Transcriptome and Metabolome Profiling Provide Insights into Flavonoid Synthesis in Acanthus ilicifolius Linn
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Transcription Experimental Methods
2.3. Metabolome Analysis Methods
2.4. UPLC Conditions
2.5. qRT-PCR
3. Results
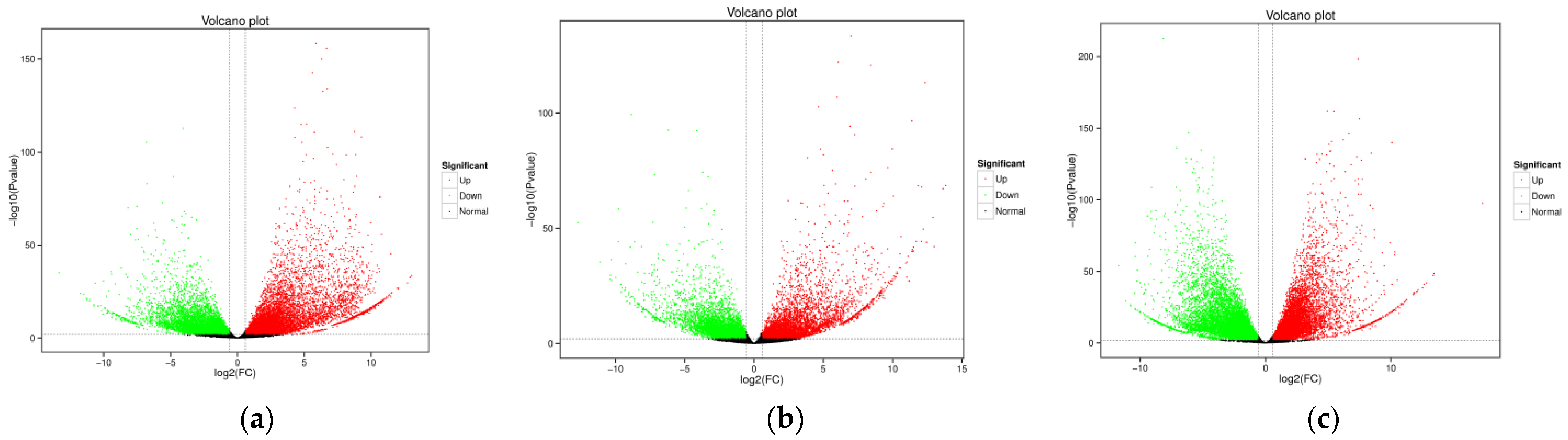
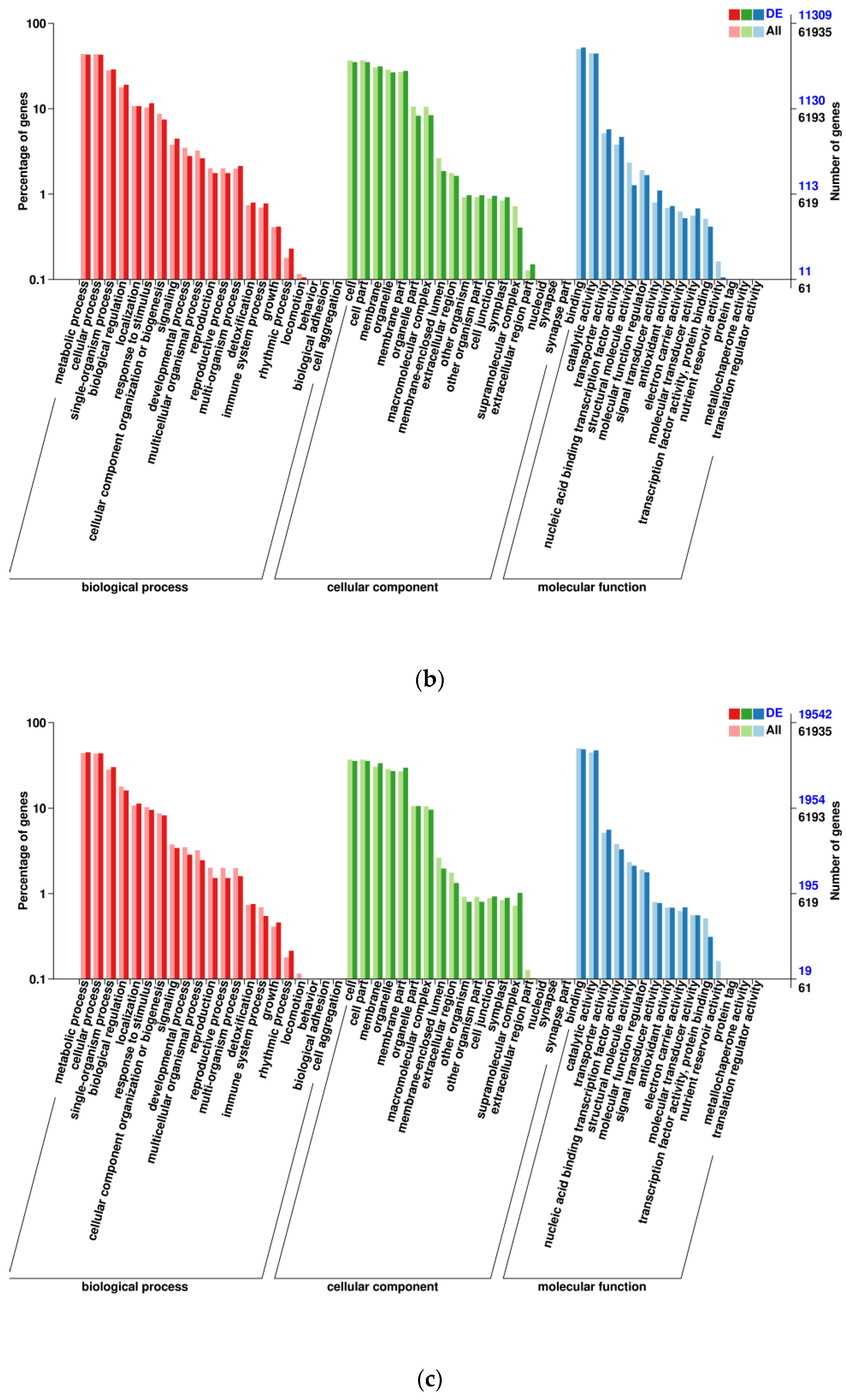
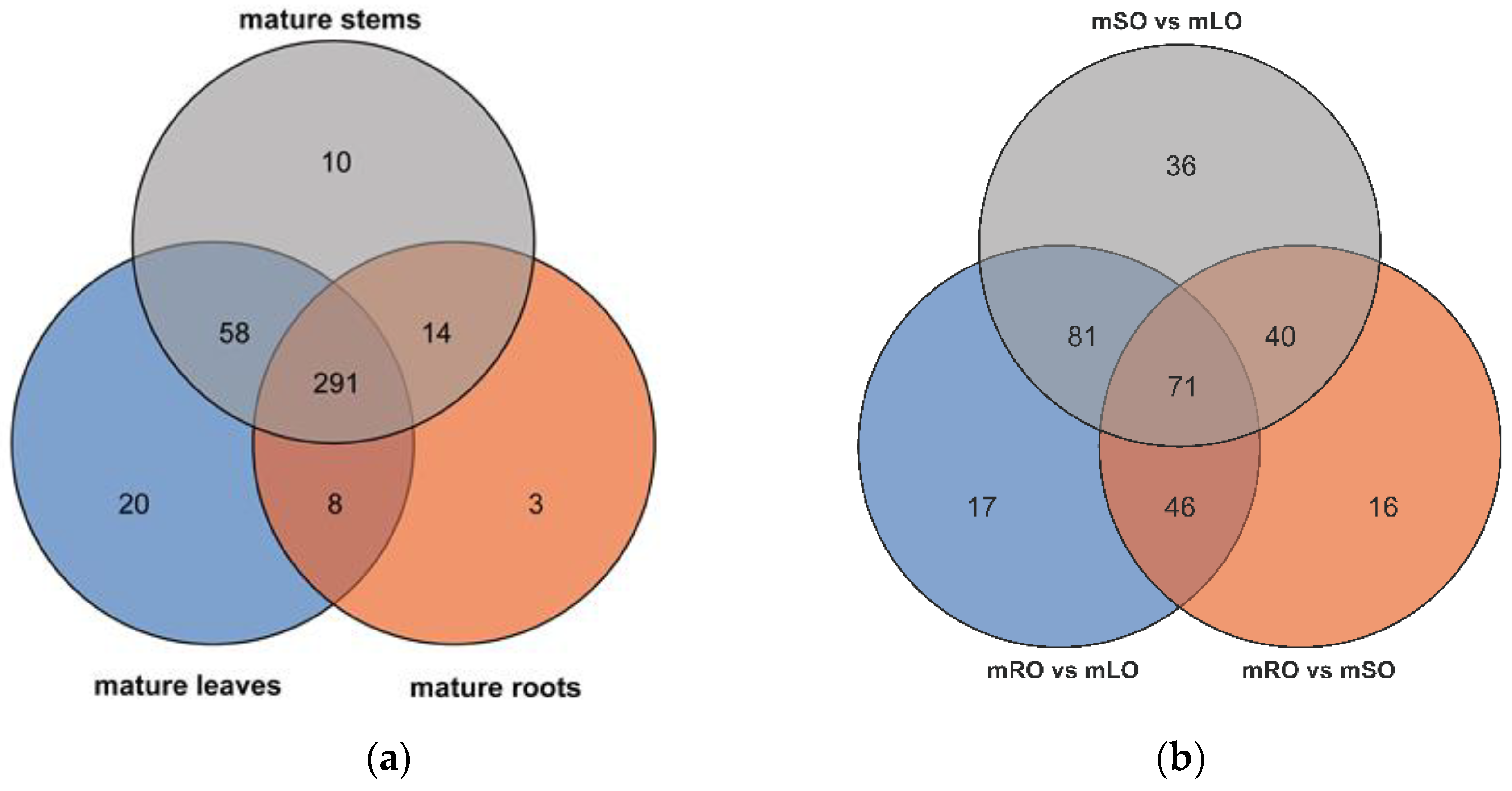
3.1. Transcriptomic Analysis and Differentially Expressed Genes
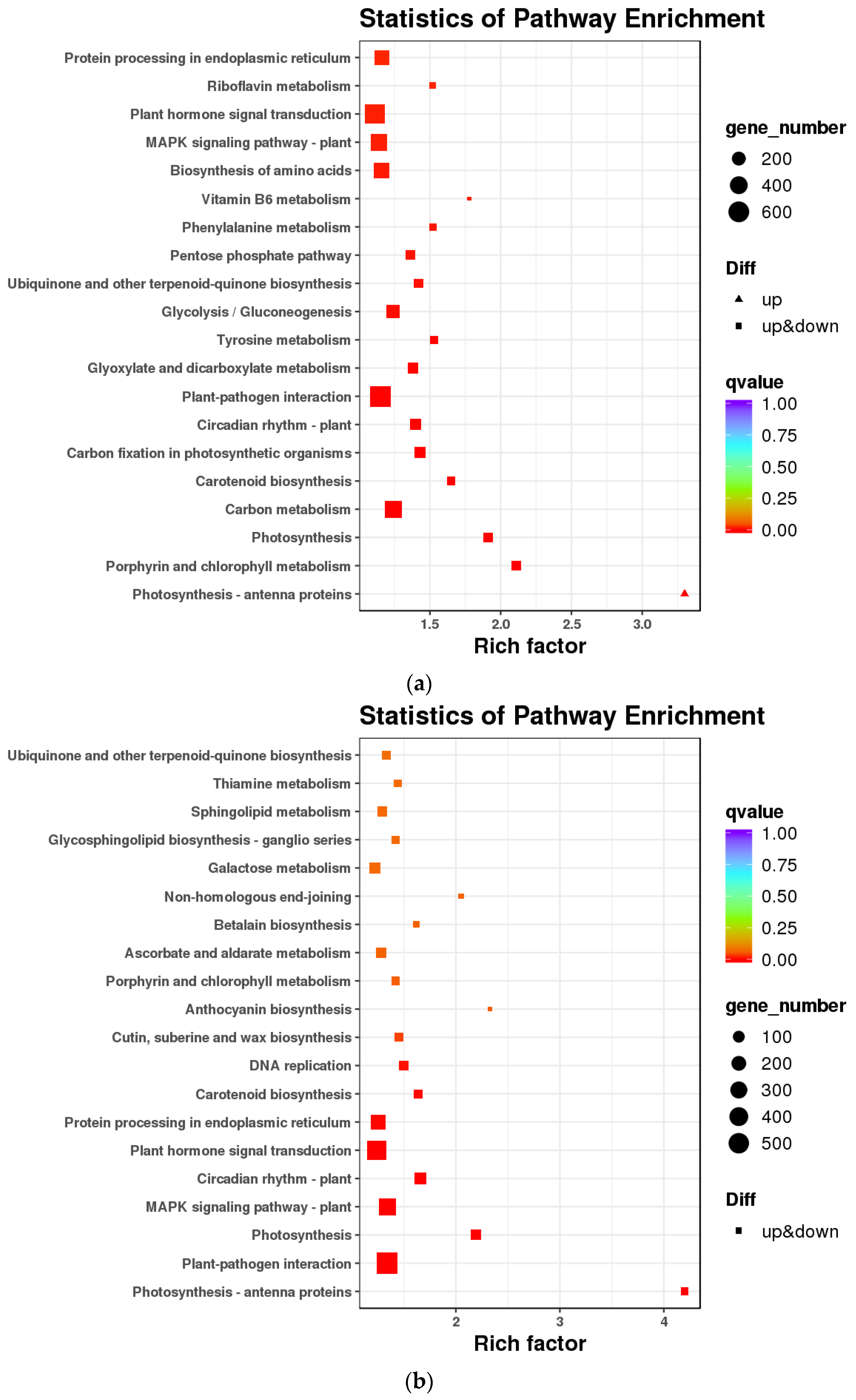
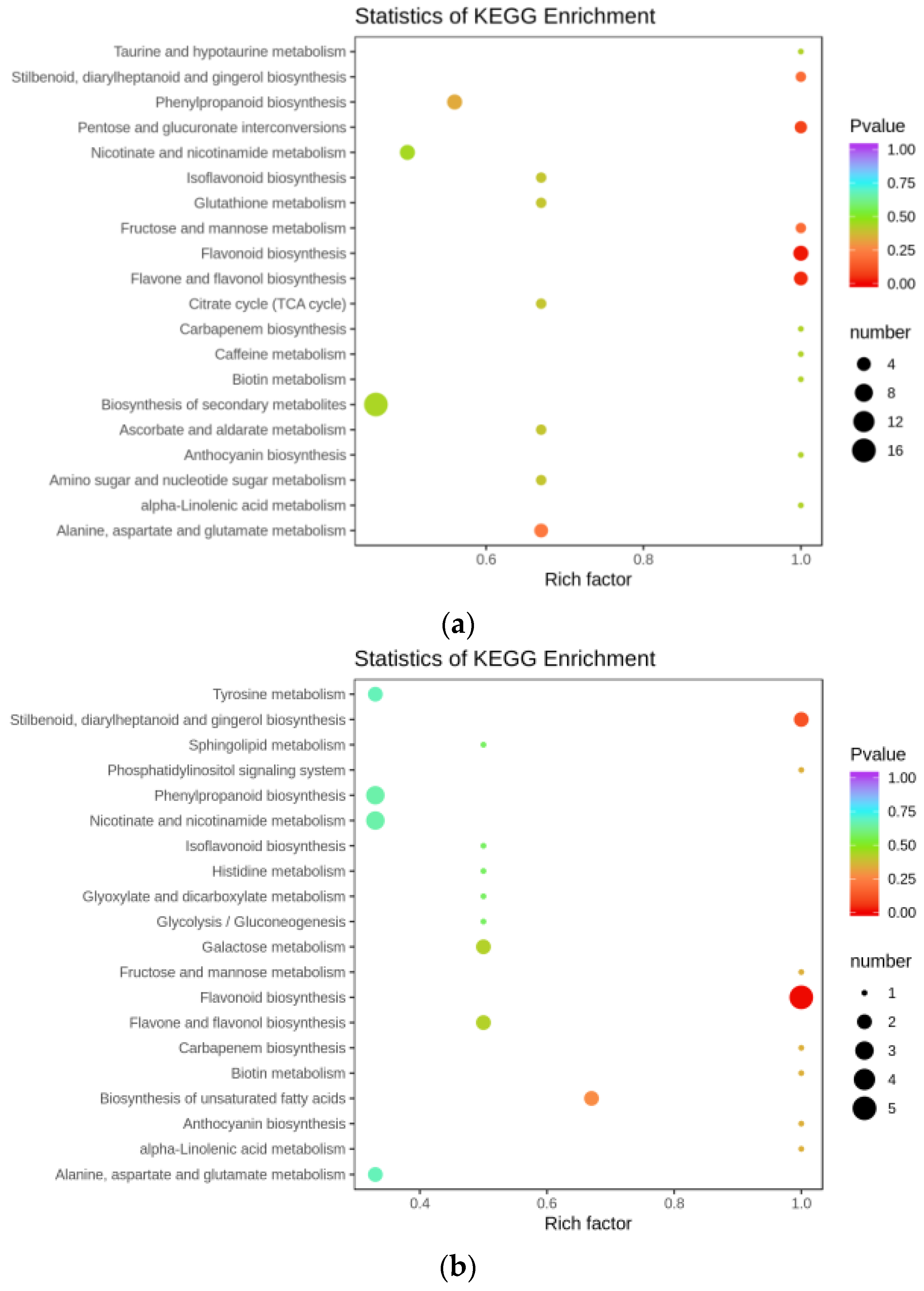
3.2. Flavonoid Biosynthesis Differential Genes
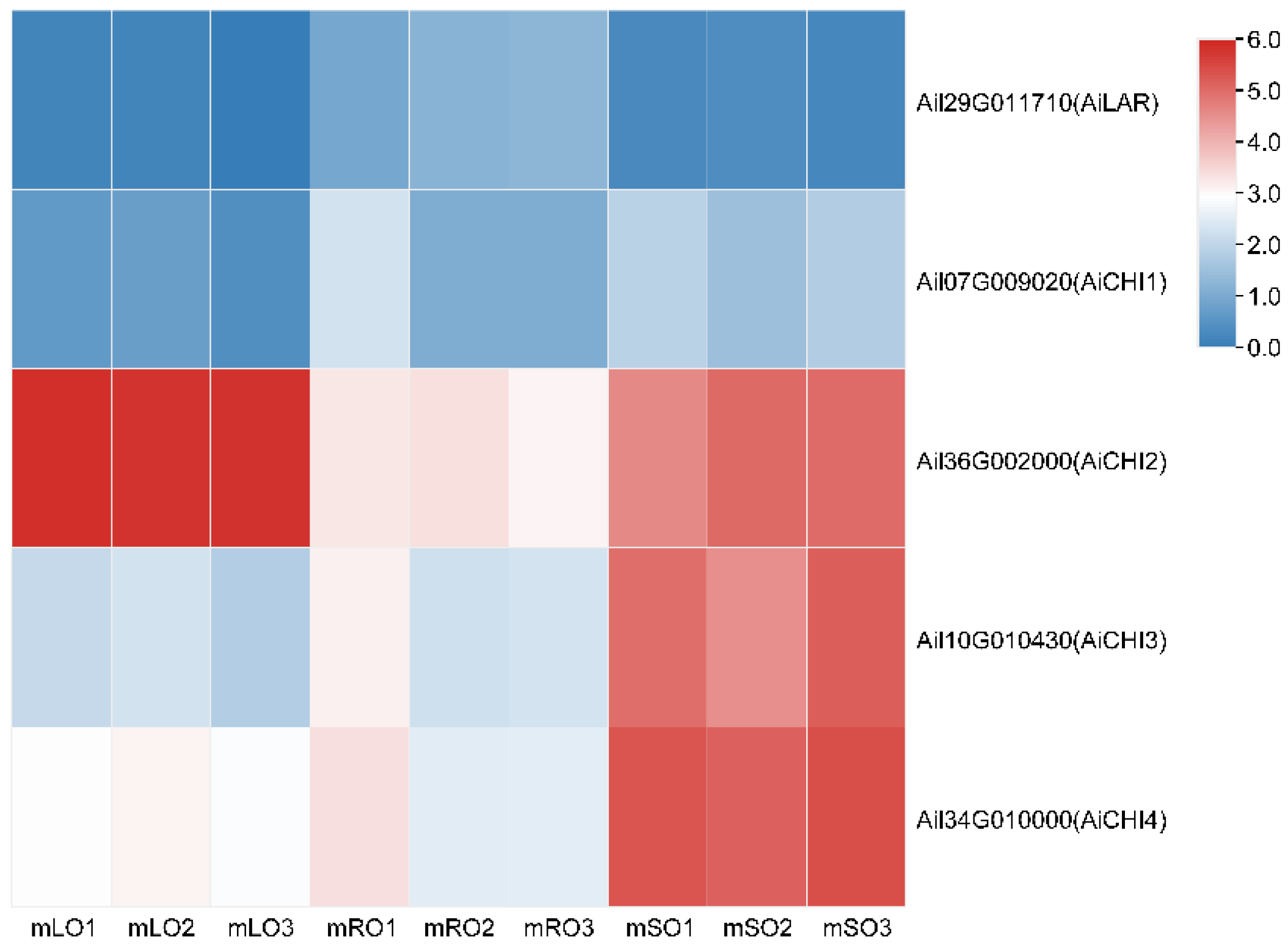
3.3. qRT-PCR
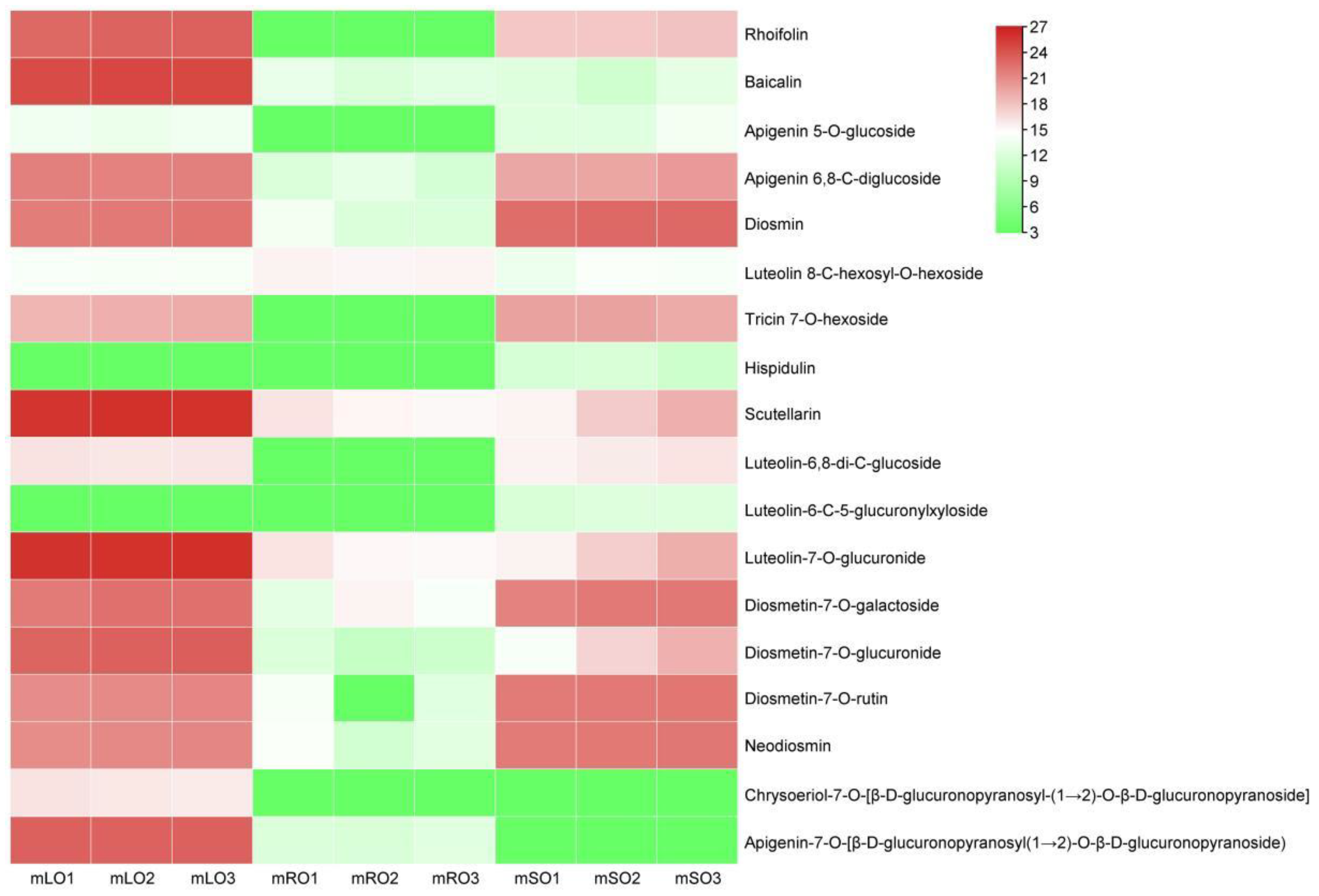
3.4. Metabolome Analysis and Differential Metabolites
3.5. Combined Analysis of Transcriptome and Metabolome Analysis
4. Discussion
4.1. Transcriptomic Analysis of Key Genes in the Flavonoid Synthesis Pathway in A. ilicifolius
4.2. Metabolomic Analysis of Flavonoid Content in A. ilicifolius
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barker, R.M. A Taxonomic Revision of Australian Acanthaceae. J. Adel. Bot. Gard. 1986, 9, 64–75. [Google Scholar]
- Wu, J.; Zhang, S.; Xiao, Q.; Li, Q.; Huang, J.; Long, L.; Huang, L. Phenylethanoid and Aliphatic Alcohol Glycosides from Acanthus ilicifolius. Phytochemistry 2003, 63, 491–495. [Google Scholar] [CrossRef] [PubMed]
- He, K.J.; Li, J.; Zhang, J.H. Compilation of Medicinal Properties of Raw Herbs; Guangdong Science and Technology Publishing House: Guangzhou, China, 2018. [Google Scholar]
- Huang, L.Q. National Compilation of Chinese Herbal Medicines, 3rd ed.; People’s Medical Publishing House: Beijing, China, 2014. [Google Scholar]
- Babu, B.H.; Shylesh, B.S.; Padikkala, J. Antioxidant and Hepatoprotective Effect of Acanthus ilicifolius. Fitoterapia 2001, 72, 272–277. [Google Scholar] [CrossRef]
- Babu, B.H.; Shylesh, B.S.; Padikkala, J. Tumour Reducing and Anticarcinogenic Activity of Acanthus ilicifolius in Mice. J. Ethnopharmacol. 2002, 79, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Huo, C.; Liang, H.; Tu, G.; Zhao, Y.; Lin, W. A New 5,11-Epoxymegastigmane Glucoside from Acanthus ilicifolius. Nat. Prod. Res. 2008, 22, 896–900. [Google Scholar] [CrossRef] [PubMed]
