ATRX/DAXX: Guarding the Genome against the Hazards of ALT
Abstract
1. Introduction
2. The ATRX/DAXX Complex
3. Localization and Function of ATRX/DAXX
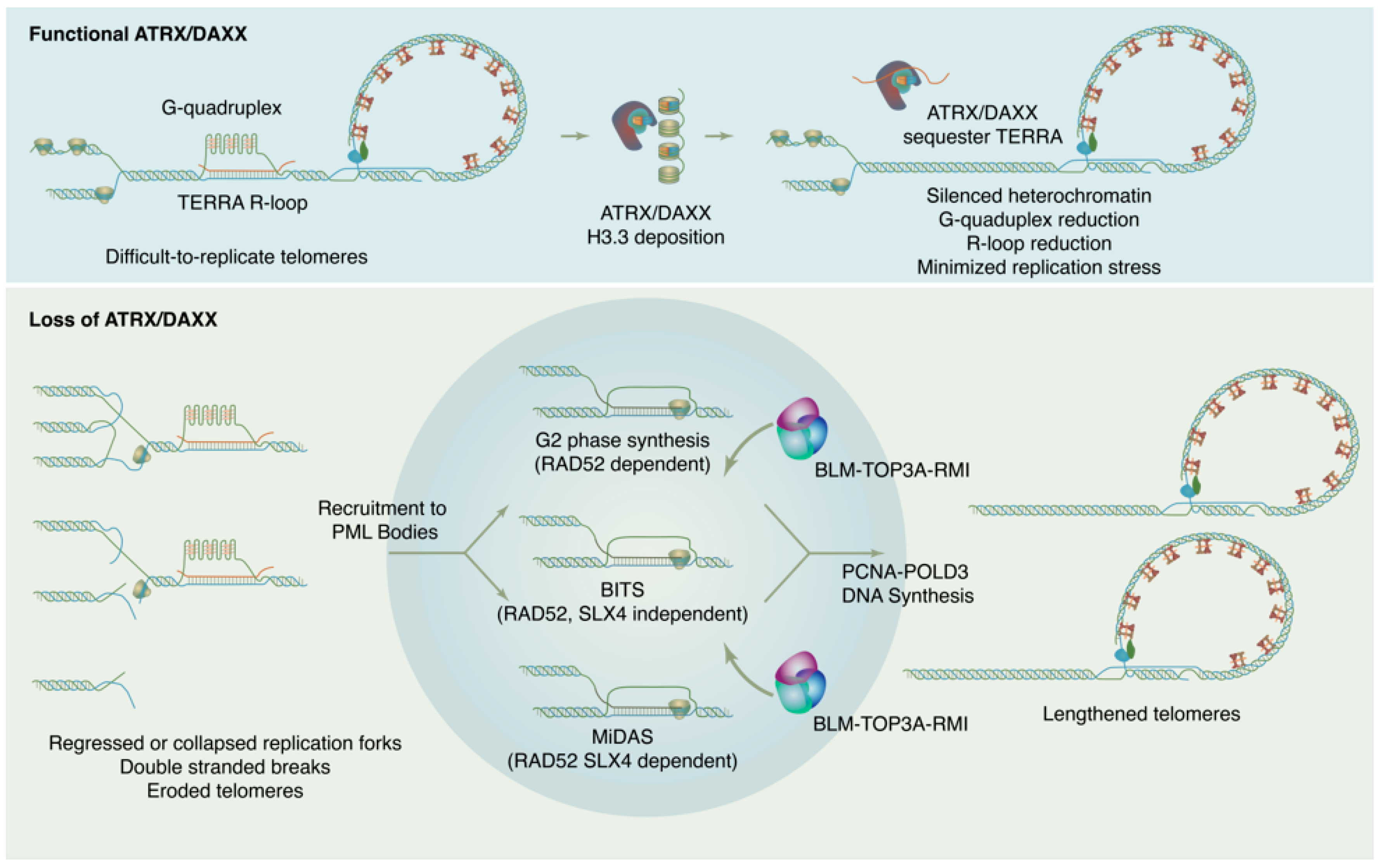
4. ATRX/DAXX and the Suppression of ALT
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, N.W.; Piatyszek, M.A.; Prowse, K.R.; Harley, C.B.; West, M.D.; Ho, P.L.; Coviello, G.M.; Wright, W.E.; Weinrich, S.L.; Shay, J.W. Specific association of human telomerase activity with immortal cells and cancer. Science 1994, 266, 2011–2015. [Google Scholar] [CrossRef]
- Shay, J.W.; Bacchetti, S. A survey of telomerase activity in human cancer. Eur. J. Cancer 1997, 33, 787–791. [Google Scholar] [CrossRef] [PubMed]
- Bryan, T.M.; Englezou, A.; Dalla-Pozza, L.; Dunham, M.A.; Reddel, R.R. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med 1997, 3, 1271–1274. [Google Scholar] [CrossRef] [PubMed]
- Bryan, T.M.; Englezou, A.; Gupta, J.; Bacchetti, S.; Reddel, R.R. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995, 14, 4240–4248. [Google Scholar] [CrossRef] [PubMed]
- Heaphy, C.M.; de Wilde, R.F.; Jiao, Y.; Klein, A.P.; Edil, B.H.; Shi, C.; Bettegowda, C.; Rodriguez, F.J.; Eberhart, C.G.; Hebbar, S.; et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science 2011, 333, 425. [Google Scholar] [CrossRef]
- Lovejoy, C.A.; Li, W.; Reisenweber, S.; Thongthip, S.; Bruno, J.; de Lange, T.; De, S.; Petrini, J.H.J.; Sung, P.A.; Jasin, M.; et al. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 2012, 8, e1002772. [Google Scholar] [CrossRef]
- Bower, K.; Napier, C.E.; Cole, S.L.; Dagg, R.A.; Lau, L.M.; Duncan, E.L.; Moy, E.L.; Reddel, R.R. Loss of wild-type ATRX expression in somatic cell hybrids segregates with activation of Alternative Lengthening of Telomeres. PLoS ONE 2012, 7, e50062. [Google Scholar] [CrossRef]
- Goldberg, A.D.; Banaszynski, L.A.; Noh, K.M.; Lewis, P.W.; Elsaesser, S.J.; Stadler, S.; Dewell, S.; Law, M.; Guo, X.; Li, X.; et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 2010, 140, 678–691. [Google Scholar] [CrossRef]
- Lewis, P.W.; Elsaesser, S.J.; Noh, K.M.; Stadler, S.C.; Allis, C.D. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. USA 2010, 107, 14075–14080. [Google Scholar] [CrossRef]
- Drane, P.; Ouararhni, K.; Depaux, A.; Shuaib, M.; Hamiche, A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010, 24, 1253–1265. [Google Scholar] [CrossRef]
- Sieverling, L.; Hong, C.; Koser, S.D.; Ginsbach, P.; Kleinheinz, K.; Hutter, B.; Braun, D.M.; Cortes-Ciriano, I.; Xi, R.; Kabbe, R.; et al. Genomic footprints of activated telomere maintenance mechanisms in cancer. Nat. Commun. 2020, 11, 733. [Google Scholar] [CrossRef] [PubMed]
- de Nonneville, A.; Reddel, R.R. Alternative lengthening of telomeres is not synonymous with mutations in ATRX/DAXX. Nat. Commun. 2021, 12, 1552. [Google Scholar] [CrossRef]
- Feuerbach, L. Formal reply to “Alternative lengthening of telomeres is not synonymous with mutations in ATRX/DAXX”. Nat. Commun. 2021, 12, 1551. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, R.J.; Picketts, D.J.; Villard, L.; Higgs, D.R. Mutations in a putative global transcriptional regulator cause X-linked mental retardation with α-thalassemia (ATR-X syndrome). Cell 1995, 80, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, R.J.; Bachoo, S.; Picketts, D.J.; Aftimos, S.; Asenbauer, B.; Bergoffen, J.; Berry, S.A.; Dahl, N.; Fryer, A.; Keppler, K.; et al. Mutations in transcriptional regulator ATRX establish the functional significance of a PHD-like domain. Nat. Genet. 1997, 17, 146–148. [Google Scholar] [CrossRef]
- McDowell, T.L.; Gibbons, R.J.; Sutherland, H.; O’Rourke, D.M.; Bickmore, W.A.; Pombo, A.; Turley, H.; Gatter, K.; Picketts, D.J.; Buckle, V.J.; et al. Localization of a putative transcriptional regulator (ATRX) at pericentromeric heterochromatin and the short arms of acrocentric chromosomes. Proc. Natl. Acad. Sci. USA 1999, 96, 13983–13988. [Google Scholar] [CrossRef] [PubMed]
- Stayton, C.L.; Dabovic, B.; Gulisano, M.; Gecz, J.; Broccoli, V.; Giovanazzi, S.; Bossolasco, M.; Monaco, L.; Rastan, S.; Boncinelli, E.; et al. Cloning and characterization of a new human Xq13 gene, encoding a putative helicase. Hum. Mol. Genet. 1994, 3, 1957–1964. [Google Scholar] [CrossRef]
