RNA-Seq Profiling between Commercial and Indigenous Iranian Chickens Highlights Differences in Innate Immune Gene Expression
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animals, RNA Extraction and Sequencing
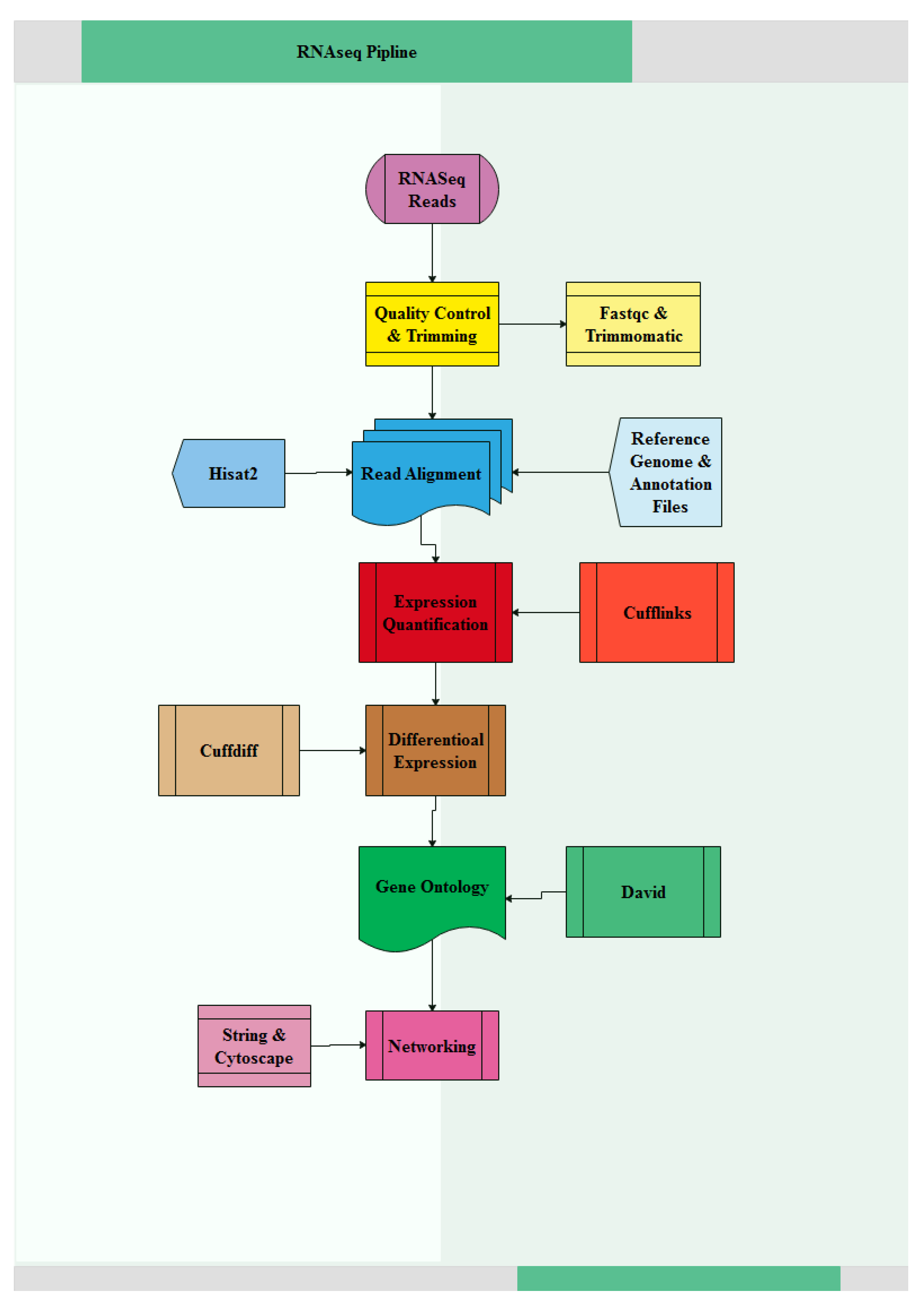
2.3. Data Quality Control, Read Mapping and Transcriptome Analysis
2.4. Functional Annotation
2.5. Protein–Protein Interaction (PPI) Network
2.6. Real-Time PCR
3. Results
3.1. Quality Control and Mapping of RNA-Seq Data
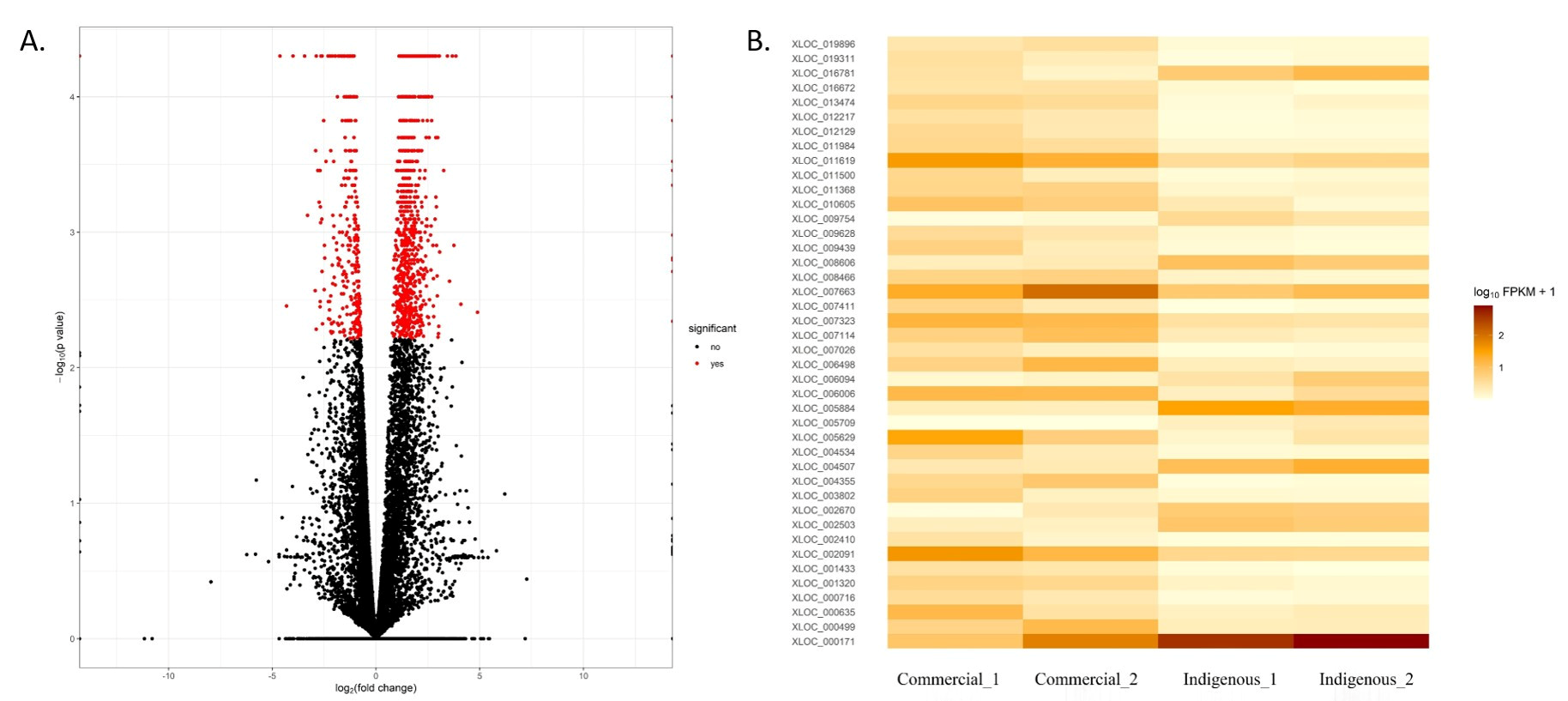
3.2. Gene Expression Analysis
3.3. Confirmation of Differential Gene Expression with Real-Time PCR
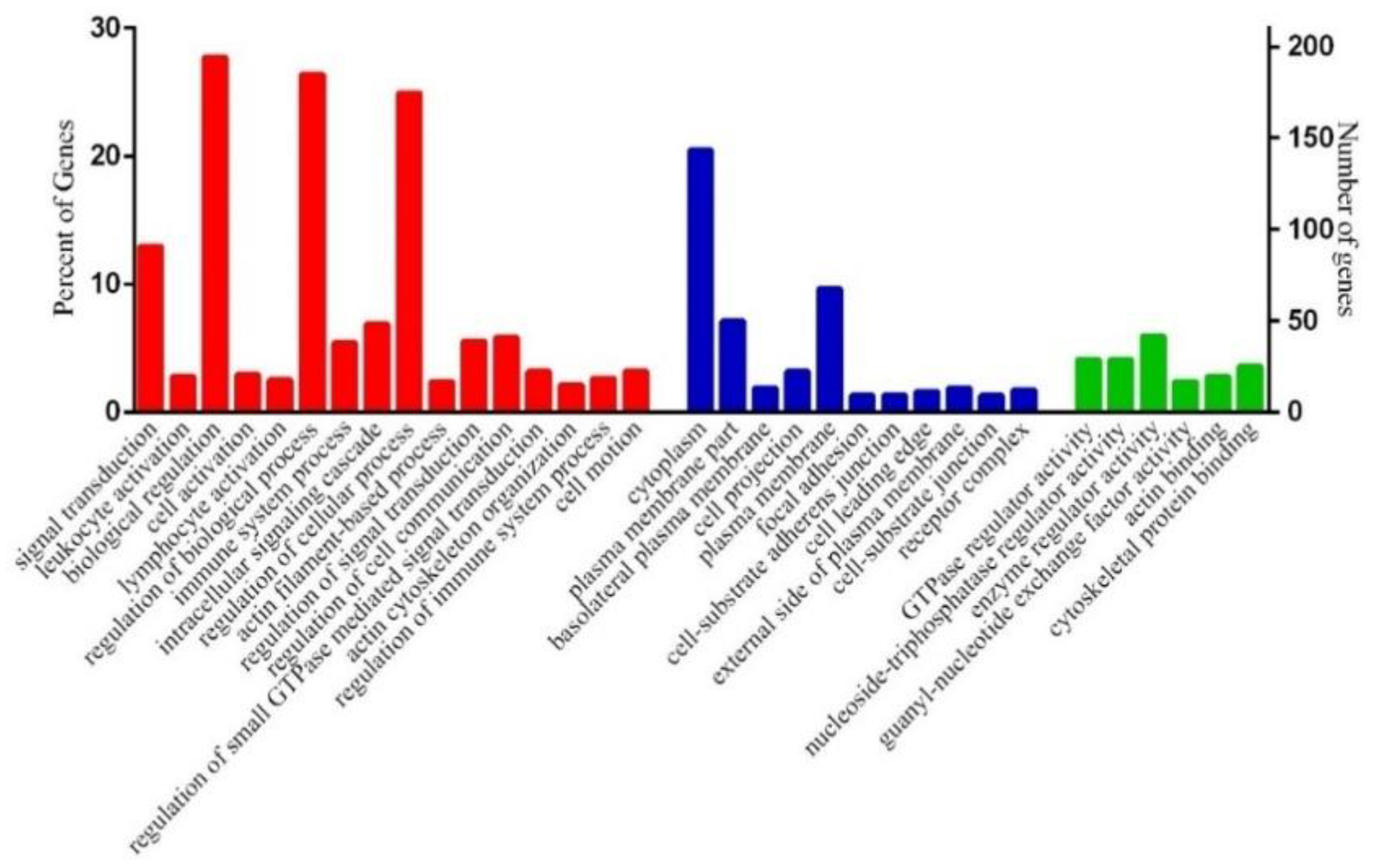
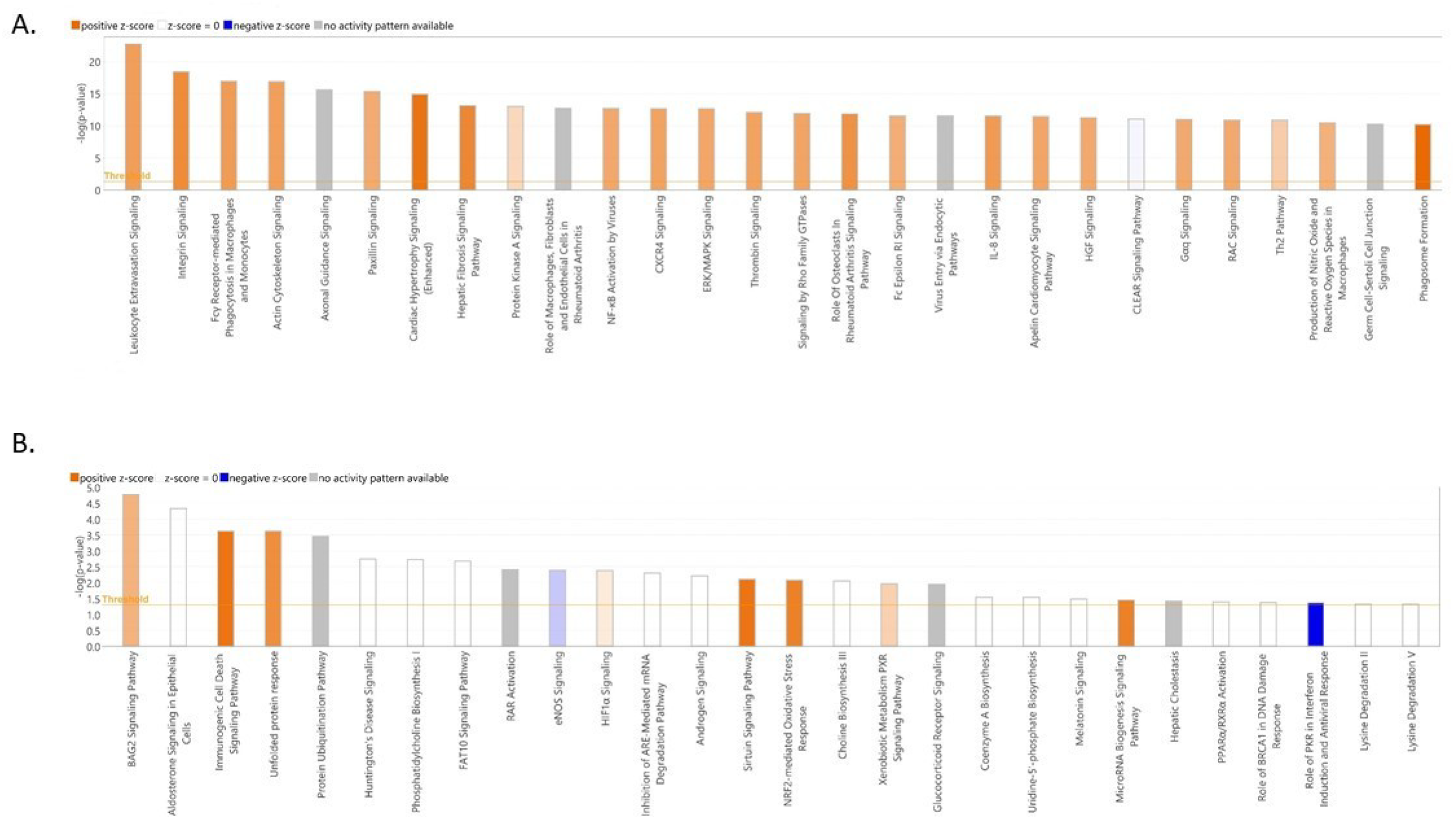
3.4. Functional Annotation of Differentially Expressed Genes
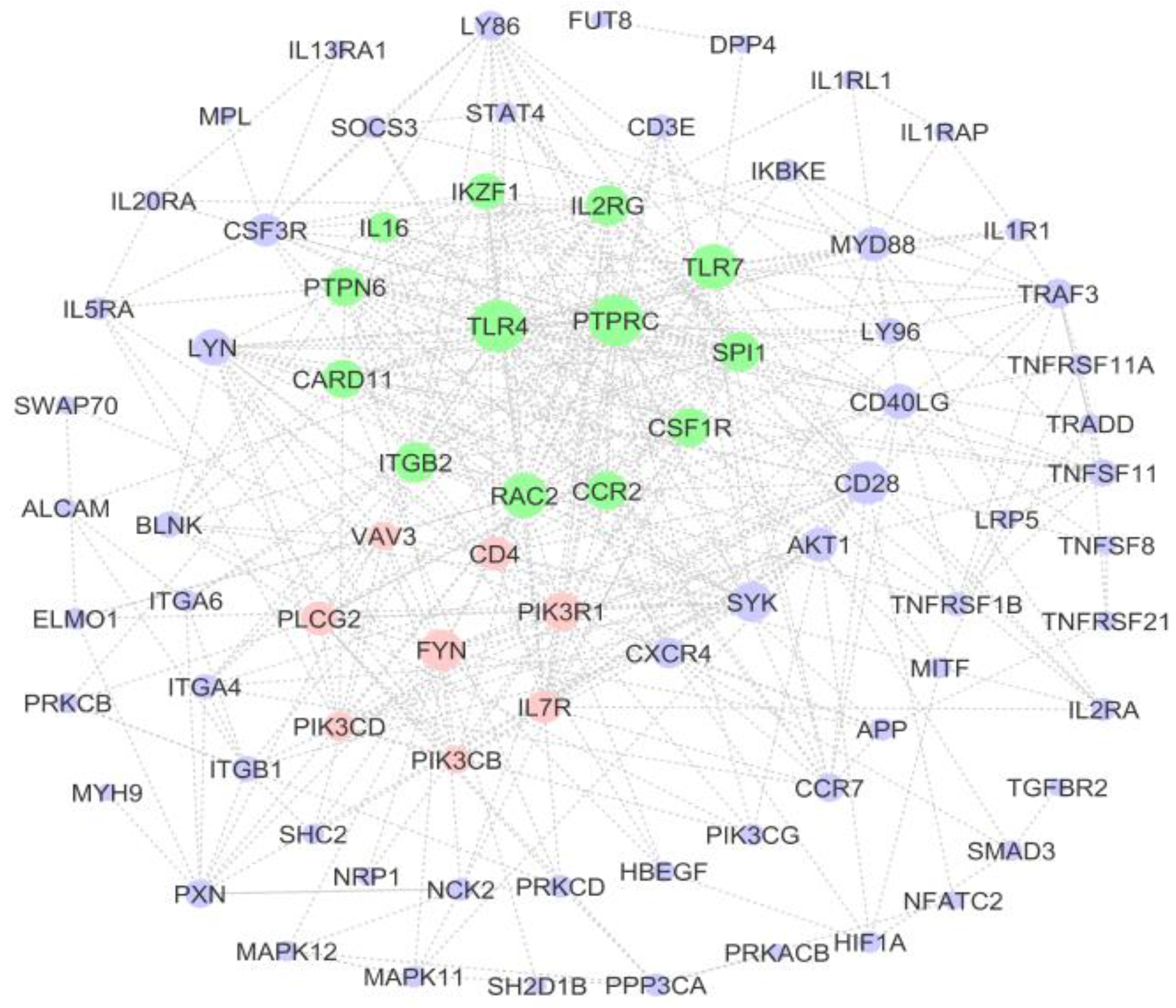
3.5. PPI Network of Immune-System-Related Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Davey, M.; Tickle, C. The chicken as a model for embryonic development. Cytogenet. Genome Res. 2007, 117, 231–239. [Google Scholar] [CrossRef]
- Mottet, A.; Tempio, G. Global poultry production: Current state and future outlook and challenges. World’s Poult. Sci. J. 2017, 73, 245–256. [Google Scholar] [CrossRef]
- Kaiser, P.B.A. Avian Immune System. In Sturkie’s Avian Physiology; Academic Press: Cambridge, MA, USA, 2015. [Google Scholar]
- Gul, H.; Habib, G.; Khan, I.M.; Rahman, S.U.; Khan, N.M.; Wang, H.; Khan, N.U.; Liu, Y. Genetic resilience in chickens against bacterial, viral and protozoal pathogens. Front. Veter. Sci. 2022, 9, 1032983. [Google Scholar] [CrossRef] [PubMed]
- Bayyari, G.; Huff, W.; Rath, N.; Balog, J.; Newberry, L.; Villines, J.; Skeeles, J.; Anthony, N.; Nestor, K. Effect of the genetic selection of turkeys for increased body weight and egg production on immune and physiological responses. Poult. Sci. 1997, 76, 289–296. [Google Scholar] [CrossRef]
