The CRISPR/Cas System: A Customizable Toolbox for Molecular Detection
Abstract
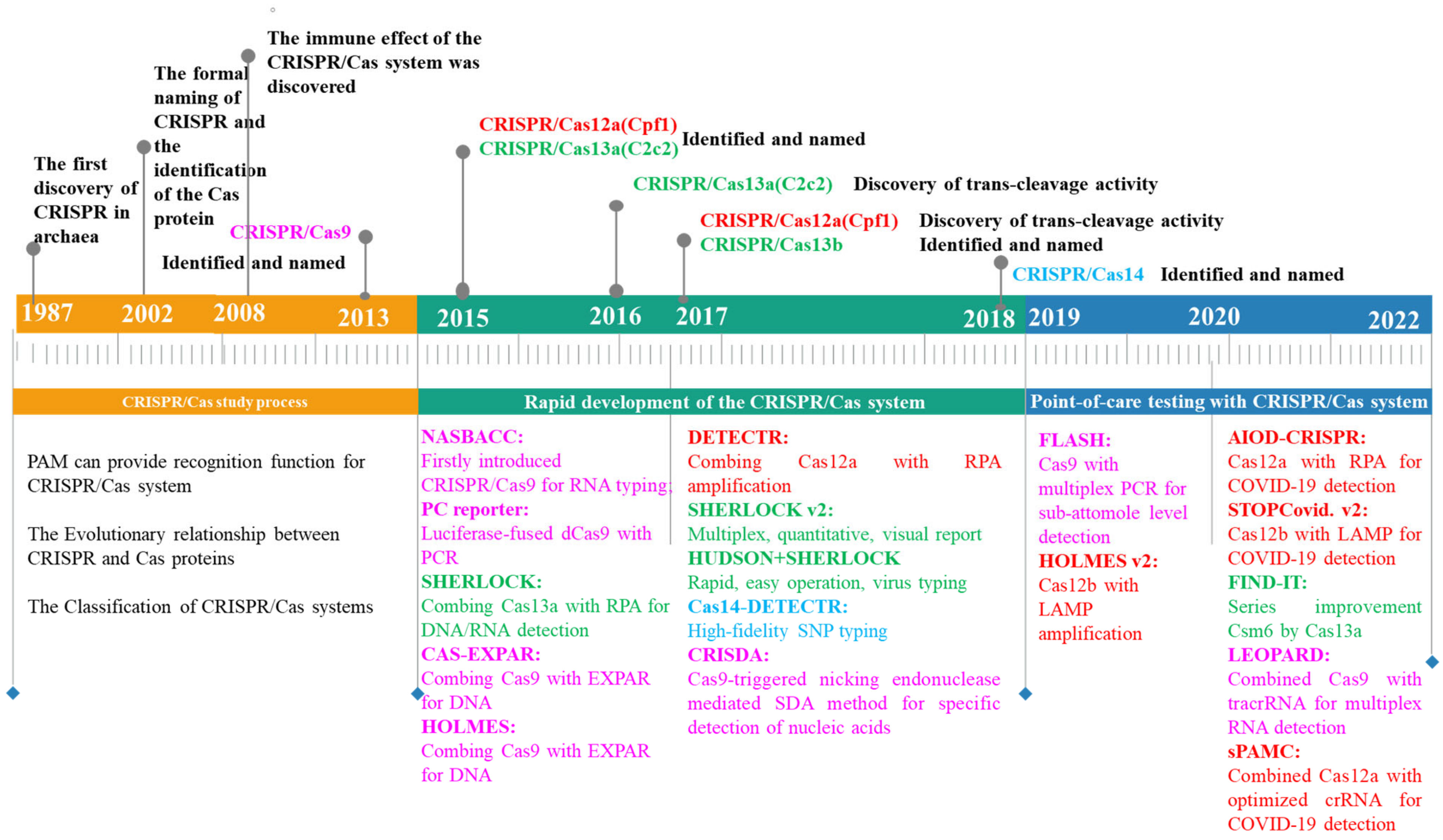
:1. Introduction
2. Classification and Characterization of CRISPR Systems
2.1. Class 1 and Its Derivatives
2.2. Class 2 and Its Derivatives
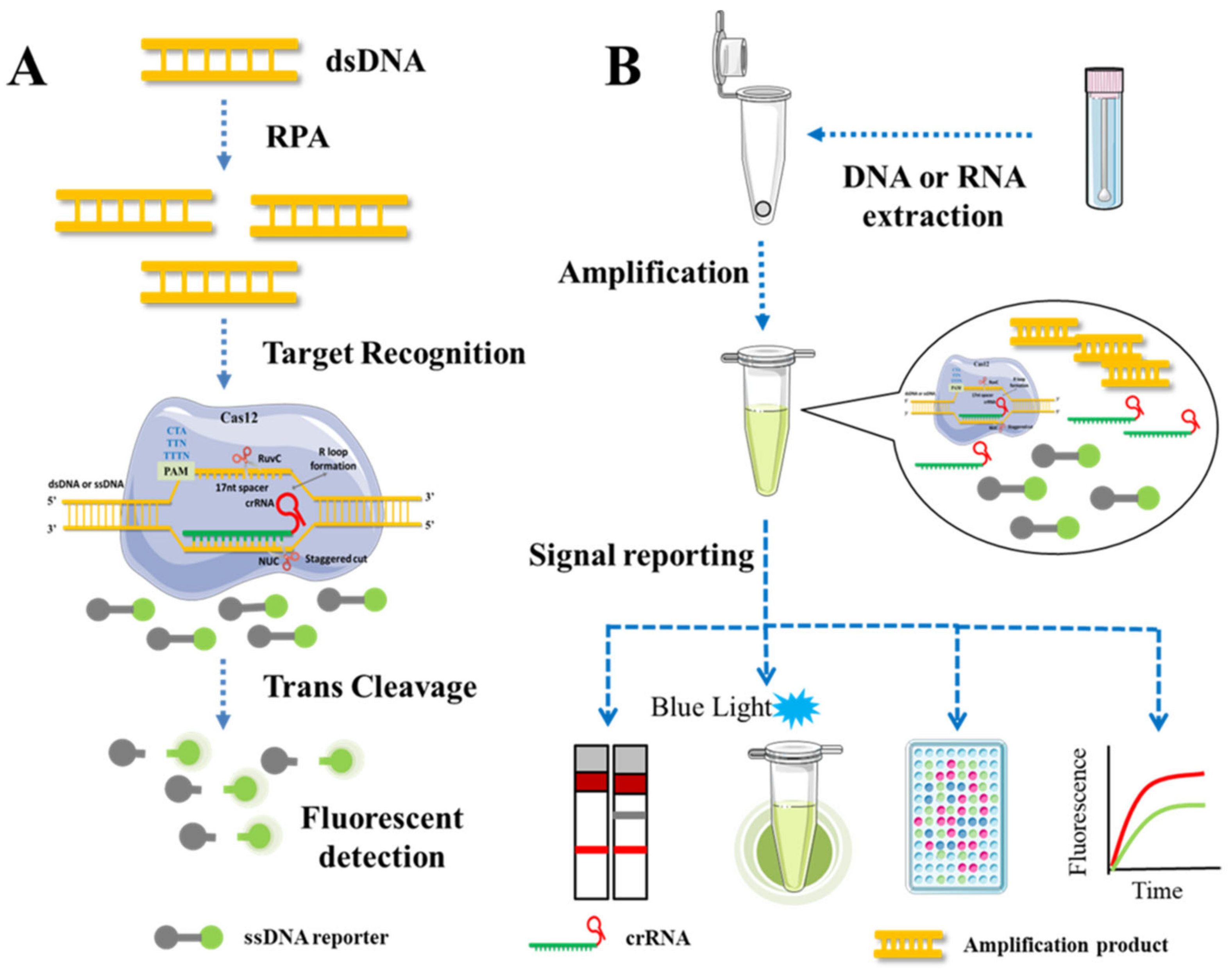
3. Mechanism of Molecular Detection Technology Based on CRISPR/Cas Systems
3.1. Guidance RNA Biogenesis
3.2. Recognition of the PAM
3.3. Target DNA Binding and Cleavage
3.4. Target RNA Binding and Cleavage
4. CRISPR/Cas Systems in Molecular Detection: Strategies and Applications beyond Genome Editing
4.1. Cas9 Protein
4.2. Cas12 Protein
4.3. Cas13 Protein
4.4. Cas14 (Cas12f) Protein
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aman, R.; Mahas, A.; Mahfouz, M. Nucleic Acid Detection Using CRISPR/Cas Biosensing Technologies. ACS Synth. Biol. 2020, 9, 1226–1233. [Google Scholar] [CrossRef]
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef] [Green Version]
- Brouns, S.J.; Jore, M.M.; Lundgren, M.; Westra, E.R.; Slijkhuis, R.J.; Snijders, A.P.; Dickman, M.J.; Makarova, K.S.; Koonin, E.V.; van der Oost, J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 2008, 321, 960–964. [Google Scholar] [CrossRef] [Green Version]
- Carter, J.; Wiedenheft, B. SnapShot: CRISPR-RNA-guided adaptive immune systems. Cell 2015, 163, 260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Haft, D.H.; Selengut, J.; Mongodin, E.F.; Nelson, K.E. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PloS Comput Biol. 2005, 1, e60. [Google Scholar] [CrossRef]
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.; Wolf, Y.I.; Yakunin, A.F.; et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011, 9, 467–477. [Google Scholar] [CrossRef] [Green Version]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Haft, D.H.; et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015, 13, 722–736. [Google Scholar] [CrossRef] [Green Version]
- Burstein, D.; Harrington, L.B.; Strutt, S.C.; Probst, A.J.; Anantharaman, K.; Thomas, B.C.; Doudna, J.A.; Banfield, J.F. New CRISPR-Cas systems from uncultivated microbes. Nature 2017, 542, 237–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, W.X.; Hunnewell, P.; Alfonse, L.E.; Carte, J.M.; Keston-Smith, E.; Sothiselvam, S.; Garrity, A.J.; Chong, S.; Makarova, K.S.; Koonin, E.V.; et al. Functionally diverse type V CRISPR-Cas systems. Science 2019, 363, 88–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.T.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef] [Green Version]
- Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.J.J.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P.; et al. Evolutionary classification of CRISPR-Cas systems: A burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020, 18, 67–83. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Makarova, K.S. Origins and evolution of CRISPR-Cas systems. Philos. T. Roy. Soc. B 2019, 374, 20180087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, D.; Li, H.; Wu, C.; Jia, T.; He, H.; Yao, S.; Yu, Y.; Chen, Q. A distinct structure of Cas1-Cas2 complex provides insights into the mechanism for the longer spacer acquisition in Pyrococcus furiosus. Int. J. Biol. Macromol. 2021, 183, 379–386. [Google Scholar] [CrossRef]
- Ma, C.-H.; Javanmardi, K.; Finkelstein, I.J.; Jayaram, M. Disintegration promotes protospacer integration by the Cas1-Cas2 complex. eLIFE 2021, 10, e65763. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Almendros, C.; Nam, K.H.; Costa, A.R.; Vink, J.N.A.; Haagsma, A.C.; Bagde, S.R.; Brouns, S.J.J.; Ke, A. Mechanism for Cas4-assisted directional spacer acquisition in CRISPR-Cas. Nature 2021, 598, 515–520. [Google Scholar] [CrossRef]
- Kieper, S.N.; Almendros, C.; Haagsma, A.C.; Barendregt, A.; Heck, A.J.; Brouns, S.J. Cas4-Cas1 Is a Protospacer Adjacent Motif-Processing Factor Mediating Half-Site Spacer Integration during CRISPR Adaptation. CRISPR J. 2021, 4, 536–548. [Google Scholar] [CrossRef]