- Garrick, D.; Samara, V.; McDowell, T.L.; Smith, A.J.; Dobbie, L.; Higgs, D.R.; Gibbons, R.J. A conserved truncated isoform of the ATR-X syndrome protein lacking the SWI/SNF-homology domain. Gene 2004, 326, 23–34. [Google Scholar] [CrossRef]
- Picketts, D.J.; Higgs, D.R.; Bachoo, S.; Blake, D.J.; Quarrell, O.W.; Gibbons, R.J. ATRX encodes a novel member of the SNF2 family of proteins: Mutations point to a common mechanism underlying the ATR-X syndrome. Hum. Mol. Genet. 1996, 5, 1899–1907. [Google Scholar] [CrossRef]
- Oates, M.E.; Romero, P.; Ishida, T.; Ghalwash, M.; Mizianty, M.J.; Xue, B.; Dosztanyi, Z.; Uversky, V.N.; Obradovic, Z.; Kurgan, L.; et al. D(2)P(2): Database of disordered protein predictions. Nucleic Acids Res. 2013, 41, D508–D516. [Google Scholar] [CrossRef]
- Flaus, A.; Martin, D.M.; Barton, G.J.; Owen-Hughes, T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006, 34, 2887–2905. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Gibbons, R.; Yan, Z.; Yang, D.; McDowell, T.L.; Sechi, S.; Qin, J.; Zhou, S.; Higgs, D.; Wang, W. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc. Natl. Acad. Sci. USA 2003, 100, 10635–10640. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Aapola, U.; Kawasaki, K.; Scott, H.S.; Ollila, J.; Vihinen, M.; Heino, M.; Shintani, A.; Kawasaki, K.; Minoshima, S.; Krohn, K.; et al. Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics 2000, 65, 293–298. [Google Scholar] [CrossRef]
- Iwase, S.; Xiang, B.; Ghosh, S.; Ren, T.; Lewis, P.W.; Cochrane, J.C.; Allis, C.D.; Picketts, D.J.; Patel, D.J.; Li, H.; et al. ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome. Nat. Struct. Mol. Biol. 2011, 18, 769–776. [Google Scholar] [CrossRef] [PubMed]
- Eustermann, S.; Yang, J.C.; Law, M.J.; Amos, R.; Chapman, L.M.; Jelinska, C.; Garrick, D.; Clynes, D.; Gibbons, R.J.; Rhodes, D.; et al. Combinatorial readout of histone H3 modifications specifies localization of ATRX to heterochromatin. Nat. Struct. Mol. Biol. 2011, 18, 777–782. [Google Scholar] [CrossRef]
- Gibbons, R.J.; Wada, T.; Fisher, C.A.; Malik, N.; Mitson, M.J.; Steensma, D.P.; Fryer, A.; Goudie, D.R.; Krantz, I.D.; Traeger-Synodinos, J. Mutations in the chromatin-associated protein ATRX. Hum. Mutat. 2008, 29, 796–802. [Google Scholar] [CrossRef]
- Cardoso, C.; Lutz, Y.; Mignon, C.; Compe, E.; Depetris, D.; Mattei, M.G.; Fontes, M.; Colleaux, L. ATR-X mutations cause impaired nuclear location and altered DNA binding properties of the XNP/ATR-X protein. J. Med. Genet. 2000, 37, 746–751. [Google Scholar] [CrossRef]
- Garrick, D.; Sharpe, J.A.; Arkell, R.; Dobbie, L.; Smith, A.J.; Wood, W.G.; Higgs, D.R.; Gibbons, R.J. Loss of Atrx affects trophoblast development and the pattern of X-inactivation in extraembryonic tissues. PLoS Genet. 2006, 2, e58. [Google Scholar] [CrossRef]
- Masliah-Planchon, J.; Levy, D.; Heron, D.; Giuliano, F.; Badens, C.; Freneaux, P.; Galmiche, L.; Guinebretierre, J.M.; Cellier, C.; Waterfall, J.J.; et al. Does ATRX germline variation predispose to osteosarcoma? Three additional cases of osteosarcoma in two ATR-X syndrome patients. Eur. J. Hum. Genet. 2018, 26, 1217–1221. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Quindipan, C.; Parham, D.; Shen, L.; Ruble, D.; Bootwalla, M.; Maglinte, D.T.; Gai, X.; Saitta, S.C.; Biegel, J.A.; et al. Inherited germline ATRX mutation in two brothers with ATR-X syndrome and osteosarcoma. Am. J. Med. Genet. A 2017, 173, 1390–1395. [Google Scholar] [CrossRef] [PubMed]
- Smolle, M.A.; Heitzer, E.; Geigl, J.B.; Al Kaissi, A.; Liegl-Atzwanger, B.; Seidel, M.G.; Holzer, L.A.; Leithner, A. A novel mutation in ATRX associated with intellectual disability, syndromic features, and osteosarcoma. Pediatr. Blood Cancer 2017, 64, e26522. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Wu, S.; Liu, H.; Stratt, R.; Barak, O.G.; Shiekhattar, R.; Picketts, D.J.; Yang, X. A novel transcription regulatory complex containing death domain-associated protein and the ATR-X syndrome protein. J. Biol. Chem. 2004, 279, 20369–20377. [Google Scholar] [CrossRef]
- Yang, X.; Khosravi-Far, R.; Chang, H.Y.; Baltimore, D. Daxx, a novel Fas-binding protein that activates JNK and apoptosis. Cell 1997, 89, 1067–1076. [Google Scholar] [CrossRef]
- Chang, H.Y.; Nishitoh, H.; Yang, X.; Ichijo, H.; Baltimore, D. Activation of apoptosis signal-regulating kinase 1 (ASK1) by the adapter protein Daxx. Science 1998, 281, 1860–1863. [Google Scholar] [CrossRef]