- Zerjal, T.; Härtle, S.; Gourichon, D.; Guillory, V.; Bruneau, N.; Laloë, D.; der Laan, M.-H.P.-V.; Trapp, S.; Bed’Hom, B.; Quéré, P. Assessment of trade-offs between feed efficiency, growth-related traits, and immune activity in experimental lines of layer chickens. Genet. Sel. Evol. 2021, 53, 44. [Google Scholar] [CrossRef]
- Swaggerty, C.L.; Pevzner, I.Y.; He, H.; Genovese, K.J.; Nisbet, D.J.; Kaiser, P.; Kogut, M.H. Selection of Broilers with Improved Innate Immune Responsiveness to Reduce On-Farm Infection by Foodborne Pathogens. Foodborne Pathog. Dis. 2009, 6, 777–783. [Google Scholar] [CrossRef]
- Padhi, M.K. Importance of Indigenous Breeds of Chicken for Rural Economy and Their Improvements for Higher Production Performance. Scientifica 2016, 2016, 2604685. [Google Scholar] [CrossRef] [PubMed]
- Pinard, M.-H.; Van Arendonk, J.; Nieuwland, M.; Van Der Zijpp, A. Divergent selection for immune responsiveness in chickens: Estimation of realized heritability with an animal model. J. Anim. Sci. 1992, 70, 2986–2993. [Google Scholar] [CrossRef]
- Cheng, H.H.; Lamont, S.J. Diseases of poultry. In Genetics of Disease Resistance, 12th ed.; Saif, Y.M., Fadly, A.M., Glisson, J.R., McDougald, L.R., Nolan, L.K., Swayne, D.E., Eds.; Iowa State University Press: Ames, IA, USA, 2008; pp. 59–72. [Google Scholar]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Griffith, M.; Walker, J.R.; Spies, N.C.; Ainscough, B.J.; Griffith, O.L. Informatics for RNA Sequencing: A Web Resource for Analysis on the Cloud. PLoS Comput. Biol. 2015, 11, e1004393. [Google Scholar] [CrossRef]
- Dar, M.A.; Ahmad, S.M.; Bhat, B.A.; Dar, T.A.; Haq, Z.U.; Wani, B.A.; Shabir, N.; Kashoo, Z.A.; Shah, R.A.; Ganai, N.A.; et al. Comparative RNA-Seq analysis reveals insights in Salmonella disease resistance of chicken; and database development as resource for gene expression in poultry. Genomics 2022, 114, 110475. