTN was defined as having a mean blood pressure of ≥140/90 mmHg or being currently treated for it. DM was classified as having fasting glucose levels of ≥126 mg/dL or non-fasting glucose levels of ≥200 mg/dL, as well as being on oral hypoglycemic medicines or insulin. Patients were admitted tothe three tertiary care (teaching) hospitals of Khyber Pakhtunkhwa, Lady Reading Hospital (LRH) Peshawar, Hayatabad Medical Complex (HMC) Peshawar, and Khyber Teaching Hospital (KTH) Peshawar, while control samples were collected from different districts. Healthy volunteers had no history of cardiovascular disease, especially MI. Inclusion criteria for cases were (i) confirmed MI patients, (ii) Patients belonging to Pakistani Pashtun origin (iii) age ≥30 years. Exclusion criteria were (i) Age ˃80 and ˂30, (ii) mentally ill patients, (iii) severe liver diseases, (iv) malignant tumor, and (v) renal dysfunction. The consent form and thorough demographic, family, and clinical history of all the participants was taken on a carefully designed Proforma. Demographic information includes age, weight, height, and residence. A family history questionnaire includes information on any CVD, MI, or other cardiac issues in the family. The clinical history section of the Proforma includes details about the current disease, co-morbid disorders, and vital signs. For illiterate participants, who have difficulty understanding English, the consent form for their understanding was read and explained in the local Pashtu language and then signed on his/her behalf by any of his/her relatives/attendants.
2.3. Blood Sampling
Following an overnight fast, blood samples were collected from each research participant through venipuncture, with 2.5 mL collected in each EDTA (Ethylene diamine tetra acetic acid) tube and plain tube (without anticoagulant). After allowing the blood in the plain tube to clot, it was centrifuged to obtain serum for biochemical examination. Following aseptic procedures, blood samples (properly labeled) were stored at −10 °C.
2.4. DNA Extraction and Biochemical Measurements
Genomic DNA (Deoxyribonucleic acid) was extracted from peripheral blood leukocytes using the WizPrep DNA extraction kit (WizPrep no. W54100). DNA measurements were carried out with the Qubit ™ dsDNA HS Assay kit (Catalog No. Q32851), and the concentration was adjusted to 10 ng/μL.The serum concentration of Total Cholesterol (TC), Triglycerides (TG), LDL-C, and high-density lipoprotein cholesterol (HDL-C) were measured by standard enzymatic methods using standard reagents on Architect Plus (Ci-4100, Germany) biochemical instrument following strictly manufacturer’s instructions in Hospital clinical laboratory.