- Ishov, A.M.; Vladimirova, O.V.; Maul, G.G. Heterochromatin and ND10 are cell-cycle regulated and phosphorylation-dependent alternate nuclear sites of the transcription repressor Daxx and SWI/SNF protein ATRX. J. Cell Sci. 2004, 117, 3807–3820. [Google Scholar] [CrossRef]
- Torii, S.; Egan, D.A.; Evans, R.A.; Reed, J.C. Human Daxx regulates Fas-induced apoptosis from nuclear PML oncogenic domains (PODs). EMBO J. 1999, 18, 6037–6049. [Google Scholar] [CrossRef]
- Michaelson, J.S.; Bader, D.; Kuo, F.; Kozak, C.; Leder, P. Loss of Daxx, a promiscuously interacting protein, results in extensive apoptosis in early mouse development. Genes Dev. 1999, 13, 1918–1923. [Google Scholar] [CrossRef]
- Kiriakidou, M.; Driscoll, D.A.; Lopez-Guisa, J.M.; Strauss, J.F., 3rd. Cloning and expression of primate Daxx cDNAs and mapping of the human gene to chromosome 6p21.3 in the MHC region. DNA Cell Biol. 1997, 16, 1289–1298. [Google Scholar] [CrossRef]
- Hoelper, D.; Huang, H.; Jain, A.Y.; Patel, D.J.; Lewis, P.W. Structural and mechanistic insights into ATRX-dependent and -independent functions of the histone chaperone DAXX. Nat. Commun. 2017, 8, 1193. [Google Scholar] [CrossRef] [PubMed]
- DeNizio, J.E.; Elsasser, S.J.; Black, B.E. DAXX co-folds with H3.3/H4 using high local stability conferred by the H3.3 variant recognition residues. Nucleic Acids Res. 2014, 42, 4318–4331. [Google Scholar] [CrossRef] [PubMed]
- Elsässer, S.J.; Huang, H.; Lewis, P.W.; Chin, J.W.; Allis, C.D.; Patel, D.J. DAXX envelops a histone H3.3-H4 dimer for H3.3-specific recognition. Nature 2012, 491, 560–565. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-P.; Xiong, C.; Wang, M.; Yu, Z.; Yang, N.; Chen, P.; Zhang, Z.; Li, G.; Xu, R.-M. Structure of the variant histone H3.3-H4 heterodimer in complex with its chaperone DAXX. Nat. Struct. Mol. Biol. 2012, 19, 1287–1292. [Google Scholar] [CrossRef]
- Wu, R.S.; Tsai, S.; Bonner, W.M. Patterns of histone variant synthesis can distinguish G0 from G1 cells. Cell 1982, 31, 367–374. [Google Scholar] [CrossRef]
- Henikoff, S.; Furuyama, T.; Ahmad, K. Histone variants, nucleosome assembly and epigenetic inheritance. Trends Genet. 2004, 20, 320–326. [Google Scholar] [CrossRef]
- Elsaesser, S.J.; Allis, C.D. HIRA and Daxx constitute two independent histone H3.3-containing predeposition complexes. Cold Spring Harb. Symp. Quant. Biol. 2010, 75, 27–34. [Google Scholar] [CrossRef]
- Tagami, H.; Ray-Gallet, D.; Almouzni, G.; Nakatani, Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 2004, 116, 51–61. [Google Scholar] [CrossRef]
- Wirbelauer, C.; Bell, O.; Schubeler, D. Variant histone H3.3 is deposited at sites of nucleosomal displacement throughout transcribed genes while active histone modifications show a promoter-proximal bias. Genes Dev. 2005, 19, 1761–1766. [Google Scholar] [CrossRef]
- Ishov, A.M.; Sotnikov, A.G.; Negorev, D.; Vladimirova, O.V.; Neff, N.; Kamitani, T.; Yeh, E.T.; Strauss, J.F., 3rd; Maul, G.G. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 1999, 147, 221–234. [Google Scholar] [CrossRef]
- Corpet, A.; Kleijwegt, C.; Roubille, S.; Juillard, F.; Jacquet, K.; Texier, P.; Lomonte, P. PML nuclear bodies and chromatin dynamics: Catch me if you can! Nucleic Acids Res. 2020, 48, 11890–11912. [Google Scholar] [CrossRef] [PubMed]
- Banani, S.F.; Rice, A.M.; Peeples, W.B.; Lin, Y.; Jain, S.; Parker, R.; Rosen, M.K. Compositional Control of Phase-Separated Cellular Bodies. Cell 2016, 166, 651–663. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.Y.; Huang, Y.S.; Jeng, J.C.; Kuo, H.Y.; Chang, C.C.; Chao, T.T.; Ho, C.C.; Chen, Y.C.; Lin, T.P.; Fang, H.I.; et al. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol. Cell 2006, 24, 341–354. [Google Scholar] [CrossRef] [PubMed]
- Santiago, A.; Godsey, A.C.; Hossain, J.; Zhao, L.Y.; Liao, D. Identification of two independent SUMO-interacting motifs in Daxx: Evolutionary conservation from Drosophila to humans and their biochemical functions. Cell Cycle 2009, 8, 76–87. [Google Scholar] [CrossRef] [PubMed]
- Delbarre, E.; Ivanauskiene, K.; Kuntziger, T.; Collas, P. DAXX-dependent supply of soluble (H3.3-H4) dimers to PML bodies pending deposition into chromatin. Genome Res. 2013, 23, 440–451. [Google Scholar] [CrossRef]