- Minocha, P.K.; Tiwari, K.P. A Triterpenoidal Saponin from Roots of Acanthus illicifolius. Phytochemistry 1981, 20, 135–137. [Google Scholar] [CrossRef]
- Kumar, K.M.S.; Puia, Z.; Barik, R.; Gorain, B.; Roy, D.; Adhikari, D.; Sen, T. The Gastroprotective Role of Acanthus ilicifolius—A Study to Unravel the Underlying Mechanism of Anti-Ulcer Activity. Sci. Pharm. 2012, 80, 701–717. [Google Scholar] [CrossRef] [Green Version]
- Hu, H.Y.; Zhu, C.; Zhang, C.C.; Lu, C.Y. Study on Optimum Technology for Extracting Total Flavonoids from Acanthus illicifolius by Orthogonal Design. Chin. J. Mar. Drugs 2009, 28, 30–34. [Google Scholar]
- Nabavi, S.M.; Šamec, D.; Tomczyk, M.; Milella, L.; Russo, D.; Habtemariam, S.; Suntar, I.; Rastrelli, L.; Daglia, M.; Xiao, J.; et al. Flavonoid Biosynthetic Pathways in Plants: Versatile Targets for Metabolic Engineering. Biotechnol. Adv. 2020, 38, 107316. [Google Scholar] [CrossRef]
- Yonekura-Sakakibara, K.; Higashi, Y.; Nakabayashi, R. The Origin and Evolution of Plant Flavonoid Metabolism. Front. Plant Sci. 2019, 10, 943. [Google Scholar] [CrossRef] [Green Version]
- García-Lafuente, A.; Guillamón, E.; Villares, A.; Rostagno, M.A.; Martínez, J.A. Flavonoids as Anti-Inflammatory Agents: Implications in Cancer and Cardiovascular Disease. Inflamm. Res. 2009, 58, 537–552. [Google Scholar] [CrossRef] [PubMed]
- Uddin, S.J.; Grice, D.; Tiralongo, E. Evaluation of Cytotoxic Activity of Patriscabratine, Tetracosane and Various Flavonoids Isolated from the Bangladeshi Medicinal Plant Acrostichum Aureum. Pharm. Biol. 2012, 50, 1276–1280. [Google Scholar] [CrossRef] [Green Version]
- Hudson, E.A.; Dinh, P.A.; Kokubun, T.; Simmonds, M.S.; Gescher, A. Characterization of Potentially Chemopreventive Phenols in Extracts of Brown Rice That Inhibit the Growth of Human Breast and Colon Cancer Cells. Cancer Epidemiol. Biomark. Prev. 2000, 9, 1163–1170. [Google Scholar]
