High-Quality Assembly and Comparative Analysis of Actinidia latifolia and A. valvata Mitogenomes
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Sequencing
2.2. Mitogenome Assembly and Annotation
2.3. Analysis of Repeat Sequences and Chloroplast-to-Mitochondrion-DNA Transfer
2.4. Phylogenetic Analyses
2.5. Substitution Rate Calculation Analysis
3. Results
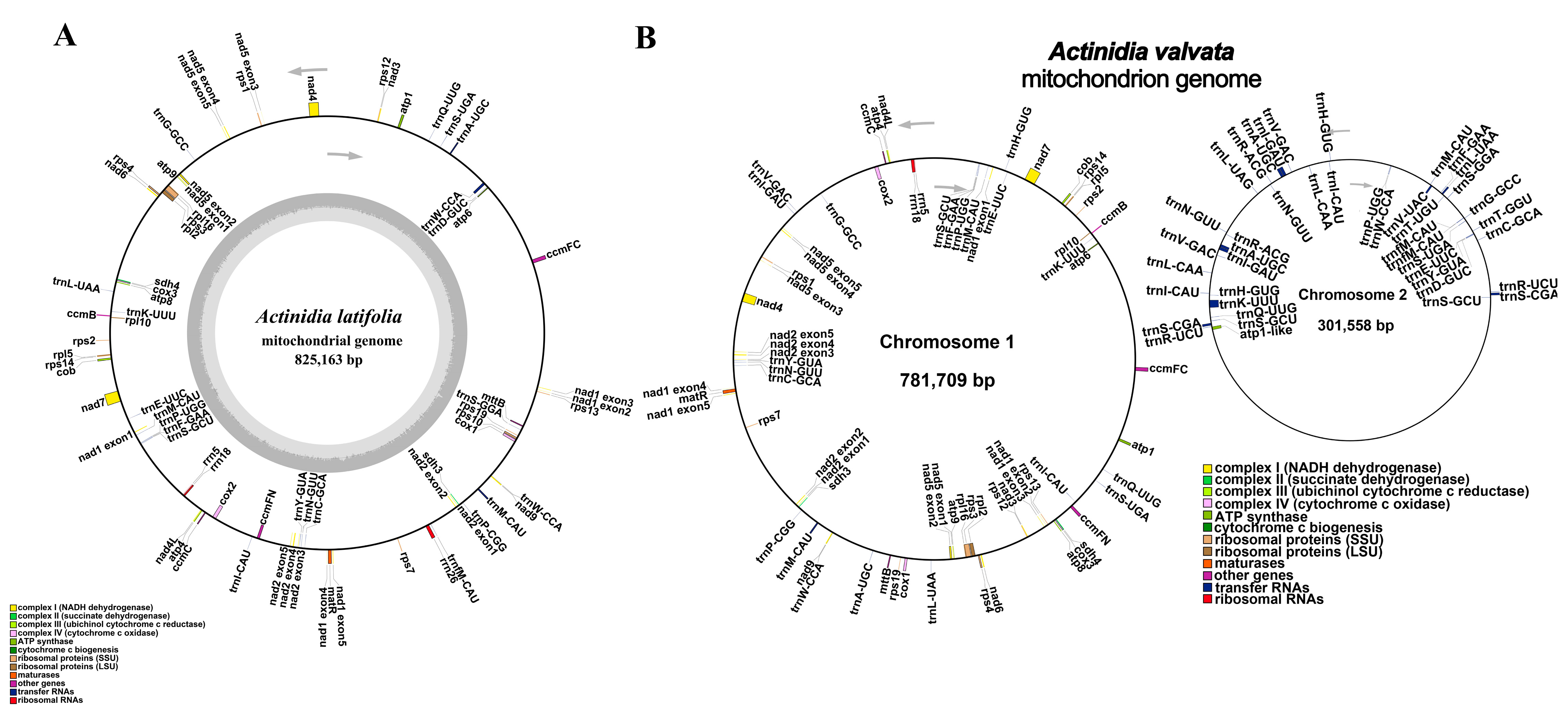
3.1. Characteristics of the Mitogenomes of Actinidia valvata and A. latifolia
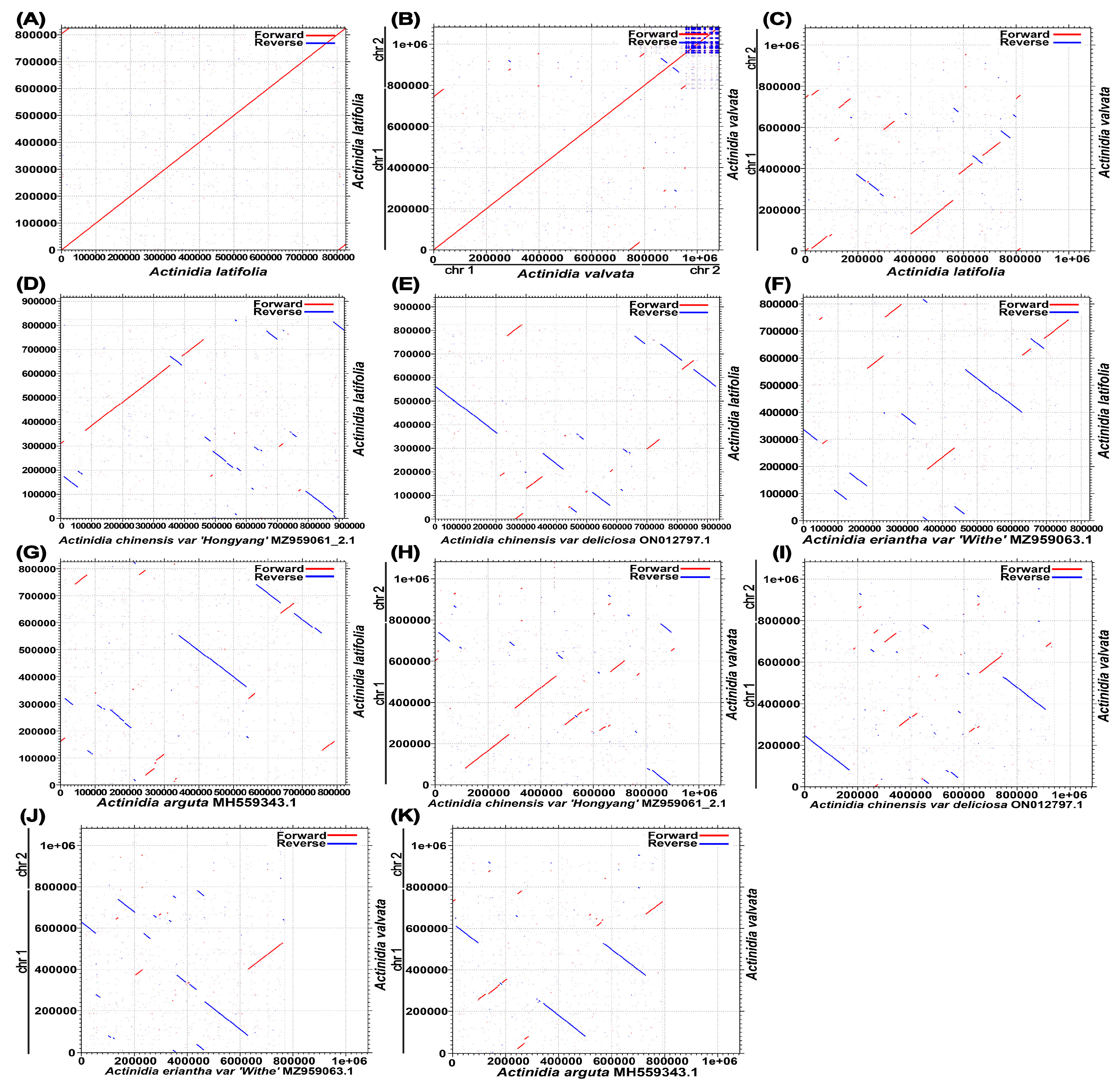
3.2. Comparative Analyses of Mitochondrial Genomes in Actinidia
3.3. Repeat Analysis
3.4. Sequence Similarity between the Mitogenome and the Cpgenome
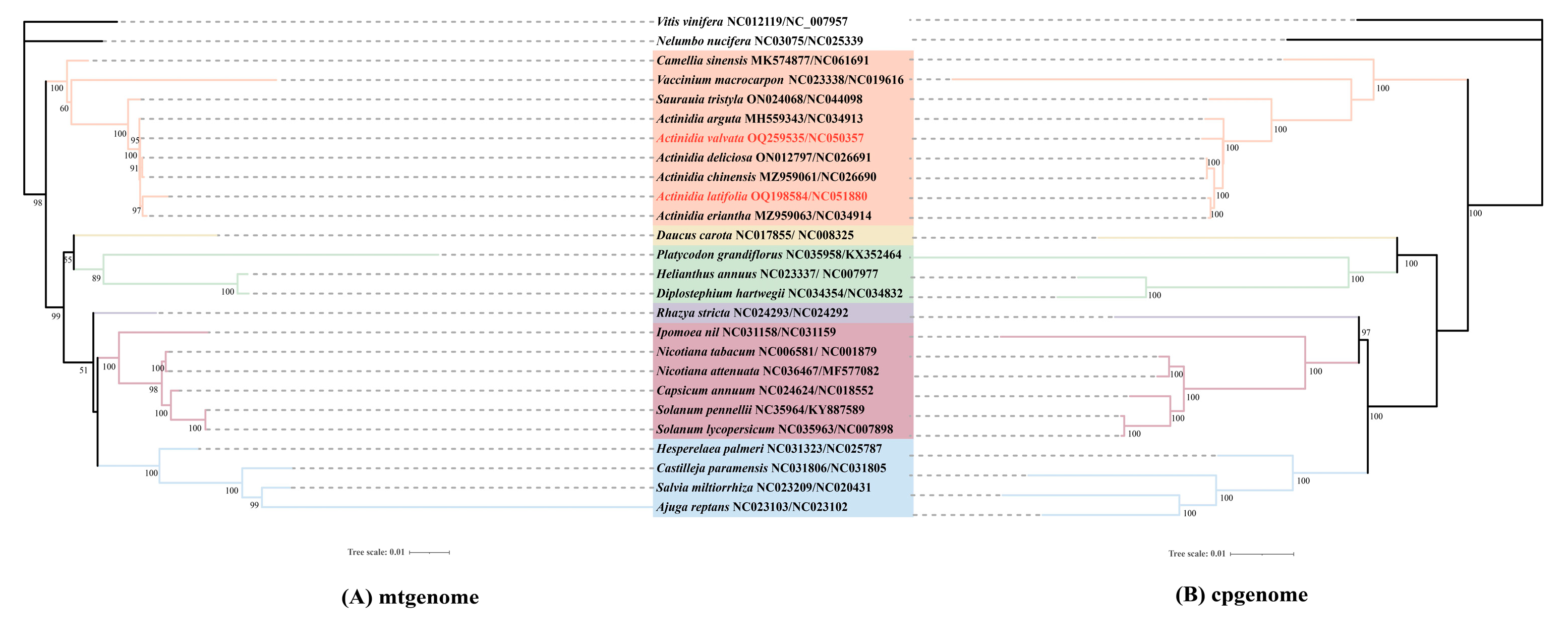
3.5. Phylogenetic Analysis
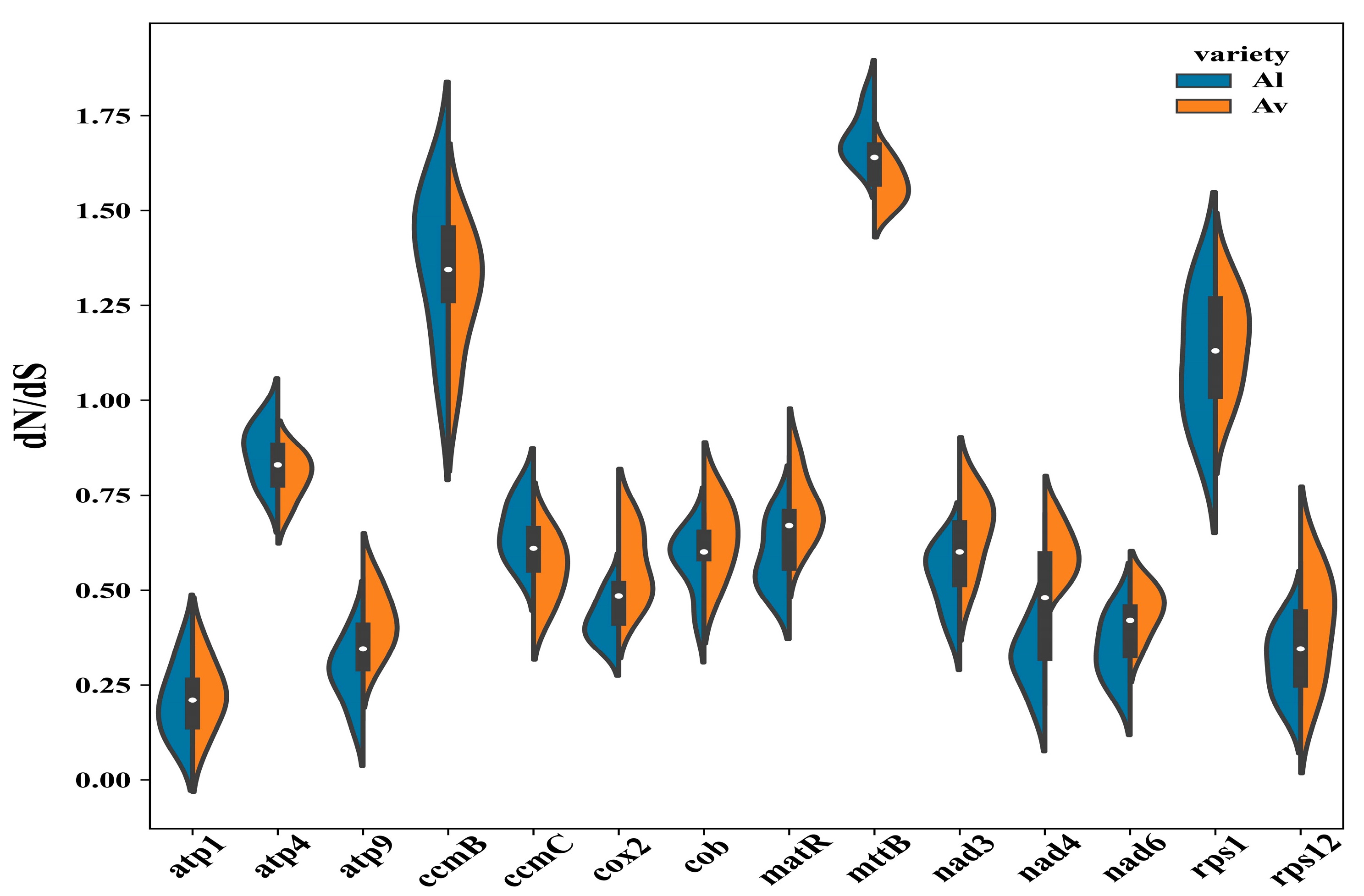
3.6. Substitution Rates of Protein-Coding Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, X.; Li, X.; Li, J.; Soejarto, D.D. Progress in the phylogeny and taxonomy of Actinidia during the past decade. Acta Hortic. 2011, 913, 71–76. [Google Scholar] [CrossRef]
- Huang, H.; Gong, J.; Wang, S.; He, Z.; Zhang, Z.; Li, J. Genetic diversity in the genus Actinidia. Biodivers. Sci. 2000, 8, 1. [Google Scholar]
- Deng, H.; Xia, H.; Guo, Y.; Liu, X.; Lin, L.; Wang, J.; Xu, K.; Lv, X.; Hu, R.; Liang, D. Dynamic Changes in Ascorbic Acid Content during Fruit Development and Ripening of Actinidia latifolia (an Ascorbate-Rich Fruit Crop) and the Associated Molecular Mechanisms. Int. J. Mol. Sci. 2022, 23, 5808. [Google Scholar] [CrossRef]