- Delbarre, E.; Ivanauskiene, K.; Spirkoski, J.; Shah, A.; Vekterud, K.; Moskaug, J.Ø.; Ove, B.S.; Wong, L.H.; Küntziger, T.; Collas, P. PML protein organizes heterochromatin domains where it regulates histone H3.3 deposition by ATRX/DAXX. Genome Res. 2017, 27, 913–921. [Google Scholar] [CrossRef] [PubMed]
- Law, M.J.; Lower, K.M.; Voon, H.P.; Hughes, J.R.; Garrick, D.; Viprakasit, V.; Mitson, M.; De Gobbi, M.; Marra, M.; Morris, A.; et al. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell 2010, 143, 367–378. [Google Scholar] [CrossRef]
- Wong, L.H.; McGhie, J.D.; Sim, M.; Anderson, M.A.; Ahn, S.; Hannan, R.D.; George, A.J.; Morgan, K.A.; Mann, J.R.; Choo, K.H.A. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 2010, 20, 351–360. [Google Scholar] [CrossRef]
- Wang, Y.; Patel, D.J. Guanine residues in d(T2AG3) and d(T2G4) form parallel-stranded potassium cation stabilized G-quadruplexes with anti glycosidic torsion angles in solution. Biochemistry 1992, 31, 8112–8119. [Google Scholar] [CrossRef]
- Bochman, M.L.; Paeschke, K.; Zakian, V.A. DNA secondary structures: Stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012, 13, 770–780. [Google Scholar] [CrossRef]
- Clynes, D.; Jelinska, C.; Xella, B.; Ayyub, H.; Taylor, S.; Mitson, M.; Bachrati, C.Z.; Higgs, D.R.; Gibbons, R.J. ATRX dysfunction induces replication defects in primary mouse cells. PloS ONE 2014, 9, e92915. [Google Scholar] [CrossRef] [PubMed]
- Watson, L.A.; Solomon, L.A.; Li, J.R.; Jiang, Y.; Edwards, M.; Shin-ya, K.; Beier, F.; Berube, N.G. Atrx deficiency induces telomere dysfunction, endocrine defects, and reduced life span. J. Clin. Investig. 2013, 123, 2049–2063. [Google Scholar] [CrossRef] [PubMed]
- Levy, M.A.; Kernohan, K.D.; Jiang, Y.; Bérubé, N.G. ATRX promotes gene expression by facilitating transcriptional elongation through guanine-rich coding regions. Hum. Mol. Genet. 2015, 24, 1824–1835. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Kim, H.; Huang, R.; Lu, W.; Tang, M.; Shi, F.; Yang, D.; Zhang, X.; Huang, J.; Liu, D.; et al. The Daxx/Atrx Complex Protects Tandem Repetitive Elements during DNA Hypomethylation by Promoting H3K9 Trimethylation. Cell Stem Cell 2015, 17, 273–286. [Google Scholar] [CrossRef]
- Voon, H.P.; Hughes, J.R.; Rode, C.; De La Rosa-Velazquez, I.A.; Jenuwein, T.; Feil, R.; Higgs, D.R.; Gibbons, R.J. ATRX Plays a Key Role in Maintaining Silencing at Interstitial Heterochromatic Loci and Imprinted Genes. Cell Rep. 2015, 11, 405–418. [Google Scholar] [CrossRef]
- Chu, H.-P.; Cifuentes-Rojas, C.; Kesner, B.; Aeby, E.; Lee, H.-G.; Wei, C.; Oh, H.J.; Boukhali, M.; Haas, W.; Lee, J.T. TERRA RNA antagonizes ATRX and protects telomeres. Cell 2017, 170, 86–101.e116. [Google Scholar] [CrossRef]
- Azzalin, C.M.; Reichenbach, P.; Khoriauli, L.; Giulotto, E.; Lingner, J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 2007, 318, 798–801. [Google Scholar] [CrossRef]
- Schoeftner, S.; Blasco, M.A. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 2008, 10, 228–236. [Google Scholar] [CrossRef]
- Nergadze, S.G.; Farnung, B.O.; Wischnewski, H.; Khoriauli, L.; Vitelli, V.; Chawla, R.; Giulotto, E.; Azzalin, C.M. CpG-island promoters drive transcription of human telomeres. RNA 2009, 15, 2186–2194. [Google Scholar] [CrossRef]
- Porro, A.; Feuerhahn, S.; Reichenbach, P.; Lingner, J. Molecular dissection of telomeric repeat-containing RNA biogenesis unveils the presence of distinct and multiple regulatory pathways. Mol. Cell. Biol. 2010, 30, 4808–4817. [Google Scholar] [CrossRef]
- Cusanelli, E.; Romero, C.A.; Chartrand, P. Telomeric noncoding RNA TERRA is induced by telomere shortening to nucleate telomerase molecules at short telomeres. Mol. Cell 2013, 51, 780–791. [Google Scholar] [CrossRef] [PubMed]
- Montero, J.J.; López de Silanes, I.; Graña, O.; Blasco, M.A. Telomeric RNAs are essential to maintain telomeres. Nat. Commun. 2016, 7, 12534. [Google Scholar] [CrossRef] [PubMed]
- Lopez de Silanes, I.; Grana, O.; De Bonis, M.L.; Dominguez, O.; Pisano, D.G.; Blasco, M.A. Identification of TERRA locus unveils a telomere protection role through association to nearly all chromosomes. Nat. Commun. 2014, 5, 4723. [Google Scholar] [CrossRef] [PubMed]