- Abbas, A.; Shah, A.; Shah, A.; Nadeem, M.; Alsaleh, A.; Javed, T.; Alotaibi, S.; Abdelsalam, N. Genome-Wide Analysis of Invertase Gene Family, and Expression Profiling under Abiotic Stress Conditions in Potato. Biology 2022, 11, 539. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Feng, Y.; Yu, S.; Fan, Z.; Li, X.; Li, J.; Yin, H. The Flavonoid Biosynthesis Network in Plants. Int. J. Mol. Sci. 2021, 22, 12824. [Google Scholar] [CrossRef]
- Yang, H.; Li, H.; Li, Q. Biosynthetic Regulatory Network of Flavonoid Metabolites in Stems and Leaves of Salvia miltiorrhiza. Sci. Rep. 2022, 12, 18212. [Google Scholar] [CrossRef]
- Huang, X.; Sun, G.; Li, Q.; Yan, H. Transcriptome Analysis Reveals Regulatory Networks and Hub Genes in the Flavonoid Metabolism of Rosa roxburghii. Horticulturae 2023, 9, 233. [Google Scholar] [CrossRef]
- Betti, M.; Garcia-Calderon, M.; Perez-Delgado, C.M.; Credali, A.; Pal’ove-Balang, P.; Estivill, G.; Repak, M.; Vega, J.M.; Galvan, F.; Marquez, A.J. Reassimilation of Ammonium in Lotus japonicus. J. Exp. Bot. 2014, 65, 5557–5566. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Kong, D.; Fu, Y.; Sussman, M.R.; Wu, H. The Effect of Developmental and Environmental Factors on Secondary Metabolites in Medicinal Plants. Plant Physiol. Biochem. 2020, 148, 80–89. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-Length Transcriptome Assembly from RNA-Seq Data without a Reference Genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R. Differential Gene and Transcript Expression Analysis of RNA-Seq Experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [Green Version]
- Florea, L.; Song, L.; Salzberg, S.L. Thousands of Exon Skipping Events Differentiate among Splicing Patterns in Sixteen Human Tissues. F1000Research 2013, 2, 188. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, A.; Greaves, I.K.; Liu, P.-C.; Wu, L.; Dennis, E.S.; Peacock, W.J. Early Changes of Gene Activity in Developing Seedlings of Arabidopsis Hybrids Relative to Parents May Contribute to Hybrid Vigour. Plant J. 2016, 88, 597–607. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Li, J.; Wu, S.; Zhu, Y.; Chen, Y.; He, F. Integrated Nr Database in Protein Annotation System and Its Localization. Comput. Eng. 2006, 32, 71–72. [Google Scholar]