- Nimkar, S.; Anand, B. Cas3/I-C mediated target DNA recognition and cleavage during CRISPR interference are independent of the composition and architecture of Cascade surveillance complex. Nucleic. Acids Res. 2020, 48, 2486–2501. [Google Scholar] [CrossRef]
- Sinkunas, T.; Gasiunas, G.; Fremaux, C.; Barrangou, R.; Horvath, P.; Siksnys, V. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 2011, 30, 1335–1342. [Google Scholar] [CrossRef] [Green Version]
- He, L.; Matošević, Z.J.; Mitić, D.; Markulin, D.; Killelea, T.; Matković, M.; Bertoša, B.; Ivančić-Baće, I.; Bolt, E.L. A Tryptophan ‘Gate’ in the CRISPR-Cas3 Nuclease Controls ssDNA Entry into the Nuclease Site, That When Removed Results in Nuclease Hyperactivity. Int. J. Mol. Sci. 2021, 22, 2848. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, C.; Feng, B. The rapidly advancing Class 2 CRISPR-Cas technologies: A customizable toolbox for molecular manipulations. J. Cell Mol. Med. 2020, 24, 3256–3270. [Google Scholar] [CrossRef] [Green Version]
- Bao, A.; Burritt, D.J.; Chen, H.; Zhou, X.; Cao, D.; Tran, L.-S.P. The CRISPR/Cas9 system and its applications in crop genome editing. Crit. Rev. Biotechnol. 2019, 39, 321–336. [Google Scholar] [CrossRef]
- Friedland, A.E.; Tzur, Y.B.; Esvelt, K.M.; Colaiácovo, M.P.; Church, G.M.; Calarco, J.A. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Method 2013, 10, 741–743. [Google Scholar] [CrossRef] [Green Version]
- Swarts, D.C.; Jinek, M. Cas9 versus Cas12a/Cpf1: Structure-function comparisons and implications for genome editing. Wiley Interdiscip. Rev. RNA 2018, 9, e1481. [Google Scholar] [CrossRef] [PubMed]
- Rhun, A.L.; Escalera-Maurer, A.; Bratovič, M.; Charpentier, E. CRISPR-Cas in Streptococcus pyogenes. RNA Biol. 2019, 16, 380–389. [Google Scholar] [CrossRef] [Green Version]
- Furuhata, Y.; Kato, Y. Asymmetric Roles of Two Histidine Residues in Streptococcus pyogenes Cas9 Catalytic Domains upon Chemical Rescue. Biochemistry 2021, 60, 194–200. [Google Scholar] [CrossRef]
- Cofsky, J.C.; Karandur, D.; Huang, C.J.; Witte, L.P.; Kuriyan, J.; Doudna, J.A. CRISPR-Cas12a exploits R-loop asymmetry to form double-strand breaks. eLIFE 2020, 9, e55143. [Google Scholar] [CrossRef] [PubMed]
- Strecker, J.; Jones, S.; Koopal, B.; Schmid-Burgk, J.; Zetsche, B.; Gao, L.; Makarova, K.S.; Koonin, E.V.; Zhang, F. Engineering of CRISPR-Cas12b for human genome editing. Nat. Commun. 2019, 10, 212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrington, L.B.; Burstein, D.; Chen, J.S.; Paez-Espino, D.; Ma, E.; Witte, I.P.; Cofsky, J.C.; Kyrpides, N.C.; Banfield, J.F.; Doudna, J.A. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science 2018, 362, 839–842. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.Z.; Haider, S.; Mansoor, S.; Amin, I. Targeting Plant ssDNA Virus with Engineered Miniature CRISPR-Cas14a. Trends Biotechnol. 2019, 37, 800–804. [Google Scholar] [CrossRef]
- Jiao, J.; Kong, K.; Han, J.; Song, S.; Bai, T.; Song, C.; Wang, M.; Yan, Z.; Zhang, H.; Zhang, R.; et al. Field detection of multiple RNA viruses/viroids in apple using a CRISPR/Cas12a-based visual assay. Plant Biotechnol. J. 2021, 19, 394–405. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, T.; Qian, S.; Peng, C.; Wang, X.; Wang, T.; Che, Y.; Ji, F.; Wu, J.; Xu, J. Multiple crRNAs-assisted CRISPR/Cas12a assay targeting cytochrome b gene for amplification-free detection of meat adulteration. Anal. Chim. Acta 2022, 1231, 340417. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, J.; Zeng, H.; Liu, X.; Jiang, W.; Wang, Y.; Ouyang, W.; Tang, X. RPA-Cas12a-FS: A frontline nucleic acid rapid detection system for food safety based on CRISPR-Cas12a combined with recombinase polymerase amplification. Food Chem. 2021, 334, 127608. [Google Scholar] [CrossRef]