2.5. DNA Samples Pooling
According to the DNA-pooling techniques previously described [
37], DNA pools were created from 200 MI patients and 100 control participants in order to cut costs and streamline the sequencing procedure. Each pool contains an equal quantity of genomic DNA (10 ng) from each subject.
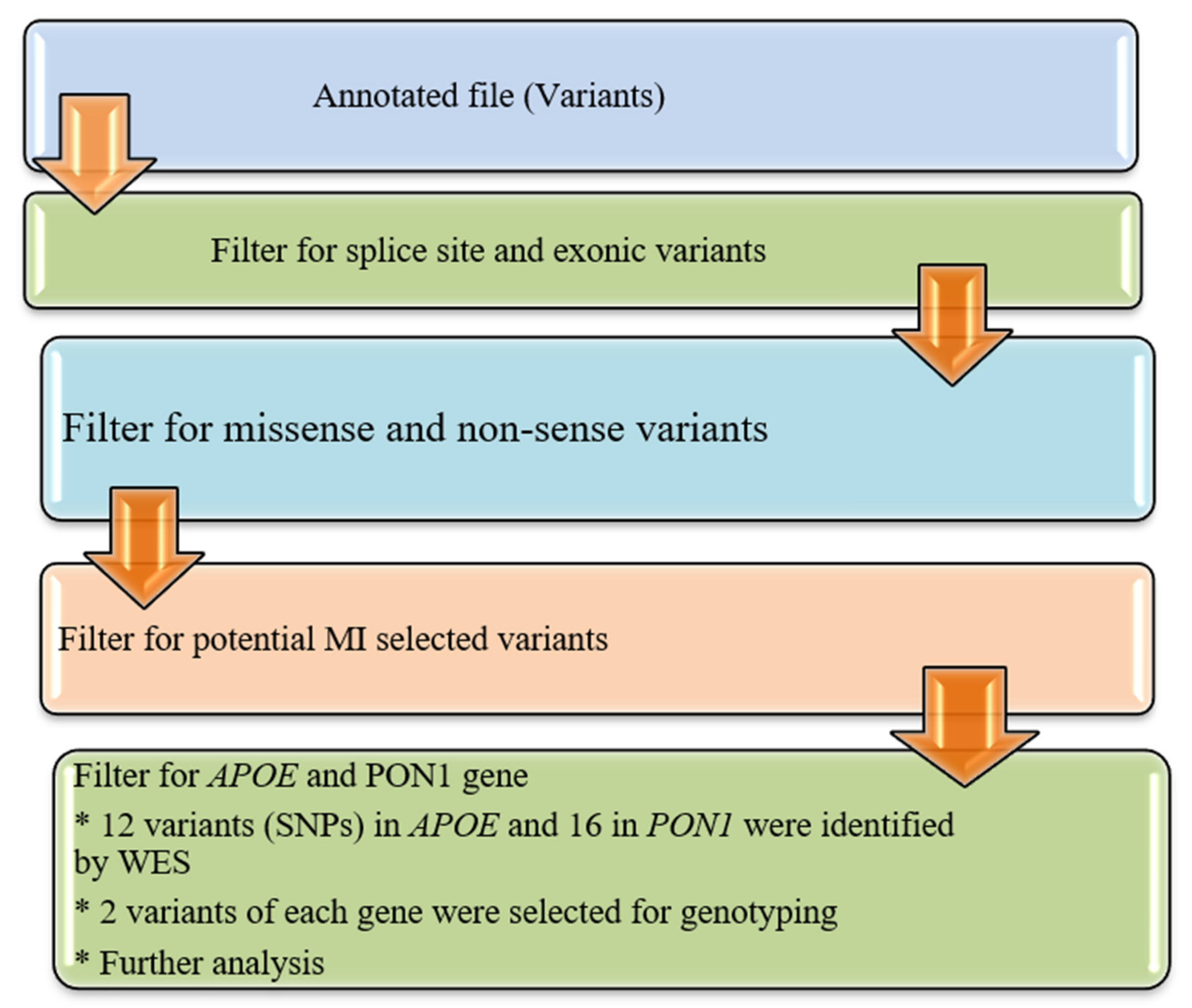
2.6. Variant Prioritization
The annotated data in the Excel file were first manually curated to screen exonic, and missense variants and synonymous variants were eliminated as shown in
Figure 1. The functional influence, biological action, and pathogenicity of the selected variants (SNPs) were checked by using prediction algorithms (PolyPhen and SIFT prediction) built within ANNOVAR.
2.7. Validation Trial and Genotyping of APOE and PON1
In the research population, Whole Exome Sequencing (WES) discovered a total of 12 variations in the APOE and 16 in the PON1 gene, respectively. The selected SNPs were genotyped to validate WES results and confirm the association with MI. Sequenom MassARRAY (Sequenom Inch., San Diego, CA, USA) platform was employed following the manufacturer’s instructions.
2.8. Statistical Analysis
The SPSS (Statistical Package for the Social Sciences) software was used to analyze statistical data. Age, gender, weight, smoking, lifestyle, exercise, PON1, and APOE gene variations were the main factors chosen for the study. W Shapiro-Wilk’s test was used to determine the normality of distribution for quantitative data. Categorical data of the cases and control individuals were reported as percentages and frequencies and analyzed with a Chi-square test., whereas continuous variables were displayed as mean standard ± deviation. Odds ratios (OR) of MI cases for each variant using a binary logistic regression model were estimated with a 95% confidence interval (CI). The difference in genotype and allelic prevalence and correlation between cases and control were assessed independently as well as adjusted for conventional risk factors. Age, gender, smoking, and family history of MI, TC, and LDL-C were included as covariates, as well as all the possible genotypes studied. Binary logistic regression was used to determine if the chosen SNP was associated with MI. A p ≤ 0.05 was statistically considered significant.