- Apweiler, R. UniProt: The Universal Protein Knowledgebase. Nucleic Acids Res. 2004, 32, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L. The COG Database: A Tool for Genome-Scale Analysis of Protein Functions and Evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef] [Green Version]
- Koonin, E.V.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Krylov, D.M.; Makarova, K.S.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; Rao, B.; et al. A Comprehensive Evolutionary Classification of Proteins Encoded in Complete Eukaryotic Genomes. Genome Biol. 2004, 5, R7. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M. The KEGG Resource for Deciphering the Genome. Nucleic Acids Res. 2004, 32, 277–280. [Google Scholar] [CrossRef] [Green Version]
- Want, E.J.; Wilson, I.D.; Gika, H.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Holmes, E.; Nicholson, J.K. Global Metabolic Profiling Procedures for Urine Using UPLC–MS. Nat. Protoc. 2010, 5, 1005–1018. [Google Scholar] [CrossRef]
- Dunn, W.B.; Broadhurst, D.; Begley, P.; Zelena, E.; Francis-McIntyre, S.; Anderson, N.; Brown, M.; Knowles, J.D.; Halsall, A.; Haselden, J.N.; et al. Procedures for Large-Scale Metabolic Profiling of Serum and Plasma Using Gas Chromatography and Liquid Chromatography Coupled to Mass Spectrometry. Nat. Protoc. 2011, 6, 1060–1083. [Google Scholar] [CrossRef]
- Chen, W.; Gong, L.; Guo, Z.; Wang, W.; Zhang, H.; Liu, X.; Yu, S.; Xiong, L.; Luo, J. A Novel Integrated Method for Large-Scale Detection, Identification, and Quantification of Widely Targeted Metabolites: Application in the Study of Rice Metabolomics. Mol. Plant 2013, 6, 1769–1780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Tu, H.; Wan, J.; Chen, W.; Liu, X.; Luo, J.; Xu, J.; Zhang, H. Spatio-Temporal Distribution and Natural Variation of Metabolites in Citrus Fruits. Food Chem. 2016, 199, 8–17. [Google Scholar] [CrossRef]
- Livak, K.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real Time Quantitative PCR and the 2−ΔCt Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Wei, L.; Sun, Z.; Gao, L.; Meng, Y.; Tang, Y.; Wu, Y. Production and Transcriptional Regulation of Proanthocyanidin Biosynthesis in Forage Legumes. Appl. Microbiol. Biotechnol. 2015, 99, 3797–3806. [Google Scholar] [CrossRef] [PubMed]