- Liu, X.; Wu, R.; Bulley, S.M.; Zhong, C.; Li, D. Kiwifruit MYBS1-like and GBF3 transcription factors influence L-ascorbic acid biosynthesis by activating transcription of GDP-L-galactose phosphorylase 3. New Phytol. 2022, 234, 1782–1800. [Google Scholar] [CrossRef]
- Qi, X.; Xie, X.; Zhong, C.; Li, D. The complete chloroplast genome of Actinidia latifolia, a species with high vitamin C content in fruit. Mitochondrial DNA Part B 2020, 5, 3425–3426. [Google Scholar] [CrossRef]
- Han, X.; Zhang, Y.; Zhang, Q.; Ma, N.; Liu, X.; Tao, W.; Lou, Z.; Zhong, C.; Deng, X.; Li, D.; et al. Two haplotype-resolved, gap-free genome assemblies of Actinidia latifolia and Actinidia chinensis shed light on regulation mechanisms of vitamin C and sucrose metabolism in kiwifruit. Mol. Plant 2022, 16, 452–470. [Google Scholar] [CrossRef]
- Zhong, Y.; Li, Z.; Bai, D.; Qi, X.; Chen, J.; Wei, C.; Lin, M.; Fang, J. In Vitro Variation of Drought Tolerance in Five Actinidia Species. J. Am. Soc. Hortic. Sci. 2018, 143, 226–234. [Google Scholar] [CrossRef]
- Abid, M.; Gu, S.; Zhang, Y.; Sun, S.; Li, Z.; Bai, D.; Sun, L.; Qi, X.; Zhong, Y.; Fang, J. Comparative transcriptome and metabolome analysis reveal key regulatory defense networks and genes involved in enhanced salt tolerance of Actinidia (kiwifruit). Hortic. Res. 2022, 9, uhac189. [Google Scholar] [CrossRef]
- Li, Z.; Zhong, Y.; Bai, D.; Lin, M.; Qi, X.; Fang, J. Comparative analysis of physiological traits of three Actinidia valvata Dunn genotypes during waterlogging and post-waterlogging recovery. Hortic. Environ. Biotechnol. 2020, 61, 825–836. [Google Scholar] [CrossRef]