- Silva, B.; Arora, R.; Bione, S.; Azzalin, C.M. TERRA transcription destabilizes telomere integrity to initiate break-induced replication in human ALT cells. Nat. Commun. 2021, 12, 3760. [Google Scholar] [CrossRef]
- Arnoult, N.; Van Beneden, A.; Decottignies, A. Telomere length regulates TERRA levels through increased trimethylation of telomeric H3K9 and HP1alpha. Nat. Struct. Mol. Biol. 2012, 19, 948–956. [Google Scholar] [CrossRef]
- Deng, Z.; Wang, Z.; Stong, N.; Plasschaert, R.; Moczan, A.; Chen, H.S.; Hu, S.; Wikramasinghe, P.; Davuluri, R.V.; Bartolomei, M.S.; et al. A role for CTCF and cohesin in subtelomere chromatin organization, TERRA transcription, and telomere end protection. EMBO J. 2012, 31, 4165–4178. [Google Scholar] [CrossRef]
- Porro, A.; Feuerhahn, S.; Delafontaine, J.; Riethman, H.; Rougemont, J.; Lingner, J. Functional characterization of the TERRA transcriptome at damaged telomeres. Nat. Commun. 2014, 5, 5379. [Google Scholar] [CrossRef]
- Arora, R.; Lee, Y.; Wischnewski, H.; Brun, C.M.; Schwarz, T.; Azzalin, C.M. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat. Commun. 2014, 5, 5220. [Google Scholar] [CrossRef]
- Feretzaki, M.; Pospisilova, M.; Valador Fernandes, R.; Lunardi, T.; Krejci, L.; Lingner, J. RAD51-dependent recruitment of TERRA lncRNA to telomeres through R-loops. Nature 2020, 587, 303–308. [Google Scholar] [CrossRef]
- Avogaro, L.; Querido, E.; Dalachi, M.; Jantsch, M.F.; Chartrand, P.; Cusanelli, E. Live-cell imaging reveals the dynamics and function of single-telomere TERRA molecules in cancer cells. RNA Biol. 2018, 15, 787–796. [Google Scholar] [CrossRef]
- Deng, Z.; Norseen, J.; Wiedmer, A.; Riethman, H.; Lieberman, P.M. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell 2009, 35, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Montero, J.J.; López-Silanes, I.; Megías, D.; Fraga, M.F.; Castells-García, Á.; Blasco, M.A. TERRA recruitment of polycomb to telomeres is essential for histone trymethylation marks at telomeric heterochromatin. Nat. Commun. 2018, 9, 1548. [Google Scholar] [CrossRef]
- Tsai, R.X.; Fang, K.C.; Yang, P.C.; Hsieh, Y.H.; Chiang, I.T.; Chen, Y.; Lee, H.G.; Lee, J.T.; Chu, H.C. TERRA regulates DNA G-quadruplex formation and ATRX recruitment to chromatin. Nucleic Acids Res. 2022, 50, 12217–12234. [Google Scholar] [CrossRef] [PubMed]
- Santos-Pereira, J.M.; Aguilera, A. R loops: New modulators of genome dynamics and function. Nat. Rev. Genet. 2015, 16, 583–597. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.T.; Voon, H.P.J.; Xella, B.; Scott, C.; Clynes, D.; Babbs, C.; Ayyub, H.; Kerry, J.; Sharpe, J.A.; Sloane-Stanley, J.A.; et al. The chromatin remodelling factor ATRX suppresses R-loops in transcribed telomeric repeats. EMBO Rep. 2017, 18, 914–928. [Google Scholar] [CrossRef] [PubMed]
- Lerner, L.K.; Sale, J.E. Replication of G Quadruplex DNA. Genes 2019, 10, 95. [Google Scholar] [CrossRef]
- Rinaldi, C.; Pizzul, P.; Longhese, M.P.; Bonetti, D. Sensing R-Loop-Associated DNA Damage to Safeguard Genome Stability. Front. Cell Dev. Biol. 2020, 8, 618157. [Google Scholar] [CrossRef]
- Leung, J.W.; Ghosal, G.; Wang, W.; Shen, X.; Wang, J.; Li, L.; Chen, J. α thalassemia/mental retardation syndrome X-linked gene product ATRX is required for proper replication restart and cellular resistance to replication stress. J. Biol. Chem. 2013, 288, 6342–6350. [Google Scholar] [CrossRef]
- Huh, M.S.; Ivanochko, D.; Hashem, L.E.; Curtin, M.; Delorme, M.; Goodall, E.; Yan, K.; Picketts, D.J. Stalled replication forks within heterochromatin require ATRX for protection. Cell Death Dis. 2016, 7, e2220. [Google Scholar] [CrossRef]
- Ritchie, K.; Seah, C.; Moulin, J.; Isaac, C.; Dick, F.; Berube, N.G. Loss of ATRX leads to chromosome cohesion and congression defects. J. Cell Biol. 2008, 180, 315–324. [Google Scholar] [CrossRef]
- Eid, R.; Demattei, M.-V.; Episkopou, H.; Augé-Gouillou, C.; Decottignies, A.; Grandin, N.; Charbonneau, M. Genetic inactivation of ATRX leads to a decrease in the amount of telomeric cohesin and level of telomere transcription in human glioma cells. Mol. Cell Biol. 2015, 35, 2818–2830. [Google Scholar] [CrossRef] [PubMed]
- Lovejoy, C.A.; Takai, K.; Huh, M.S.; Picketts, D.J.; de Lange, T. ATRX affects the repair of telomeric DSBs by promoting cohesion and a DAXX-dependent activity. PLoS Biol. 2020, 18, e3000594. [Google Scholar] [CrossRef]