- Ngaki, M.N.; Louie, G.V.; Philippe, R.N.; Manning, G.; Pojer, F.; Bowman, M.E.; Li, L.; Larsen, E.; Wurtele, E.S.; Noel, J.P. Evolution of the Chalcone-Isomerase Fold from Fatty-Acid Binding to Stereospecific Catalysis. Nature 2012, 485, 530–533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Calderón, M.; Pérez-Delgado, C.M.; Palove-Balang, P.; Betti, M.; Márquez, A.J. Flavonoids and Isoflavonoids Biosynthesis in the Model Legume Lotus japonicus; Connections to Nitrogen Metabolism and Photorespiration. Plants 2020, 9, 774. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. It Takes a Garden. How Work on Diverse Plant Species Has Contributed to an Understanding of Flavonoid Metabolism. Plant Physiol. 2001, 127, 1399–1404. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the Anthocyanin Biosynthetic Pathway by the TTG1/BHLH/Myb Transcriptional Complex in Arabidopsis Seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef]
- Matsui, K.; Walker, A.R. Biosynthesis and Regulation of Flavonoids in Buckwheat. Breed. Sci. 2020, 70, 74–84. [Google Scholar] [CrossRef] [Green Version]
- Deng, X.; Bashandy, H.; Ainasoja, M.; Kontturi, J.; Pietiäinen, M.; Laitinen, R.A.E.; Albert, V.A.; Valkonen, J.P.T.; Elomaa, P.; Teeri, T.H. Functional Diversification of Duplicated Chalcone Synthase Genes in Anthocyanin Biosynthesis of Gerbera Hybrida. New Phytol. 2014, 201, 1469–1483. [Google Scholar] [CrossRef] [PubMed]
- Jaakola, L. New Insights into the Regulation of Anthocyanin Biosynthesis in Fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef] [Green Version]
- Koskela, S.; Söderholm, P.P.; Ainasoja, M.; Wennberg, T.; Klika, K.D.; Ovcharenko, V.V.; Kylänlahti, I.; Auerma, T.; Yli-Kauhaluoma, J.; Pihlaja, K.; et al. Polyketide Derivatives Active against Botrytis Cinerea in Gerbera Hybrida. Planta 2011, 233, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, J.J.; Nakajima, J.; Welford, R.W.D.; Yamazaki, M.; Saito, K.; Schofield, C.J. Mechanistic Studies on Three 2-Oxoglutarate-Dependent Oxygenases of Flavonoid Biosynthesis. J. Biol. Chem. 2004, 279, 1206–1216. [Google Scholar] [CrossRef] [Green Version]