- Mayuramart, O.; Nimsamer, P.; Rattanaburi, S.; Chantaravisoot, N.; Khongnomnan, K.; Chansaenroj, J.; Puenpa, J.; Suntronwong, N.; Vichaiwattana, P.; Poovorawan, Y.; et al. Detection of severe acute respiratory syndrome coronavirus 2 and influenza viruses based on CRISPR-Cas12a. Exp. Biol. Med. 2021, 246, 400–405. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Lin, C.; Mo, G.; Xi, B.; Li, A.A.; Huang, D.; Wan, Y.; Chen, F.; Liang, Y.; Zuo, Q.; et al. Rapid and accurate detection of SARS-CoV-2 mutations using a Cas12a-based sensing platform. Biosens. Bioelectron. 2022, 198, 113857. [Google Scholar] [CrossRef]
- Chi, Z.; Wu, Y.; Chen, L.; Yang, H.; Khan, M.R.; Busquets, R.; Huang, N.; Lin, X.; Deng, R.; Yang, W.; et al. CRISPR-Cas14a-integrated strand displacement amplification for rapid and isothermal detection of cholangiocarcinoma associated circulating microRNAs. Anal. Chim. Acta 2022, 1205, 339763. [Google Scholar] [CrossRef]
- Arizti-Sanz, J.; Freije, C.A.; Stanton, A.C.; Petros, B.A.; Boehm, C.K.; Siddiqui, S.; Shaw, B.M.; Adams, G.; Kosoko-Thoroddsen, T.-S.F.; Kemball, M.E.; et al. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat. Commun. 2020, 11, 5921. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Qiu, M.; Li, X.; Yang, J.; Li, J. CRISPR-Cas13a system: A novel tool for molecular diagnostics. Front. Microbiol. 2022, 13, 1060947. [Google Scholar] [CrossRef]
- Wang, F.; Wang, L.; Zou, X.; Duan, S.; Li, Z.; Deng, Z.; Luo, J.; Lee, S.Y.; Chen, S. Advances in CRISPR-Cas systems for RNA targeting, tracking and editing. Biotechnol. Adv. 2019, 37, 708–729. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xue, T.; Wang, M.; Ledesma-Amaro, R.; Lu, Y.; Hu, X.; Zhang, T.; Yang, M.; Li, Y.; Xiang, J.; et al. CRISPR-Cas13a cascade-based viral RNA assay for detecting SARS-CoV-2 and its mutations in clinical samples. Sens. Actuators. B Chem. 2022, 362, 131765. [Google Scholar] [CrossRef]
- Yan, W.X.; Chong, S.; Zhang, H.; Makarova, K.S.; Koonin, E.V.; Cheng, D.R.; Scott, D.A. Cas13d Is a Compact RNA-Targeting Type VI CRISPR Effector Positively Modulated by a WYL-Domain-Containing Accessory Protein. Mol. Cell 2018, 70, 327–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, W.A.; Barney, R.E.; Tsongalis, G.J. CRISPR-cas13 enzymology rapidly detects SARS-CoV-2 fragments in a clinical setting. J. Clin. Virol. 2021, 145, 105019. [Google Scholar] [CrossRef]
- Li, S.; Zhang, H.; Zhang, L.; Liu, K.; Liu, B.; Mathews, D.H.; Huang, L. LinearTurboFold: Linear-time global prediction of conserved structures for RNA homologs with applications to SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2021, 118, e2116269118. [Google Scholar] [CrossRef] [PubMed]
- Van Dongen, J.E.; Berendsen, J.T.W.; Steenbergen, R.D.M.; Wolthuis, R.M.F.; Eijkel, J.C.T.; Segerink, L.I. Point-of-care CRISPR/Cas nucleic acid detection: Recent advances, challenges and opportunities. Biosens. Bioelectron. 2020, 166, 112445. [Google Scholar] [CrossRef] [PubMed]
- Safari, F.; Zare, K.; Negahdaripour, M.; Barekati-Mowahed, M.; Ghasemi, Y. CRISPR Cpf1 proteins: Structure, function and implications for genome editing. Cell Biosci. 2019, 9, 36. [Google Scholar] [CrossRef] [Green Version]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 is a single RNA-Guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef] [Green Version]
- Hajian, R.; Balderston, S.; Tran, T.; Deboer, T.; Etienne, J.; Sandhu, M.; Wauford, N.A.; Chung, J.-Y.; Nokes, J.; Athaiya, M.; et al. Detection of unamplified target genes via CRISPR-Cas9 immobilized on a graphene field-effect transistor. Nat. Biomed. Eng. 2019, 3, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Gao, L.; Feng, W.; Guo, C.; Yang, Q.; Li, F.; Le, X.C. The CRISPR–Cas toolbox for analytical and diagnostic assay development. Chem. Soc. Rev. 2021, 20, 11844–11869. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, M.; Zheng, T.; Hou, Y.; Zhang, P.; Tang, T.; Wei, J.; Du, Q. Specificity profiling of CRISPR system reveals greatly enhanced off-target gene editing. Sci. Rep. 2020, 10, 2269. [Google Scholar] [CrossRef] [Green Version]