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Liu, X.; Mu, J.; Yan, W.; Wang, M.; Chai, H.; Sha, Y.; Jiang, S.; Wang, S.; Ren, Y.; et al. Intense Innate Immune Responses and Severe Metabolic Disorders in Chicken Embryonic Visceral Tissues Caused by Infection with Highly Virulent Newcastle Disease Virus Compared to the Avirulent Virus: A Bioinformatics Analysis. Viruses 2022, 14, 911. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016, 17, 333–351. [Google Scholar] [CrossRef] [PubMed]
- Kumar, H.; Choo, H.; Iskender, A.U.; Srikanth, K.; Kim, H.; Zhunushov, A.T.; Jang, G.W.; Lim, Y.; Song, K.-D.; Park, J.-E. RNA seq analyses of chicken reveals biological pathways involved in acclimation into different geographical locations. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 17 May 2018).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Dennis, G.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003, 4, 1. [Google Scholar] [CrossRef]
- Zhang, B.; Kirov, S.; Snoddy, J. WebGestalt: An integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005, 33, W741–W748. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Kuhn, M.; Simonovic, M.; Roth, A.; Minguez, P.; Doerks, T.; Stark, M.; Muller, J.; Bork, P. The STRING database in 2011: Functional interaction networks of proteins, globally inte-grated and scored. Nucleic Acids Res. 2011, 39, D561–D568. [Google Scholar] [CrossRef]
- Tang, Y.; Li, M.; Wang, J.; Pan, Y.; Wu, F.-X. CytoNCA: A cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems 2015, 127, 67–72. [Google Scholar] [CrossRef]
- Bader, G.D.; Hogue, C.W.V. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2–27. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Emam, M.; Mehrabani-Yeganeh, H.; Barjesteh, N.; Nikbakht, G.; Thompson-Crispi, K.; Charkhkar, S.; Mallard, B. The influence of genetic background versus commercial breeding programs on chicken immunocom-petence. Poult. Sci. 2013, 93, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Zhang, Y.; Chen, J.; Hua, G.; Li, J.; Deng, X.; Deng, X. Transcriptome analyses of differential gene expression in the bursa of Fabricius between Silky Fowl and White Leghorn. Sci. Rep. 2017, 7, srep45959. [Google Scholar] [CrossRef]
- Takeda, K.; Tanaka, T.; Shi, W.; Matsumoto, M.; Minami, M.; Kashiwamura, S.-I.; Nakanishi, K.; Yoshida, N.; Kishimoto, T.; Akira, S. Essential role of Stat6 in IL-4 signalling. Nature 1996, 380, 627–630. [Google Scholar] [CrossRef]
- Liu, Y.; Cheng, Y.; Shan, W.; Ma, J.; Wang, H.; Sun, J.; Yan, Y. Chicken interferon regulatory factor 1 (IRF1) involved in antiviral innate immunity via regulating IFN-β production. Dev. Comp. Immunol. 2018, 88, 77–82. [Google Scholar] [CrossRef]
- Wang, P.; Hou, J.; Lin, L.; Wang, C.; Liu, X.; Li, D.; Ma, F.; Wang, Z.; Cao, X. Inducible microRNA-155 Feedback Promotes Type I IFN Signaling in Antiviral Innate Immunity by Targeting Suppressor of Cytokine Signaling 1. J. Immunol. 2010, 185, 6226–6233. [Google Scholar] [CrossRef] [PubMed]
- Mantuano, E.; Brifault, C.; Lam, M.S.; Azmoon, P.; Gilder, A.S.; Gonias, S.L. LDL receptor-related protein-1 regulates NFκB and microRNA-155 in macrophages to control the inflammatory response. Proc. Natl. Acad. Sci. USA 2016, 113, 1369–1374. [Google Scholar] [CrossRef]
- Berghof, T.; Parmentier, H.; Lammers, A. Transgenerational epigenetic effects on innate immunity in broilers: An underestimated field to be explored? Poult. Sci. 2013, 92, 2904–2913. [Google Scholar] [CrossRef]
- Swaggerty, C.L.; Callaway, T.R.; Kogut, M.H.; Piva, A.; Grilli, E. Modulation of the Immune Response to Improve Health and Reduce Foodborne Pathogens in Poultry. Microorganisms 2019, 7, 65. [Google Scholar] [CrossRef]