- Gao, Y.; Jiang, Z.; Shi, M.; Zhou, Y.; Huo, L.; Li, X.; Xu, K. Comparative transcriptome provides insight into responding mechanism of waterlogging stress in Actinidia valvata Dunn. Gene 2022, 845, 146843. [Google Scholar] [CrossRef]
- Bai, D.; Li, Z.; Hu, C.; Zhang, Y.; Muhammad, A.; Zhong, Y.; Fang, J. Transcriptome-wide identification and expression analysis of ERF family genes in Actinidia valvata during waterlogging stress. Sci. Hortic. 2021, 281, 109994. [Google Scholar] [CrossRef]
- Gu, S.; Abid, M.; Bai, D.; Chen, C.; Sun, L.; Qi, X.; Zhong, Y.; Fang, J. Transcriptome-Wide Identification and Functional Characterization of CIPK Gene Family Members in Actinidia valvata under Salt Stress. Int. J. Mol. Sci. 2023, 24, 805. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Bai, D.; Zhong, Y.; Abid, M.; Qi, X.; Hu, C.; Fang, J. Physiological Responses of Two Contrasting Kiwifruit (Actinidia spp.) Rootstocks against Waterlogging Stress. Plants 2021, 10, 2586. [Google Scholar] [CrossRef] [PubMed]
- Bai, D.; Li, Z.; Gu, S.; Li, Q.; Sun, L.; Qi, X.; Fang, J.; Zhong, Y.; Hu, C. Effects of Kiwifruit Rootstocks with Opposite Tolerance on Physiological Responses of Grafting Combinations under Waterlogging Stress. Plants 2022, 11, 2098. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Xu, Y.; Bai, D. The complete chloroplast genome of Actinidia valvata (Actinidiaceae). Mitochondrial DNA Part B 2020, 5, 1607–1608. [Google Scholar] [CrossRef]