- Nishimasu, H.; Shi, X.; Ishiguro, S.; Gao, L.; Hirano, S.; Okazaki, S.; Noda, T.; Abudayyeh, O.O.; Gootenberg, J.S.; Mori, H.; et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 2018, 361, 1259–1262. [Google Scholar] [CrossRef]
- Hu, J.H.; Miller, S.M.; Geurts, M.H.; Tang, W.; Chen, L.; Sun, N.; Zeina, C.M.; Gao, X.; Rees, H.A.; Lin, Z.; et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 2018, 556, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Walton, R.T.; Christie, K.A.; Whittaker, M.N.; Kleinstiver, B.P. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 2020, 368, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Kleinstiver, B.P.; Sousa, A.A.; Walton, R.T.; Tak, Y.E.; Hsu, J.Y.; Clement, K.; Welch, M.M.; Horng, J.E.; Malagon-Lopez, J.; Scarfò, I.; et al. Engineered CRISPR-Cas12a Variants with Increased Activities and Improved Targeting Ranges for Gene, Epigenetic and Base Editing. Nat. Biotechnol. 2019, 37, 276–282. [Google Scholar] [CrossRef]
- Li, L.; Li, S.; Wu, N.; Wu, J.; Wang, G.; Zhao, G.; Wang, J. HOLMESv2: A CRISPR-Cas12b-Assisted Platform for Nucleic Acid Detection and DNA Methylation Quantitation. ACS Synth. Biol. 2019, 8, 2228–2237. [Google Scholar] [CrossRef]
- Li, S.-Y.; Cheng, Q.-X.; Wang, J.-M.; Li, X.-Y.; Zhang, Z.-L.; Gao, S.; Cao, R.-B.; Zhao, G.-P.; Wang, J. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 2018, 4, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hillary, V.E.; Ceasar, S.A. A Review on the Mechanism and Applications of CRISPR/Cas9/Cas12/Cas13/Cas14 Proteins Utilized for Genome Engineering. Mol. Biotechnol. 2022, 65, 311–325. [Google Scholar] [CrossRef]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef] [Green Version]
- Ma, P.; Meng, Q.; Sun, B.; Zhao, B.; Dang, L.; Zhong, M.; Liu, S.; Xu, H.; Mei, H.; Liu, J.; et al. MeCas12a, a Highly Sensitive and Specific System for COVID-19 Detection. Adv. Sci. 2020, 7, 2001300. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhao, W.; Ma, S.; Li, Z.; Yao, Y.; Fei, T. A chemical-enhanced system for CRISPR-based nucleic acid detection. Biosens. Bioelectron. 2021, 192, 113493. [Google Scholar] [CrossRef]
- Qiu, M.; Zhou, X.M.; Liu, L. Improved Strategies for CRISPR-Cas12-based Nucleic Acids Detection. J. Anal. Test. 2022, 6, 44–52. [Google Scholar] [CrossRef]
- Naqvi, M.M.; Lee, L.; Montaguth, O.E.T.; Diffin, F.M.; Szczelkun, M.D. CRISPR-Cas12a-mediated DNA clamping triggers target-strand cleavage. Nat. Chem. Biol. 2022, 18, 1014–1022. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Smith, B.M.; Jain, P.K. Enhancement of trans-cleavage activity of Cas12a with engineered crRNA enables amplified nucleic acid detection. Nat. Commun. 2020, 11, 4906. [Google Scholar] [CrossRef]
- Kim, D.; Kim, J.; Hur, J.K.; Been, K.W.; Yoon, S.-H.; Kim, J.-S. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 2016, 34, 863–874. [Google Scholar] [CrossRef] [PubMed]
- Swarts, D.C.; Jinek, M. Mechanistic Insights into the cis- and trans-Acting DNase Activities of Cas12a. Mol. Cell 2019, 73, 589–600.e4. [Google Scholar] [CrossRef] [Green Version]
- Stella, S.; Mesa, P.; Thomsen, J.; Paul, B.; Alcón, P.; Jensen, S.B.; Saligram, B.; Moses, M.E.; Hatzakis, N.S.; Montoya, G. Conformational Activation Promotes CRISPR-Cas12a Catalysis and Resetting of the Endonuclease Activity. Cell 2018, 175, 1856–1871.e21. [Google Scholar] [CrossRef] [Green Version]
- Saha, A.; Arantes, P.R.; Hsu, R.V.; Narkhede, Y.B.; Jinek, M.; Palermo, G. Molecular Dynamics Reveals a DNA-Induced Dynamic Switch Triggering Activation of CRISPR-Cas12a. J. Chem. Inf. Model. 2020, 60, 6427–6437. [Google Scholar] [CrossRef]