- Van Der Most, P.J.; De Jong, B.; Parmentier, H.K.; Verhulst, S. Trade-off between growth and immune function: A meta-analysis of selection experiments. Funct. Ecol. 2011, 25, 74–80. [Google Scholar] [CrossRef]
- Beere, H.M. ‘The stress of dying’: The role of heat shock proteins in the regulation of apoptosis. J. Cell Sci. 2004, 117, 2641–2651. [Google Scholar] [CrossRef]
- Wang, L.; Yang, S.; Zhao, K.; Han, L. Expression Profiles of the Heat Shock Protein 70 Gene in Response to Heat Stress in Agrotis c-nigrum (Lepidoptera: Noctuidae). J. Insect Sci. 2015, 15, 9. [Google Scholar] [CrossRef]
- Carrettiero, D.C.; Almeida, M.C.; Longhini, A.P.; Rauch, J.N.; Han, D.; Zhang, X.; Najafi, S.; Gestwicki, J.E.; Kosik, K.S. Stress routes clients to the proteasome via a BAG2 ubiquitin-independent degradation condensate. Nat. Commun. 2022, 13, 3074. [Google Scholar] [CrossRef]
- Xue, B.; Song, J.; Liu, L.; Luo, J.; Tian, G.; Yang, Y. Effect of epigallocatechin gallate on growth performance and antioxidant capacity in heat-stressed broilers. Arch. Anim. Nutr. 2017, 71, 362–372. [Google Scholar] [CrossRef] [PubMed]
- Nawab, A.; An, L.; Wu, J.; Li, G.; Liu, W.; Zhao, Y.; Wu, Q.; Xiao, M. Chicken toll-like receptors and their significance in immune response and disease resistance. Int. Rev. Immunol. 2019, 38, 284–306. [Google Scholar] [CrossRef] [PubMed]
- Ramasamy, K.T.; Reddy, M.R.; Raveendranathan, D.N.; Murugesan, S.; Chatterjee, R.N.; Ullengala, R.; Haunshi, S. Differential expression of Toll-like receptor mRNA in White Leghorn and indigenous chicken of India. Veter. Res. Commun. 2010, 34, 633–639. [Google Scholar] [CrossRef]
- Oppenheim, J.J. Cytokines: Past, present, and future. Int. J. Hematol. 2001, 74, 3–8. [Google Scholar] [CrossRef]
- Rogers, S.L.; Viertlboeck, B.C.; Göbel, T.W.; Kaufman, J. Avian NK activities, cells and receptors. Semin. Immunol. 2008, 20, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Vainchenker, W.; Constantinescu, S.N. JAK/STAT signaling in hematological malignancies. Oncogene 2012, 32, 2601–2613. [Google Scholar] [CrossRef]