- Maréchal, A.; Brisson, N. Recombination and the maintenance of plant organelle genome stability. New Phytol. 2010, 186, 299–317. [Google Scholar] [CrossRef]
- Johnston, I.G. Tension and resolution: Dynamic, evolving populations of organelle genomes within plant cells. Mol. Plant 2019, 12, 764–783. [Google Scholar] [CrossRef]
- Spinelli, J.B.; Haigis, M.C. The multifaceted contributions of mitochondria to cellular metabolism. Nat. Cell Biol. 2018, 20, 745–754. [Google Scholar] [CrossRef]
- Chen, Z.; Zhao, N.; Li, S.; Grover, C.E.; Nie, H.; Wendel, J.F.; Hua, J. Plant mitochondrial genome evolution and cytoplasmic male sterility. Crit. Rev. Plant Sci. 2017, 36, 55–69. [Google Scholar] [CrossRef]
- Gualberto, J.M.; Newton, K.J. Plant mitochondrial genomes: Dynamics and mechanisms of mutation. Annu. Rev. Plant Biol. 2017, 68, 225–252. [Google Scholar] [CrossRef]
- Wolfe, K.H.; Li, W.H.; Sharp, P.M. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. USA 1987, 84, 9054–9058. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Ling, C.; Zhang, H.; Hussain, Q.; Lyu, S.; Zheng, G.; Liu, Y. A Comparative Genomics Approach for Analysis of Complete Mitogenomes of Five Actinidiaceae Plants. Genes 2022, 13, 1827. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wu, S.; Li, A.; Ruan, J. SMARTdenovo: A de novo assembler using long noisy reads. GigaByte 2021, 2021, gigabyte15. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Vaser, R.; Sović, I.; Nagarajan, N.; Šikić, M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 2017, 27, 737–746. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Ypung, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSe-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, w6–w11. [Google Scholar] [CrossRef]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, w59–w64. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ye, W.; Zhang, Y.; Xu, Y. High speed BLASTN: An accelerated MegaBLAST search tool. Nucleic Acids Res. 2015, 43, 7762–7768. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlic, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Rozewicki, J.; Li, S.; Amada, K.M.; Standley, D.M.; Katoh, K. MAFFT-DASH: Integrated protein sequence and structural alignment. Nucleic Acids Res. 2019, 47, w5–w10. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, w242–w245. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Powell, W.; Machray, G.C.; Provan, J. Polymorphism revealed by simple sequence repeats. Trends Plant Sci. 1996, 1, 215–222. [Google Scholar] [CrossRef]
- Smyth, D.R. Dispersed repeats in plant genomes. Chromosoma 1991, 100, 355–359. [Google Scholar] [CrossRef]
- Ni, Y.; Li, J.; Chen, H.; Yue, J.; Chen, P.; Liu, C. Comparative analysis of the chloroplast and mitochondrial genomes of Saposhnikovia divaricata revealed the possible transfer of plastome repeat regions into the mitogenome. BMC Genom. 2022, 23, 570. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, D.; Yao, X.; Song, Q.; Wang, Z.; Zhang, Q.; Zhong, C.; Liu, Y.; Huang, H. Evolution and Diversification of Kiwifruit Mitogenomes through Extensive Whole-Genome Rearrangement and Mosaic Loss of Intergenic Sequences in a Highly Variable Region. Genome Biol. Evol. 2019, 11, 1192–1206. [Google Scholar] [CrossRef]
- Wynn, E.L.; Christensen, A.C. Repeats of Unusual Size in Plant Mitochondrial Genomes: Identification, Incidence and Evolution. G3 2019, 9, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.R. Extending the limited transfer window hypothesis to inter-organelle DNA migration. Genome Biol. Evol. 2011, 3, 743–748. [Google Scholar] [CrossRef]
- Hazkani-Covo, E.; Zeller, R.M.; Martin, W. Molecular poltergeists: Mitochondrial DNA copies (numts) in sequenced nuclear genomes. PLoS Genet. 2010, 6, e1000834. [Google Scholar] [CrossRef]
- Niu, Y.; Gao, C.; Liu, J. Complete mitochondrial genomes of three Mangifera species, their genomic structure and gene transfer from chloroplast genomes. BMC Genom. 2022, 23, 147. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, B.; Yang, Y.; Zhuang, Q.; Chen, S.; Liu, Y.; Huang, S. The comparative studies of complete chloroplast genomes in Actinidia (Actinidiaceae): Novel insights into heterogenous variation, clpP gene annotation and phylogenetic relationships. Mol. Genet. Genom. 2022, 297, 535–551. [Google Scholar] [CrossRef]