- Ramamoorthy, M.; Smith, S. Loss of ATRX Suppresses Resolution of Telomere Cohesion to Control Recombination in ALT Cancer Cells. Cancer Cell 2015, 28, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Juhasz, S.; Elbakry, A.; Mathes, A.; Lobrich, M. ATRX Promotes DNA Repair Synthesis and Sister Chromatid Exchange during Homologous Recombination. Mol. Cell 2018, 71, 11–24.e17. [Google Scholar] [CrossRef] [PubMed]
- Elbakry, A.; Juhasz, S.; Chan, K.C.; Lobrich, M. ATRX and RECQ5 define distinct homologous recombination subpathways. Proc. Natl. Acad. Sci. USA 2021, 118, e2010370118. [Google Scholar] [CrossRef]
- Hein, M.Y.; Hubner, N.C.; Poser, I.; Cox, J.; Nagaraj, N.; Toyoda, Y.; Gak, I.A.; Weisswange, I.; Mansfeld, J.; Buchholz, F.; et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 2015, 163, 712–723. [Google Scholar] [CrossRef]
- Li, R.; Pei, H.; Watson, D.K.; Papas, T.S. EAP1/Daxx interacts with ETS1 and represses transcriptional activation of ETS1 target genes. Oncogene 2000, 19, 745–753. [Google Scholar] [CrossRef]
- Croxton, R.; Puto, L.A.; de Belle, I.; Thomas, M.; Torii, S.; Hanaii, F.; Cuddy, M.; Reed, J.C. Daxx represses expression of a subset of antiapoptotic genes regulated by nuclear factor-kappaB. Cancer Res. 2006, 66, 9026–9035. [Google Scholar] [CrossRef]
- Zhao, L.Y.; Liu, J.; Sidhu, G.S.; Niu, Y.; Liu, Y.; Wang, R.; Liao, D. Negative regulation of p53 functions by Daxx and the involvement of MDM2. J. Biol. Chem. 2004, 279, 50566–50579. [Google Scholar] [CrossRef]
- Bouchard, J.J.; Otero, J.H.; Scott, D.C.; Szulc, E.; Martin, E.W.; Sabri, N.; Granata, D.; Marzahn, M.R.; Lindorff-Larsen, K.; Salvatella, X.; et al. Cancer Mutations of the Tumor Suppressor SPOP Disrupt the Formation of Active, Phase-Separated Compartments. Mol. Cell 2018, 72, 19–36.e18. [Google Scholar] [CrossRef]
- Huang, L.; Agrawal, T.; Zhu, G.; Yu, S.; Tao, L.; Lin, J.; Marmorstein, R.; Shorter, J.; Yang, X. DAXX represents a new type of protein-folding enabler. Nature 2021, 597, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.S.; Zakian, V.A. Telomere-telomere recombination provides an express pathway for telomere acquisition. Nature 1990, 345, 456–458. [Google Scholar] [CrossRef] [PubMed]
- Lundblad, V.; Blackburn, E.H. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell 1993, 73, 347–360. [Google Scholar] [CrossRef]
- Biessmann, H.; Mason, J.M.; Ferry, K.; d’Hulst, M.; Valgeirsdottir, K.; Traverse, K.L.; Pardue, M.L. Addition of telomere-associated HeT DNA sequences “heals” broken chromosome ends in Drosophila. Cell 1990, 61, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Dunham, M.A.; Neumann, A.A.; Fasching, C.L.; Reddel, R.R. Telomere maintenance by recombination in human cells. Nat. Genet. 2000, 26, 447–450. [Google Scholar] [CrossRef] [PubMed]
- Cives, M.; Partelli, S.; Palmirotta, R.; Lovero, D.; Mandriani, B.; Quaresmini, D.; Pelle, E.; Andreasi, V.; Castelli, P.; Strosberg, J.; et al. DAXX mutations as potential genomic markers of malignant evolution in small nonfunctioning pancreatic neuroendocrine tumors. Sci. Rep. 2019, 9, 18614. [Google Scholar] [CrossRef]
- Singhi, A.D.; Liu, T.C.; Roncaioli, J.L.; Cao, D.; Zeh, H.J.; Zureikat, A.H.; Tsung, A.; Marsh, J.W.; Lee, K.K.; Hogg, M.E.; et al. Alternative Lengthening of Telomeres and Loss of DAXX/ATRX Expression Predicts Metastatic Disease and Poor Survival in Patients with Pancreatic Neuroendocrine Tumors. Clin. Cancer Res. 2017, 23, 600–609. [Google Scholar] [CrossRef]
- Jiao, Y.; Shi, C.; Edil, B.H.; de Wilde, R.F.; Klimstra, D.S.; Maitra, A.; Schulick, R.D.; Tang, L.H.; Wolfgang, C.L.; Choti, M.A.; et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science 2011, 331, 1199–1203. [Google Scholar] [CrossRef]
- de Wilde, R.F.; Heaphy, C.M.; Maitra, A.; Meeker, A.K.; Edil, B.H.; Wolfgang, C.L.; Ellison, T.A.; Schulick, R.D.; Molenaar, I.Q.; Valk, G.D.; et al. Loss of ATRX or DAXX expression and concomitant acquisition of the alternative lengthening of telomeres phenotype are late events in a small subset of MEN-1 syndrome pancreatic neuroendocrine tumors. Mod. Pathol. 2012, 25, 1033–1039. [Google Scholar] [CrossRef]
- Minasi, S.; Baldi, C.; Gianno, F.; Antonelli, M.; Buccoliero, A.M.; Pietsch, T.; Massimino, M.; Buttarelli, F.R. Alternative lengthening of telomeres in molecular subgroups of paediatric high-grade glioma. Child’s Nerv. Syst. 2021, 37, 809–818. [Google Scholar] [CrossRef]