- Hesani, Z.; Nasiry, M.R.; Bakhtiarizadeh, M.R.; Tahmoorespur, M.; Javadmanesh, A. Gene Expression Analysis on Apoptosis network and design it in Esfahani and Ross Breeds. Iran. J. Anim. Sci. Res. 2018, 10, 117–130. [Google Scholar]
- Tonks, N.K. Protein tyrosine phosphatases: From genes, to function, to disease. Nat. Rev. Mol. Cell Biol. 2006, 7, 833–846. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Fu, Y. FYN: Emerging biological roles and potential therapeutic targets in cancer. J. Transl. Med. 2023, 21, 84. [Google Scholar] [CrossRef] [PubMed]
- Peacock, A.J.; Pickett, C.; Morris, K.; Reeves, J.T. The Relationship between Rapid Growth and Pulmonary Hemodynamics in the Fast-growing Broiler Chicken. Am. Rev. Respir. Dis. 1989, 139, 1524–1530. [Google Scholar] [CrossRef] [PubMed]
- Xing, T.; Pan, X.; Zhang, L.; Gao, F. Hepatic Oxidative Stress, Apoptosis, and Inflammation in Broiler Chickens with Wooden Breast Myopathy. Front. Physiol. 2021, 12, 659777. [Google Scholar] [CrossRef]
- Psifidi, A.; Banos, G.; Matika, O.; Desta, T.T.; Bettridge, J.; Hume, D.A.; Dessie, T.; Christley, R.; Wigley, P.; Hanotte, O.; et al. Genome-wide association studies of immune, disease and production traits in indigenous chicken ecotypes. Genet. Sel. Evol. 2016, 48, 74. [Google Scholar] [CrossRef]

| Gene Name | Primer Sequence | Product Size |
|---|---|---|
| STAT4 |
F-ttggcaaacactacagctgtc R-atggagaatgtgggtctgtag | 132 |
| TLR7 |
F-gaatgggtgatgacagaattgg R-gctgaatgctctgggaaagg | 130 |
| IL2RG |
F-caaccccagcaagaacttcg R-tggcagatgctttcactgtag | 123 |
| AKT1 |
F-gtacctccatttaagccacaag R-acaatccatgctgtcatcttgg | 117 |
| ITGB2 |
F-acaacagctcagtcatctgc R-gttgtcacagtcgcagaagg | 116 |
| HSPD1 |
F-aagttggctatgatgcgatgc R-ttcagtcactactgcttctgc | 146 |
| JMJD6 |