- Takeda, S.N.; Nakagawa, R.; Okazaki, S.; Hirano, H.; Kobayashi, K.; Kusakizako, T.; Nishizawa, T.; Yamashita, K.; Nishimasu, H.; Nureki, O. Structure of the miniature type V-F CRISPR−Cas effector enzyme. Mol. Cell 2021, 81, 558–570.e3. [Google Scholar] [CrossRef]
- Liu, J.-J.; Orlova, N.; Oakes, B.L.; Ma, E.; Spinner, H.B.; Baney, K.L.M.; Chuck, J.; Tan, D.; Knott, G.J.; Harrington, L.B.; et al. CasX enzymes comprise a distinct family of RNA-guided genome editors. Nature 2019, 566, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Azevedo, C.A.F.; Li, X.; Kamali, E.; Nielsen, O.H.; Sørensen, C.S.; Frödin, M. Multiparametric and accurate functional analysis of genetic sequence variants using CRISPR-Select. Nat. Genet. 2022, 54, 1983–1993. [Google Scholar] [CrossRef] [PubMed]
- Kick, L.M.; von Wrisberg, M.-K.; Runtsch, L.S.; Schneider, S. Structure and mechanism of the RNA dependent RNase Cas13a from Rhodobacter capsulatus. Commun. Biol. 2022, 5, 71. [Google Scholar] [CrossRef]
- Smargon, A.A.; Cox, D.B.; Pyzocha, N.K.; Zheng, K.; Slaymaker, I.M.; Gootenberg, J.; Abudayyeh, O.A.; Essletzbichler, P.; Shmakov, S.; Makarova, K.S.; et al. Cas13b Is a Type VI-B CRISPR-Associated RNA-Guided RNase Differentially Regulated by Accessory Proteins Csx27 and Csx28. Mol. Cell 2017, 65, 618–630.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makarova, K.S.; Gao, L.; Zhang, F.; Koonin, E.V. Unexpected connections between type VI-B CRISPR-Cas systems, bacterial natural competence, ubiquitin signaling network and DNA modification through a distinct family of membrane proteins. FEMS Microbiol. Lett. 2019, 366, fnz088. [Google Scholar] [CrossRef] [Green Version]
- Konermann, S.; Lotfy, P.; Brideau, N.J.; Oki, J.; Shokhirev, M.N.; Hsu, P.D. Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell 2018, 173, 665–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, R.; Ghosh, A.; Chakravarti, R.; Singh, R.; Ravichandiran, V.; Swarnakar, S.; Ghosh, D. Cas13d: A New Molecular Scissor for Transcriptome Engineering. Front. Cell Dev. Biol. 2022, 10, 866800. [Google Scholar] [CrossRef] [PubMed]
- Safari, F.; Farajnia, S.; Arya, M.; Zarredar, H.; Nasrolahi, A. CRISPR and personalized Treg therapy: New insights into the treatment of rheumatoid arthritis. Immunopharmacol. Immunotoxicol. 2018, 40, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Zhang, F. Genome engineering using CRISPR-Cas9 system. Methods Mol. Biol. 2015, 1239, 197–217. [Google Scholar]
- Yue, H.; Huang, M.; Tian, T.; Xiong, E.; Zhou, X. Advances in Clustered, Regularly Interspaced Short Palindromic Repeats (CRISPR)-Based Diagnostic Assays Assisted by Micro/Nanotechnologies. ACS Nano. 2021, 15, 7848–7859. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Crawford, E.D.; O’Donovan, B.D.; Wilson, M.R.; Chow, E.D.; Retallack, H.; DeRisi, J.L. Depletion of abundant sequences by hybridization (DASH): Using Cas9 to remove unwanted high-abundance species in sequencing libraries and molecular counting applications. Genome Biol. 2016, 17, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.H.; Yu, J.; Hwang, G.H.; Kim, S.; Kim, H.S.; Ye, S.; Kim, K.; Park, J.; Park, D.Y.; Cho, Y.K.; et al. CUT-PCR: CRISPR-mediated, ultrasensitive detection of target DNA using PCR. Oncogene 2017, 36, 6823–6829. [Google Scholar] [CrossRef] [Green Version]
- Quan, J.; Langelier, C.; Kuchta, A.; Batson, J.; Teyssier, N.; Lyden, A.; Caldera, S.; McGeever, A.; Dimitrov, B.; King, R.; et al. FLASH: A next-generation CRISPR diagnostic for multiplexed detection of antimicrobial resistance sequences. Nucleic Acids Res. 2019, 47, e83. [Google Scholar] [CrossRef] [Green Version]
- Pardee, K.; Green, A.A.; Takahashi, M.K.; Braff, D.; Lambert, G.; Lee, J.W.; Ferrante, T.; Ma, D.; Donghia, N.; Fan, M.; et al. Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell 2016, 165, 1255–1266. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Wang, Q.; Xu, X.; Xia, Q.; Long, F.; Li, W.; Shui, Y.; Xia, X.; Wang, J. Detection of target DNA with a novel Cas9/sgRNAs-associated reverse PCR (CARP) technique. Anal. Bioanal. Chem. 2018, 410, 2889–2900. [Google Scholar] [CrossRef]