- Petrussa, E.; Braidot, E.; Zancani, M.; Peresson, C.; Bertolini, A.; Patui, S.; Vianello, A. Plant Flavonoids—Biosynthesis, Transport and Involvement in Stress Responses. Int. J. Mol. Sci. 2013, 14, 14950–14973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, M.-Z.; Xie, D.-Y. Biosynthesis and Metabolic Engineering of Anthocyanins in Arabidopsis Thaliana. Recent Pat. Biotechnol. 2014, 8, 47–60. [Google Scholar] [CrossRef] [Green Version]
- Cheng, A.-X.; Han, X.-J.; Wu, Y.-F.; Lou, H.-X. The Function and Catalysis of 2-Oxoglutarate-Dependent Oxygenases Involved in Plant Flavonoid Biosynthesis. Int. J. Mol. Sci. 2014, 15, 1080–1095. [Google Scholar] [CrossRef] [Green Version]
- Huo, C.H.; Zhao, Y.Y.; Liang, H.; Lin, W.H. Studies on chemical constituents in herbs of Acanthus ilicifolius. China J. Chin. Mater. Med. 2005, 30, 763–765. [Google Scholar]
- Saranya, A.; Ramanathan, T.; Kesavanarayanan, K.S.; Adam, A. Traditional Medicinal Uses, Chemical Constituents and Biological Activities of a Mangrove Plant, Acanthus ilicifolius Linn.: A Brief Review. Am Eurasian J. Agric. Environ. Sci. 2015, 15, 243–250. [Google Scholar]
- Nair, A.G.R.; Pouchaname, V.J. Methylapigenin 7-O-β-D-Glucuronate-A New Flavone Glycoside from Acanthus ilicifolius. Indian Chem. Soc. 1987, 64, 228–229. [Google Scholar]
- Wu, J.; Zhang, S.; Xiao, Q.; Li, Q.; Huang, J.; Long, L.; Huang, L. Megastigmane and Flavone Glycosides from Acanthus ilicifolius. Pharmazie 2003, 58, 363–364. [Google Scholar]
- Zeng, G.; Li, D.; Han, Y.; Teng, W.; Wang, J.; Qiu, L.; Li, W. Identification of QTL Underlying Isoflavone Contents in Soybean Seeds among Multiple Environments. Theor. Appl. Genet. 2009, 118, 1455–1463. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Yonekura-Sakakibara, K.; Nakabayashi, R.; Higashi, Y.; Yamazaki, M.; Tohge, T.; Fernie, A.R. The Flavonoid Biosynthetic Pathway in Arabidopsis: Structural and Genetic Diversity. Plant Physiol. Biochem. 2013, 72, 21–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, J.; Li, J.; Dai, C.; Li, L. Transcriptome and Metabolome Analyses Revealed the Response Mechanism of Sugar Beet to Salt Stress of Different Durations. Int. J. Mol. Sci. 2022, 23, 9599. [Google Scholar] [CrossRef]