F-ttccagctcctcgagttcc R-tatcaccgttacccatcatgc | 125 |
| β–actin |
F-gaactccctgatggtcagg R-catggataccacaggactcc | 106 |
| Sample Name | Raw Fragments | Trimmed Fragments | Overall Alignment Rate (%) |
|---|---|---|---|
| Indigenous Breed 1 | 18768307 | 18509420 | 86.68 |
| Indigenous Breed 2 | 17995632 | 17650558 | 84.89 |
| Commercial Breed 1 | 15923838 | 15677357 | 84.36 |
| Commercial Breed 2 | 15621164 | 15313649 | 84.17 |
| Upregulated DEG | Downregulated DEG | ||||
|---|---|---|---|---|---|
| Gene Name | Fold Change | FDR | Gene Name | Fold Change | FDR |
| SPARC | 4.88976 | 0.0039 | PAPPA | −4.31828 | 0.03474 |
| ATP6V0D2 | 4.09016 | 0.0034 | DUSP1 | −3.44519 | 0.001678 |
| IL4I1 | 3.8447 | 5.00 × 10−5 | PSMD12 | −2.91163 | 0.00565 |
| SMPDL3A | 3.75038 | 0.00125 | LHX8 | −2.88599 | 0.001678 |
| ADAM7 | 3.67512 | 5.00 × 10−5 | IL8 | −2.70154 | 0.007142 |
| TMCC3 | 3.54063 | 0.0023 | TRPM2 | −2.64277 | 0.001678 |
| ULK2 | 3.43567 | 5.00 × 10−5 | GDAP1L1 | −2.61655 | 0.012906 |
| MYO6 | 3.25879 | 0.00035 | FAM161A | −2.60569 | 0.001678 |
| THG1L | 3.06427 | 0.0014 | ABCC2 | −2.47906 | 0.020111 |
| IRG1 | 3.02656 | 5.00 × 10−5 | ASAH2 | −2.41558 | 0.006498 |
| Pathway | Pathway Definition | Count | p-Value | List of Genes |
|---|---|---|---|---|
| gga04650 | Natural killer cell mediated cytotoxicity | 16 | 2.73 × 10−4 | PIK3CG, PTPN6, VAV3, PIK3CB, PIK3CD, ITGB2, PRKCB, RAC2, FYN, PLCG2, PPP3CA, NFATC2, SH2D1B, SHC2, PIK3R1, SYK |
| gga04060 | Cytokine–cytokine receptor interaction | 23 | 0.00287 | TNFRSF21, IL1R1, IL2RA, TGFBR2, IL7R, TNFSF8, CCR7, TNFRSF1B, TNFRSF11A, TNFSF11, CCR5, CD40LG, IL20RA, CXCR4, CCR2, IL1RAP, CSF3R, CSF2RB, IL2RG, IL5RA, MPL, IL13RA1, CSF1R |
| gga04620 | Toll-like receptor signaling pathway | 15 | 0.00404 | PIK3CG, PIK3CB, LY96, PIK3CD, MAPK11, TLR4, TLR7, TLR2-1, AKT1, IKBKE, STAT4, MYD88, MAPK12, PIK3R1, TRAF3 |