4. Discussion
The current study investigated the relationship between
APOE and
PON1 polymorphism and the risk of MI in Pakistan’s Pashtun ethnic population. The genes were selected for genotype validation due to their prominent association with other ethnicities along with data absence in the study population. The selected variants of APOE (rs7412 and rs429358) were genotyped and validated by MassARRAY to confirm the association with MI. The notable variant among the 12 identified variants of
APOE was rs429358 (
p.Cys130Arg), located on the 4th exon of chromosome 19. SIFT and PolyPhen predicted the variant rs429358 as deleterious and probably damaging, respectively. Likewise, another exonic missense SNP reported was rs7412 (
p.Arg176Cys). SIFT and PolyPhen labeled them deleterious and benign, respectively. Furthermore, a significant association between the ɛ4 allele (rs429358) and the risk of MI has been found in the study population, which is in broad agreement with other ethnic populations [
27,
39,
40]. This association remained significant when adjusted for several MI confounding factors.
The
APOE gene polymorphisms are associated with many diseases such as dementia, Parkinson’s disease, epilepsy, and CAD [
41]. Its association with MI or CAD has been extensively studied in the last two decades, and the ϵ4 allele has been found to have a link with it in many studies [
42]. Moreover, the same allele was associated with an increased risk of developing HTN [
43]. A large-scale genomic study comprising 32,965 controls and 15,492 cases showed that individuals with the ϵ4 allele had a higher risk for coronary heart disease (CHD) compared to individuals with the ϵ3/ϵ3 genotype [
44]. However, another study has shown no association of
APOE gene polymorphism with the development of CAD in the study on the relationship between
APOE gene polymorphism and blood lipid and CAD in African Caribbean people [
45]. These inconsistencies may be because of regional and ethnic variability. This study found the ɛ3 allele to be the most common isoform of the
APOE gene accounting for 73% of cases and 81% in controls, respectively, which was consistent with most of the previous studies [
40,
46]. The findings of our study regarding the frequencies of
APOE allele are consistent with that of other ethnicities [
47,
48].
Similarly, this study has also assessed the association of
Q192R and
L55M variants. Findings suggested the missense SNP
Q192R (rs662), located on the short arm of chromosome 7 as significant. The frequency of the RR genotype of
Q192R was found to be higher in the MI cases compared to the control. The
Q192R (rs662) polymorphism cases with MI revealed a higher frequency of the R allele compared to the control. Both the SIFT and PolyPhen scores predicted it as pathogenic and damaging, respectively. The second missense, exonic SNP, was
L55M (rs854560). It was shown tolerable and benign by SIFT and PolyPhen score, respectively, and was found not associated with MI (
p ˃ 0.05). This finding is supported by other studies [
23]. Studies conducted in different ethnic populations have shown interesting results of the association of
Q192R polymorphism of
PON1 with MI [
49]. Many studies have revealed the RR genotype and R allele of
PON Q192R with susceptibility to MI [
23]. A study conducted on the Colombian ethnic population proposed
Q192R polymorphism of
PON1 as a useful biomarker of CAD [
50]. Another study also showed an association of the
PON Q192R variant with CAD [
51]. In line with these findings, a significant association was observed for Q192R with CAD by Liu and colleagues [
52]. Similar findings were also found in a Chinese ethnic population, south Indian Tamil, and Asian Indians. [
53,
54,
55] Conversely, many other studies have demonstrated conflicting findings and found no association of
PON1 Q192R polymorphism with CAD [
23]. In particular, a genetic study conducted on 120 CAD and 102 healthy volunteers revealed that
PON1 192R allele frequency was the same among the cases and control [
56]. Furthermore, no link was found between the Q192R polymorphism and CAD in a Turkish population [
57]. Similarly no association was observed in Taiwan ethnic population [
58].
Furthermore, Sociodemographic analysis of cases and controls revealed a higher incidence of DM and HTN in cases compared to the control. Moreover, the results showed an increased prevalence of MI in males compared to females (70% vs. 30%). Most of the MI patients were smokers compared to controls (58.5% vs. 26%). Furthermore, a family history of MI and other heart diseases was more prevalent in some cases. Physically activity (exercise) was found to be very poor in cases compared to controls (30% vs. 70%).