- Chen, Z.; Hong, X.; Zeng, Z.; Zeng, Y.; Lin, Q.; Tan, Q. Study on Natural Product of Quercetin from Flos Sophorae Immaturus against Activity of Human Nasopharyngeal Carcinoma CNE1 Cells. Genom. Appl. Biol. 2019, 38, 3305–3311. [Google Scholar]
- Skalski, B.; Lis, B.; Pecio, Ł.; Kontek, B.; Olas, B.; Żuchowski, J.; Stochmal, A. Isorhamnetin and Its New Derivatives Isolated from Sea Buckthorn Berries Prevent H2O2/Fe—Induced Oxidative Stress and Changes in Hemostasis. Food Chem. Toxicol. 2019, 125, 614–620. [Google Scholar] [CrossRef]
- Boubaker, J.; Bhouri, W.; Sghaier, M.B.; Ghedira, K.; Franca, M.G.D.; Chekir-Ghedira, L. Ethyl Acetate Extract and Its Major Constituent, Isorhamnetin 3-O-Rutinoside, from Nitraria Retusa Leaves, Promote Apoptosis of Human Myelogenous Erythroleukaemia Cells: N. Retusa Leaf Extracts Promote Apoptosis. Cell Prolif. 2011, 44, 453–461. [Google Scholar] [CrossRef]
- Canarini, A.; Kaiser, C.; Merchant, A.; Richter, A.; Wanek, W. Root Exudation of Primary Metabolites: Mechanisms and Their Roles in Plant Responses to Environmental Stimuli. Front. Plant Sci. 2019, 10, 157. [Google Scholar] [CrossRef] [Green Version]
- Delli-Ponti, R.; Shivhare, D.; Mutwil, M. Using Gene Expression to Study Specialized Metabolism—A Practical Guide. Front. Plant Sci. 2021, 11, 625035. [Google Scholar] [CrossRef]









| Index | Formula | Compounds | VIP | Log2FC | Type |
|---|---|---|---|---|---|
| GQ512006 | C27H30O16 | Quercetin-3-O-glucoside-7-O-rhamnoside | 1.24 | 16.60 | up |
| mws1329 | C21H20O12 | Gossypitrin | 1.17 | 1.24 | up |
| pmb3894 | C17H14O7 | Di-O-methylquercetin | 1.15 | −1.59 | down |
| pmp000580 | C21H20O12 | Isoquercitrin(Quercetin 3-O-β-D-glucoside) | 1.24 | 19.40 | up |
| pmp000596 | C27H30O17 | Quercetin 3,7-bis-O-β-D-glucoside | 1.15 | 3.12 | up |
| mws0066 | C16H12O7 | Isorhamnetin | 1.21 | −1.87 | down |
| pmn001645 | C24H22O14 | Isorhamnetin 3-O-β-(2″-O-acetyl-β-D-glucuronide) | 1.23 | 10.50 | up |