- Huang, M.; Zhou, X.; Wang, H.; Xing, D. Clustered Regularly Interspaced Short Palindromic Repeats/Cas9 Triggered Isothermal Amplification for Site-Specific Nucleic Acid Detection. Anal. Chem. 2018, 90, 2193–2200. [Google Scholar] [CrossRef]
- Wang, T.; Liu, Y.; Sun, H.-H.; Yin, B.-C.; Ye, B.-C. An RNA-Guided Cas9 Nickase-Based Method for Universal Isothermal DNA Amplification. Angew. Chem. Int. Ed. Engl. 2019, 58, 5382–5386. [Google Scholar] [CrossRef]
- Moradpour, M.; Abdulah, S.N.A. CRISPR/dCas9 platforms in plants: Strategies and applications beyond genome editing. Plant Biotechnol. J. 2020, 18, 32–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Qian, L.; Wei, W.; Wang, Y.; Wang, B.; Lin, P.; Liu, W.; Yixuan, Y.; Li, X.; Liu, D.; et al. Paired Design of dCas9 as a Systematic Platform for the Detection of Featured Nucleic Acid Sequences in Pathogenic Strains. ACS Synth. Biol. 2017, 6, 211–216. [Google Scholar] [CrossRef]
- Barber, K.W.; Shrock, E.; Elledge, S.J. CRISPR-based peptide library display and programmable microarray self-assembly for rapid quantitative protein binding assays. Mol. Cell 2021, 81, 3650–3658.e5. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.-Y.; Zhu, L.-Y.; Zhu, C.-S.; Ma, J.-X.; Hou, T.; Wu, X.-M.; Xie, S.-S.; Min, L.; Tan, D.-A.; Zhang, D.-Y.; et al. Highly Effective and Low-Cost MicroRNA Detection with CRISPR-Cas9. ACS Synth. Biol. 2018, 7, 807–813. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Yang, L.; Zou, Y.; Liang, J.-Y.; Liu, P.; Gao, G.; Yang, A.; Tang, H.; Xie, X. Long non-coding RNA HUMT hypomethylation promotes lymphangiogenesis and metastasis via activating FOXK1 transcription in triple-negative breast cancer. J. Hematol. Oncol. 2020, 13, 17. [Google Scholar] [CrossRef] [Green Version]
- Luo, N.; Li, J.; Chen, Y.; Xu, Y.; Wei, Y.; Lu, J.; Dong, R. Hepatic stellate cell reprogramming via exosome-mediated CRISPR/dCas9-VP64 delivery. Drug Deliv. 2021, 28, 10–18. [Google Scholar] [CrossRef]
- Strohkendl, I.; Saifuddin, F.A.; Rybarski, J.R.; Finkelstein, I.J.; Russell, R. Kinetic Basis for DNA Target Specificity of CRISPR-Cas12a. Mol. Cell 2018, 71, 816–824. [Google Scholar] [CrossRef]
- Li, Z.; Wei, J.; Di, D.; Wang, X.; Li, C.; Li, B.; Qiu, Y.; Liu, K.; Gu, F.; Tong, M.; et al. Rapid and accurate detection of African swine fever virus by DNA endonuclease-targeted CRISPR trans reporter assay. Acta Biochim. Biophys. Sin. 2020, 52, 1413–1419. [Google Scholar] [CrossRef]
- Wang, B.; Wang, R.; Wang, D.; Wu, J.; Li, J.; Wang, J.; Liu, H.; Wang, Y. Cas12aVDet: A CRISPR/Cas12a-Based Platform for Rapid and Visual Nucleic Acid Detection. Anal. Chem. 2019, 91, 12156–12161. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Qian, C.; Pang, Y.; Li, M.; Yang, Y.; Ma, H.; Zhao, M.; Qian, F.; Yu, H.; Liu, Z.; et al. opvCRISPR: One-pot visual RT-LAMP-CRISPR platform for SARS-CoV-2 detection. Biosens. Bioelectron. 2021, 172, 112766. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Yin, K.; Li, Z.; Lalla, R.V.; Ballesteros, E.; Sfeir, M.M.; Liu, C. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 2020, 11, 4711. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Peng, H.; Xu, J.; Liu, Y.; Pabbaraju, K.; Tipples, G.; Joyce, M.A.; Saffran, H.A.; Tyrrell, D.L.; Babiuk, S.; et al. Integrating Reverse Transcription Recombinase Polymerase Amplification with CRISPR Technology for the One-Tube Assay of RNA. Anal. Chem. 2021, 93, 12808–12816. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Tong, X.; Han, Y.; Zhang, K.; Zhang, Y.; Chen, Q.; Duan, J.; Lei, X.; Huang, M.; Qiu, Y.; et al. Fast and sensitive detection of SARS-CoV-2 RNA using suboptimal protospacer adjacent motifs for Cas12a. Nat. Biomed. Eng. 2022, 6, 286–297. [Google Scholar] [CrossRef]