| gga04630 | Jak-STAT signaling pathway | 17 | 0.026406 | PIK3CG, PTPN6, IL2RA, PIK3CB, SOCS3, PIK3CD, IL7R, AKT1, STAT4, IL20RA, CSF3R, CSF2RB, IL2RG, IL5RA, MPL, IL13RA1, PIK3R1 |
| gga04210 | Apoptosis | 12 | 0.034505 | PIK3CG, AKT1, IL1R1, MYD88, PIK3CB, IL1RAP, PIK3CD, CSF2RB, PPP3CA, PRKACB, PIK3R1, TRADD |
| Gene | Degree | Betweenness | Closeness |
|---|---|---|---|
| TLR4 | 32 | 669.87 | 0.59 |
| PTPRC | 32 | 665.01 | 0.59 |
| TLR7 | 26 | 478.68 | 0.54 |
| RAC2 | 25 | 338.41 | 0.54 |
| CD28 | 24 | 479.34 | 0.55 |
| IL2RG | 22 | 295.60 | 0.52 |
| FYN | 22 | 612.12 | 0.52 |
| ITGB2 | 22 | 279.22 | 0.52 |
| SYK | 21 | 264.84 | 0.53 |
| SPI1 | 21 | 127.23 | 0.52 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sadr, A.S.; Nassiri, M.; Ghaderi-Zefrehei, M.; Heidari, M.; Smith, J.; Muhaghegh Dolatabady, M. RNA-Seq Profiling between Commercial and Indigenous Iranian Chickens Highlights Differences in Innate Immune Gene Expression. Genes 2023, 14, 793. https://doi.org/10.3390/genes14040793
Sadr AS, Nassiri M, Ghaderi-Zefrehei M, Heidari M, Smith J, Muhaghegh Dolatabady M. RNA-Seq Profiling between Commercial and Indigenous Iranian Chickens Highlights Differences in Innate Immune Gene Expression. Genes. 2023; 14(4):793. https://doi.org/10.3390/genes14040793
Chicago/Turabian StyleSadr, Ayeh Sadat, Mohammadreza Nassiri, Mostafa Ghaderi-Zefrehei, Maryam Heidari, Jacqueline Smith, and Mustafa Muhaghegh Dolatabady. 2023. "RNA-Seq Profiling between Commercial and Indigenous Iranian Chickens Highlights Differences in Innate Immune Gene Expression" Genes 14, no. 4: 793. https://doi.org/10.3390/genes14040793
APA StyleSadr, A. S., Nassiri, M., Ghaderi-Zefrehei, M., Heidari, M., Smith, J., & Muhaghegh Dolatabady, M. (2023). RNA-Seq Profiling between Commercial and Indigenous Iranian Chickens Highlights Differences in Innate Immune Gene Expression. Genes, 14(4), 793. https://doi.org/10.3390/genes14040793