- Agalarov, S.C.; Kalinichenko, A.A.; Kommer, A.A.; Spirin, A.S. Ribosomal protein S1 induces a conformational change of the 30S ribosomal subunit. FEBS Lett. 2006, 580, 6797–6799. [Google Scholar] [CrossRef]
- Giegé, P.; Grienenberger, J.M.; Bonnard, G. Cytochrome c biogenesis in mitochondria. Mitochondrion 2008, 8, 61–73. [Google Scholar] [CrossRef]
- Li, J.; Xu, Y.; Shan, Y.; Pei, X.; Yong, S.; Liu, C.; Yu, J. Assembly of the complete mitochondrial genome of an endemic plant, Scutellaria tsinyunensis, revealed the existence of two conformations generated by a repeat mediated recombination. Planta 2021, 254, 36. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, W.; Wang, L.; Feng, G.; Tao, C.; Liu, Y.; Yang, J. High-Quality Assembly and Comparative Analysis of Actinidia latifolia and A. valvata Mitogenomes. Genes 2023, 14, 863. https://doi.org/10.3390/genes14040863
Ren W, Wang L, Feng G, Tao C, Liu Y, Yang J. High-Quality Assembly and Comparative Analysis of Actinidia latifolia and A. valvata Mitogenomes. Genes. 2023; 14(4):863. https://doi.org/10.3390/genes14040863
Chicago/Turabian StyleRen, Wangmei, Liying Wang, Guangcheng Feng, Cheng Tao, Yongsheng Liu, and Jun Yang. 2023. "High-Quality Assembly and Comparative Analysis of Actinidia latifolia and A. valvata Mitogenomes" Genes 14, no. 4: 863. https://doi.org/10.3390/genes14040863
APA StyleRen, W., Wang, L., Feng, G., Tao, C., Liu, Y., & Yang, J. (2023). High-Quality Assembly and Comparative Analysis of Actinidia latifolia and A. valvata Mitogenomes. Genes, 14(4), 863. https://doi.org/10.3390/genes14040863