- Hexiang, G.; Yulin, W.; Ruijin, Z.; Yongyi, Z.; Xiaolong, L.; Dianping, T. CRISPR/Cas12a-mediated liposome-amplified strategy for the photoelectrochemical detection of nucleic acid. Chem. Commun. 2021, 57, 8977–8980. [Google Scholar]
- Kang, X.; Lei, C.; Shi, J.; Liu, X.; Ren, W.; Liu, C. A versatile CRISPR/Cas12a-based biosensing platform coupled with a target-protected transcription strategy. Biosens. Bioelectron. 2022, 219, 114801. [Google Scholar] [CrossRef]
- Liang, M.; Li, Z.; Wang, W.; Liu, J.; Liu, L.; Zhu, G.; Karthik, L.; Wang, M.; Wang, K.-F.; Wang, Z.; et al. A CRISPR-Cas12a-derived biosensing platform for the highly sensitive detection of diverse small molecules. Nat. Commun. 2019, 10, 3672. [Google Scholar] [CrossRef] [Green Version]
- Mahas, A.; Wang, Q.; Marsic, T.; Mahfouz, M.M. Development of Cas12a-Based Cell-Free Small-Molecule Biosensors via Allosteric Regulation of CRISPR Array Expression. Anal. Chem. 2022, 94, 4617–4626. [Google Scholar] [CrossRef] [PubMed]
- Qiao, B.; Xu, J.; Yin, W.; Xin, W.; Ma, L.; Qiao, J.; Liu, Y. "Aptamer-locker" DNA coupling with CRISPR/Cas12a-guided biosensing for high-efficiency melamine analysis. Biosens. Bioelectron. 2021, 183, 113233. [Google Scholar] [CrossRef]
- Kellner, M.J.; Koob, J.G.; Gootenberg, J.S.; Abudayyeh, O.O.; Zhang, F. SHERLOCK: Nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019, 14, 2986–3012. [Google Scholar] [CrossRef] [PubMed]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fozouni, P.; Son, S.; Diaz de Leon Derby, M.; Knott, G.J.; Gray, C.N.; D’Ambrosio, M.V.; Zhao, C.; Switz, N.A.; Kumar, G.R.; Stephens, S.I.; et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell 2021, 184, 323–333.e9. [Google Scholar] [CrossRef]
- Shinoda, H.; Taguchi, Y.; Nakagawa, R.; Makino, A.; Okazaki, S.; Nakano, M.; Muramoto, Y.; Takahashi, C.; Takahashi, I.; Ando, J.; et al. Amplification-free RNA detection with CRISPR-Cas13. Commun. Biol. 2021, 4, 476. [Google Scholar] [CrossRef] [PubMed]
- Aquino-Jarquin, G. CRISPR-Cas14 is now part of the artillery for gene editing and molecular diagnostic. Nanomedicine 2019, 18, 428–431. [Google Scholar] [CrossRef]
- Li, C.; Liu, C.; Liu, R.; Wang, Y.; Li, A.; Tian, S.; Cheng, W.; Ding, S.; Li, W.; Zhao, M.; et al. A novel CRISPR/Cas14a-based electrochemical biosensor for ultrasensitive detection of Burkholderia pseudomallei with PtPd@PCN-224 nanoenzymes for signal amplification. Biosens. Bioelectron. 2023, 225, 115098. [Google Scholar] [CrossRef]
- Hu, M.; Zhu, D.; Zhou, X. M-CDC: Magnetic pull-down-assisted colorimetric method based on the CRISPR/Cas12a system. Methods 2022, 203, 259–267. [Google Scholar] [CrossRef]
- Joung, J.; Ladha, A.; Saito, M.; Kim, N.-G.; Woolley, A.E.; Segel, M.; Barretto, R.P.J.; Ranu, A.; Macrae, R.K.; Faure, G.; et al. Detection of SARS-CoV-2 with SHERLOCK One-Pot Testing. N. Engl. J. Med. 2020, 383, 1492–1494. [Google Scholar] [CrossRef]
- Myhrvold, C.; Freije, C.A.; Gootenberg, J.S.; Abudayyeh, O.O.; Metsky, H.C.; Durbin, A.F.; Kellner, M.J.; Tan, A.L.; Paul, L.M.; Parham, L.A.; et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science 2018, 360, 444–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Class | Types | Subtypes | Effector Module | Spacer Integration |
|---|---|---|---|---|
| Class 1 | I | I-A,I-B,I-C,I-G,I-D,I-E,I-F1,I-F3,I-F2, | Cas3″,Cas5,Cas6,Cas7, Cas8, Cas11, Cas10 | Cas1,Cas2,Cas4 |
| III | III-A,III-D,III-E,III-F,III-C,III-B | Cas5,Cas7,Cas10,Cas11, Csx19 | Cas1,Cas2 | |
| IV | IV-A,IV-B,IV-C | Cas5,Cas7,Cas8, Cas11 | Cas1,Cas2 | |
| Class 2 | II | II-A,II-B,II-C1,II-C2 | Cas9 | Cas1,Cas2,Cas4 |
| V | V-A,V-E,V-B1,V-B2,V-I,V-H,V-C, V-D,V-F1,V-F1(V-U3),V-F2,V-U2, V-U4,V-F3,V-U1,V-G,V-K(V-U5) | Cas12 | Cas1,Cas2,Cas4 | |
| VI | VI-A,VI-D,VI-C,VI-B1,VI-B2 | Cas13 | Cas1,Cas2 |
| Categories | Efficient Characterizations | Inefficient Characterizations |
|---|---|---|
| Nucleotide content | A count | U, G count |
| A in the middle of sgRNA | GG, GGG count | |