- Ogino, H.; Nakabayashi, K.; Suzuki, M.; Takahashi, E.; Fujii, M.; Suzuki, T.; Ayusawa, D. Release of telomeric DNA from chromosomes in immortal human cells lacking telomerase activity. Biochem. Biophys. Res. Commun. 1998, 248, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Yeager, T.R.; Neumann, A.A.; Englezou, A.; Huschtscha, L.I.; Noble, J.R.; Reddel, R.R. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 1999, 59, 4175–4179. [Google Scholar] [PubMed]
- Draskovic, I.; Arnoult, N.; Steiner, V.; Bacchetti, S.; Lomonte, P.; Londono-Vallejo, A. Probing PML body function in ALT cells reveals spatiotemporal requirements for telomere recombination. Proc. Natl. Acad. Sci. USA 2009, 106, 15726–15731. [Google Scholar] [CrossRef] [PubMed]
- Murnane, J.P.; Sabatier, L.; Marder, B.A.; Morgan, W.F. Telomere dynamics in an immortal human cell line. EMBO J. 1994, 13, 4953–4962. [Google Scholar] [CrossRef]
- Rivera, T.; Haggblom, C.; Cosconati, S.; Karlseder, J. A balance between elongation and trimming regulates telomere stability in stem cells. Nat. Struct. Mol. Biol. 2017, 24, 30–39. [Google Scholar] [CrossRef]
- Mazzucco, G.; Huda, A.; Galli, M.; Piccini, D.; Giannattasio, M.; Pessina, F.; Doksani, Y. Telomere damage induces internal loops that generate telomeric circles. Nat. Commun. 2020, 11, 5297. [Google Scholar] [CrossRef] [PubMed]
- Henson, J.D.; Lau, L.M.; Koch, S.; Martin La Rotta, N.; Dagg, R.A.; Reddel, R.R. The C-Circle Assay for alternative-lengthening-of-telomeres activity. Methods 2016, 114, 74–84. [Google Scholar] [CrossRef]
- Chen, Y.A.; Shen, Y.L.; Hsia, H.Y.; Tiang, Y.P.; Sung, T.L.; Chen, L.Y. Extrachromosomal telomere repeat DNA is linked to ALT development via cGAS-STING DNA sensing pathway. Nat. Struct. Mol. Biol. 2017, 24, 1124–1131. [Google Scholar] [CrossRef]
- Zhang, J.M.; Yadav, T.; Ouyang, J.; Lan, L.; Zou, L. Alternative Lengthening of Telomeres through Two Distinct Break-Induced Replication Pathways. Cell Rep. 2019, 26, 955–968.e953. [Google Scholar] [CrossRef]
- Loe, T.K.; Li, J.S.Z.; Zhang, Y.; Azeroglu, B.; Boddy, M.N.; Denchi, E.L. Telomere length heterogeneity in ALT cells is maintained by PML-dependent localization of the BTR complex to telomeres. Genes Dev. 2020, 34, 650–662. [Google Scholar] [CrossRef]
- Potts, P.R.; Yu, H. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat. Struct. Mol. Biol. 2007, 14, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.M.; Genois, M.M.; Ouyang, J.; Lan, L.; Zou, L. Alternative lengthening of telomeres is a self-perpetuating process in ALT-associated PML bodies. Mol. Cell 2021, 81, 1027–1042.e1024. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhao, R.; Tones, J.; Liu, M.; Dilley, R.L.; Chenoweth, D.M.; Greenberg, R.A.; Lampson, M.A. Nuclear body phase separation drives telomere clustering in ALT cancer cells. Mol. Biol. Cell 2020, 31, 2048–2056. [Google Scholar] [CrossRef]
- Napier, C.E.; Huschtscha, L.I.; Harvey, A.; Bower, K.; Noble, J.R.; Hendrickson, E.A.; Reddel, R.R. ATRX represses alternative lengthening of telomeres. Oncotarget 2015, 6, 16543–16558. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Shi, G.; Zhang, L.; Li, F.; Jiang, Y.; Jiang, S.; Ma, W.; Zhao, Y.; Songyang, Z.; Huang, J. Switch telomerase to ALT mechanism by inducing telomeric DNA damages and dysfunction of ATRX and DAXX. Sci. Rep. 2016, 6, 32280. [Google Scholar] [CrossRef]
- O’Sullivan, R.J.; Arnoult, N.; Lackner, D.H.; Oganesian, L.; Haggblom, C.; Corpet, A.; Almouzni, G.; Karlseder, J. Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1. Nat. Struct. Mol. Biol. 2014, 21, 167–174. [Google Scholar] [CrossRef]
- Lee, S.B.; Segura-Bayona, S.; Villamor-Paya, M.; Saredi, G.; Todd, M.A.M.; Attolini, C.S.; Chang, T.Y.; Stracker, T.H.; Groth, A. Tousled-like kinases stabilize replication forks and show synthetic lethality with checkpoint and PARP inhibitors. Sci. Adv. 2018, 4, eaat4985. [Google Scholar] [CrossRef]
- Hoang, S.M.; Kaminski, N.; Bhargava, R.; Barroso-Gonzalez, J.; Lynskey, M.L.; Garcia-Exposito, L.; Roncaioli, J.L.; Wondisford, A.R.; Wallace, C.T.; Watkins, S.C.; et al. Regulation of ALT-associated homology-directed repair by polyADP-ribosylation. Nat. Struct. Mol. Biol. 2020, 27, 1152–1164. [Google Scholar] [CrossRef]