| Index | Formula | Compounds | VIP | Log2FC | Type |
|---|---|---|---|---|---|
| GQ512006 | C27H30O16 | Quercetin-3-O-glucoside-7-O-rhamnoside | 1.32 | 12.80 | up |
| Li512117 | C27H30O16 | Quercetin 3-O-rhanosylgalactoside | 1.19 | −1.01 | down |
| pmn001583 | C27H30O16 | Bioquercetin | 1.20 | −1.20 | down |
| pmp000580 | C21H20O12 | Isoquercitrin(Quercetin 3-O-β-D-glucoside) | 1.32 | 14.20 | up |
| pmp000596 | C27H30O17 | Quercetin 3,7-bis-O-β-D-glucoside | 1.28 | 4.81 | up |
| Index | Formula | Compounds | VIP | Log2FC | Type |
|---|---|---|---|---|---|
| GQ512006 | C27H30O16 | Quercetin-3-O-glucoside-7-O-rhamnoside | 1.16 | 3.84 | up |
| mws0045 | C21H20O11 | Quercitrin | 1.07 | −1.85 | down |
| pmb3894 | C17H14O7 | Di-O-methylquercetin | 1.11 | −2.21 | down |
| pmp000580 | C21H20O12 | Isoquercitrin(Quercetin 3-O-β-D-glucoside) | 1.16 | 5.22 | up |
| pmp000596 | C27H30O17 | Quercetin 3,7-bis-O-β-D-glucoside | 1.08 | −1.69 | down |
| mws0066 | C16H12O7 | Isorhamnetin | 1.14 | −1.09 | down |
| pmn001645 | C24H22O14 | Isorhamnetin 3-O-β-(2″-O-acetyl-β-D-glucuronide) | 1.17 | 10.5 | up |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Z.; Wang, Z.; Xie, Y.; Liu, G.; Shang, X.; Zhan, N. Transcriptome and Metabolome Profiling Provide Insights into Flavonoid Synthesis in Acanthus ilicifolius Linn. Genes 2023, 14, 752. https://doi.org/10.3390/genes14030752
Wu Z, Wang Z, Xie Y, Liu G, Shang X, Zhan N. Transcriptome and Metabolome Profiling Provide Insights into Flavonoid Synthesis in Acanthus ilicifolius Linn. Genes. 2023; 14(3):752. https://doi.org/10.3390/genes14030752
Chicago/Turabian StyleWu, Zhihua, Zhen Wang, Yaojian Xie, Guo Liu, Xiuhua Shang, and Ni Zhan. 2023. "Transcriptome and Metabolome Profiling Provide Insights into Flavonoid Synthesis in Acanthus ilicifolius Linn" Genes 14, no. 3: 752. https://doi.org/10.3390/genes14030752
APA StyleWu, Z., Wang, Z., Xie, Y., Liu, G., Shang, X., & Zhan, N. (2023). Transcriptome and Metabolome Profiling Provide Insights into Flavonoid Synthesis in Acanthus ilicifolius Linn. Genes, 14(3), 752. https://doi.org/10.3390/genes14030752