| AG, CA, AC, UA count | UU, GC count | |
| Position-specific nucleotides | G in position 20 | C in position 20 |
| A in position 20 | U in position 17–20 | |
| Purines in position 19 | G in position 16 | |
| C in position 18 | T in PAM (TGG) | |
| C in position 16 | G in position + 1 (NGGG) | |
| C in PAM | ||
| Structural characterizations | GC in first ten bases | GC content > 80% or GC content < 35% |
| GC in position 4–8 | Stable self-folding | |
| GC in position 15–20 | Stable DNA/RNA duplex | |
| * Low Tm in the middle of sgRNA | ||
| Accessibility in positions 18–20 | ||
| GG at 5′ end of sgRNA | ||
| Extension at 5′ end of tracrRNA | ||
| Genomic context | Target near N-terminus of coding seq | Target in 5′ or 3′ UTR |
| High Cas9-sgRNA concentration | Low Cas9-sgRNA concentration | |
| Target chromatin more open | Target in introns | |
| Motifs | NGG PAM | Poly-N,TT-motif, GCC-motif |
| Experimental Method | Effector | Amplification | Sensitivity | Specificity | Quantitative Detection | Multiple Detection | Read Out | Time | Sample | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| RCA | Sp-dCas9 | RCA | fM | 1 nt | Yes | No | Colorimetric | <4 h | RNA | [88] |
| NASBACC | SpCas9 | NASBA | fM | 1 nt | No | No | Colorimetric | ≈3 h | RNA | [81] |
| PC reporter | Sp-dCas9 | PCR | 1 copy | / | No | No | Bioluminescent | 10 min after PCR | DNA | [86] |
| CAS-EXPAR | SpCas9 | EXPAR | aM | 1 nt | No | No | SYBR Green I | <1 h | DNA/RNA | [83] |
| Cas12aVDet | Cas12a | RPA | 10 aM | 1 nt | No | No | Fluorescent signal (FAM) | 30 min | DNA | [93] |
| RPA-Cas12a-FS | Cas12a | RPA | 10 copies | 1 nt | No | No | Fluorescent signal (HEX) | 45 min | DNA | [33] |
| HOLMES | LbCas12a | PCR, RT-PCR | aM | 1 nt | No | No | Fluorescent signal (HEX) | ≈1 h | DNA/RNA | [55] |
| DETECTR | LbCas12a | RPA | aM | 6 nt | No | No | Fluorescent signal (FAM) | ≈2 h | DNA | [92] |
| MeCas12a | LbCas12a | RT-RAA, qPCR | 5 copies | 1 nt | No | No | Fluorescent signal | 30 min | DNA/RNA | [58] |
| HOLMES v2 | AacCas12b | LAMP, RT-LAMP, Asymmetric PCR | aM | 1 nt | Yes | No | Fluorescent signal (HEX, FAM) | ≈1 h | DNA/RNA | [54] |
| STOPCovid.V2 | AapCas12b | LAMP | 100 copies | Fluorescent | ≈1 h | RNA | [110] | |||
| SHERLOCK | LwCas13a | RPA | aM | 1 nt | No | No | Fluorescent signal (FAM) | 2–5 h | DNA/RNA | [103] |
| SHERLOCK v2 | LwCas13a CcaCas13b PsmCas13b | RPA | zM | 1 nt | Yes | Yes | Fluorescent signal (FAM, TEX, Cy5, HEX); Colorimetric (Gold-NP, anti-FAM, Abs) | 0.5–43 h | DNA/RNA | [104] |
| HUDSON + SHERLOCK | LwCas13a | RPA | aM | 1 nt | No | No | Fluorescent signal (FAM); Colorimetric (Gold-NP, anti-FAM, Abs) | <2 h | DNA/RNA | [111] |
| Cas14SDA | Cas14a | SDA | 680 fM | 1 nt | Yes | No | Fluorescent signal (FAM) | <1 h | RNA | [36] |
| DETECTR-Cas14 | Cas14a | RPA | aM | 2 nt | No | No | Λex: 485 nm; Λex: 535 nm | ≈2 h | DNA | [29] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Y.; Yan, W.; Long, L.; Dong, L.; Ma, Y.; Li, C.; Xie, Y.; Liu, N.; Xing, Z.; Xia, W.; et al. The CRISPR/Cas System: A Customizable Toolbox for Molecular Detection. Genes 2023, 14, 850. https://doi.org/10.3390/genes14040850
He Y, Yan W, Long L, Dong L, Ma Y, Li C, Xie Y, Liu N, Xing Z, Xia W, et al. The CRISPR/Cas System: A Customizable Toolbox for Molecular Detection. Genes. 2023; 14(4):850. https://doi.org/10.3390/genes14040850
Chicago/Turabian StyleHe, Yuxuan, Wei Yan, Likun Long, Liming Dong, Yue Ma, Congcong Li, Yanbo Xie, Na Liu, Zhenjuan Xing, Wei Xia, and et al. 2023. "The CRISPR/Cas System: A Customizable Toolbox for Molecular Detection" Genes 14, no. 4: 850. https://doi.org/10.3390/genes14040850
APA StyleHe, Y., Yan, W., Long, L., Dong, L., Ma, Y., Li, C., Xie, Y., Liu, N., Xing, Z., Xia, W., & Li, F. (2023). The CRISPR/Cas System: A Customizable Toolbox for Molecular Detection. Genes, 14(4), 850. https://doi.org/10.3390/genes14040850