- Dilley, R.L.; Verma, P.; Cho, N.W.; Winters, H.D.; Wondisford, A.R.; Greenberg, R.A. Break-induced telomere synthesis underlies alternative telomere maintenance. Nature 2016, 539, 54–58. [Google Scholar] [CrossRef]
- Min, J.; Wright, W.E.; Shay, J.W. Alternative Lengthening of Telomeres Mediated by Mitotic DNA Synthesis Engages Break-Induced Replication Processes. Mol. Cell Biol 2017, 37, e00226-17. [Google Scholar] [CrossRef]
- Ozer, O.; Bhowmick, R.; Liu, Y.; Hickson, I.D. Human cancer cells utilize mitotic DNA synthesis to resist replication stress at telomeres regardless of their telomere maintenance mechanism. Oncotarget 2018, 9, 15836–15846. [Google Scholar] [CrossRef]
- Verma, P.; Dilley, R.L.; Zhang, T.; Gyparaki, M.T.; Li, Y.; Greenberg, R.A. RAD52 and SLX4 act nonepistatically to ensure telomere stability during alternative telomere lengthening. Genes Dev. 2019, 33, 221–235. [Google Scholar] [CrossRef]
- Sobinoff, A.P.; Allen, J.A.; Neumann, A.A.; Yang, S.F.; Walsh, M.E.; Henson, J.D.; Reddel, R.R.; Pickett, H.A. BLM and SLX4 play opposing roles in recombination-dependent replication at human telomeres. EMBO J. 2017, 36, 2907–2919. [Google Scholar] [CrossRef]
- Min, J.; Wright, W.E.; Shay, J.W. Clustered telomeres in phase-separated nuclear condensates engage mitotic DNA synthesis through BLM and RAD52. Genes Dev. 2019, 33, 814–827. [Google Scholar] [CrossRef]
- Episkopou, H.; Draskovic, I.; Van Beneden, A.; Tilman, G.; Mattiussi, M.; Gobin, M.; Arnoult, N.; Londoño-Vallejo, A.; Decottignies, A. Alternative Lengthening of Telomeres is characterized by reduced compaction of telomeric chromatin. Nucleic Acids Res. 2014, 42, 4391–4405. [Google Scholar] [CrossRef]
- Kaminski, N.; Wondisford, A.R.; Kwon, Y.; Lynskey, M.L.; Bhargava, R.; Barroso-Gonzalez, J.; Garcia-Exposito, L.; He, B.; Xu, M.; Mellacheruvu, D.; et al. RAD51AP1 regulates ALT-HDR through chromatin-directed homeostasis of TERRA. Mol. Cell 2022, 82, 4001–4017.e4007. [Google Scholar] [CrossRef]
- Yadav, T.; Zhang, J.M.; Ouyang, J.; Leung, W.; Simoneau, A.; Zou, L. TERRA and RAD51AP1 promote alternative lengthening of telomeres through an R- to D-loop switch. Mol. Cell 2022, 82, 3985–4000.e3984. [Google Scholar] [CrossRef]
- Clynes, D.; Jelinska, C.; Xella, B.; Ayyub, H.; Scott, C.; Mitson, M.; Taylor, S.; Higgs, D.R.; Gibbons, R.J. Suppression of the alternative lengthening of telomere pathway by the chromatin remodelling factor ATRX. Nat. Commun. 2015, 6, 7538. [Google Scholar] [CrossRef]
- Mason-Osann, E.; Dai, A.; Floro, J.; Lock, Y.J.; Reiss, M.; Gali, H.; Matschulat, A.; Labadorf, A.; Flynn, R.L. Identification of a novel gene fusion in ALT positive osteosarcoma. Oncotarget 2018, 9, 32868–32880. [Google Scholar] [CrossRef]
- Yost, K.E.; Clatterbuck Soper, S.F.; Walker, R.L.; Pineda, M.A.; Zhu, Y.J.; Ester, C.D.; Showman, S.; Roschke, A.V.; Waterfall, J.J.; Meltzer, P.S. Rapid and reversible suppression of ALT by DAXX in osteosarcoma cells. Sci. Rep. 2019, 9, 4544. [Google Scholar] [CrossRef]
- Lu, R.; O’Rourke, J.J.; Sobinoff, A.P.; Allen, J.A.M.; Nelson, C.B.; Tomlinson, C.G.; Lee, M.; Reddel, R.R.; Deans, A.J.; Pickett, H.A. The FANCM-BLM-TOP3A-RMI complex suppresses alternative lengthening of telomeres (ALT). Nat. Commun. 2019, 10, 2252. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Drosopoulos, W.C.; Sethi, L.; Madireddy, A.; Schildkraut, C.L.; Zhang, D. FANCM, BRCA1, and BLM cooperatively resolve the replication stress at the ALT telomeres. Proc. Natl. Acad. Sci. USA 2017, 114, E5940–E5949. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clatterbuck Soper, S.F.; Meltzer, P.S. ATRX/DAXX: Guarding the Genome against the Hazards of ALT. Genes 2023, 14, 790. https://doi.org/10.3390/genes14040790
Clatterbuck Soper SF, Meltzer PS. ATRX/DAXX: Guarding the Genome against the Hazards of ALT. Genes. 2023; 14(4):790. https://doi.org/10.3390/genes14040790
Chicago/Turabian StyleClatterbuck Soper, Sarah F., and Paul S. Meltzer. 2023. "ATRX/DAXX: Guarding the Genome against the Hazards of ALT" Genes 14, no. 4: 790. https://doi.org/10.3390/genes14040790
APA StyleClatterbuck Soper, S. F., & Meltzer, P. S. (2023). ATRX/DAXX: Guarding the Genome against the Hazards of ALT. Genes, 14(4), 790. https://doi.org/10.3390/genes14040790




