Genome-Wide Identification and Bioinformatics Analyses of Host Defense Peptides Snakin/GASA in Mangrove Plants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Gene Identification of Snakin/GASA Family in K. obovata, A. corniculatum, and A. marina
2.2. Physicochemical Characterization, Chromosomal Location, and Sequence Alignments
2.3. Phylogenetic, Gene Structure, Motif, and Promoter Region Analyses
2.4. Subcellular Localization, Protein Structure and Gene Duplication
2.5. Sample Collection
2.6. RNA Extraction, cDNA Synthesis, and qRT-PCR Analysis
2.7. Identification of Pathogenic Microorganism
2.8. Statistical Analysis
3. Results
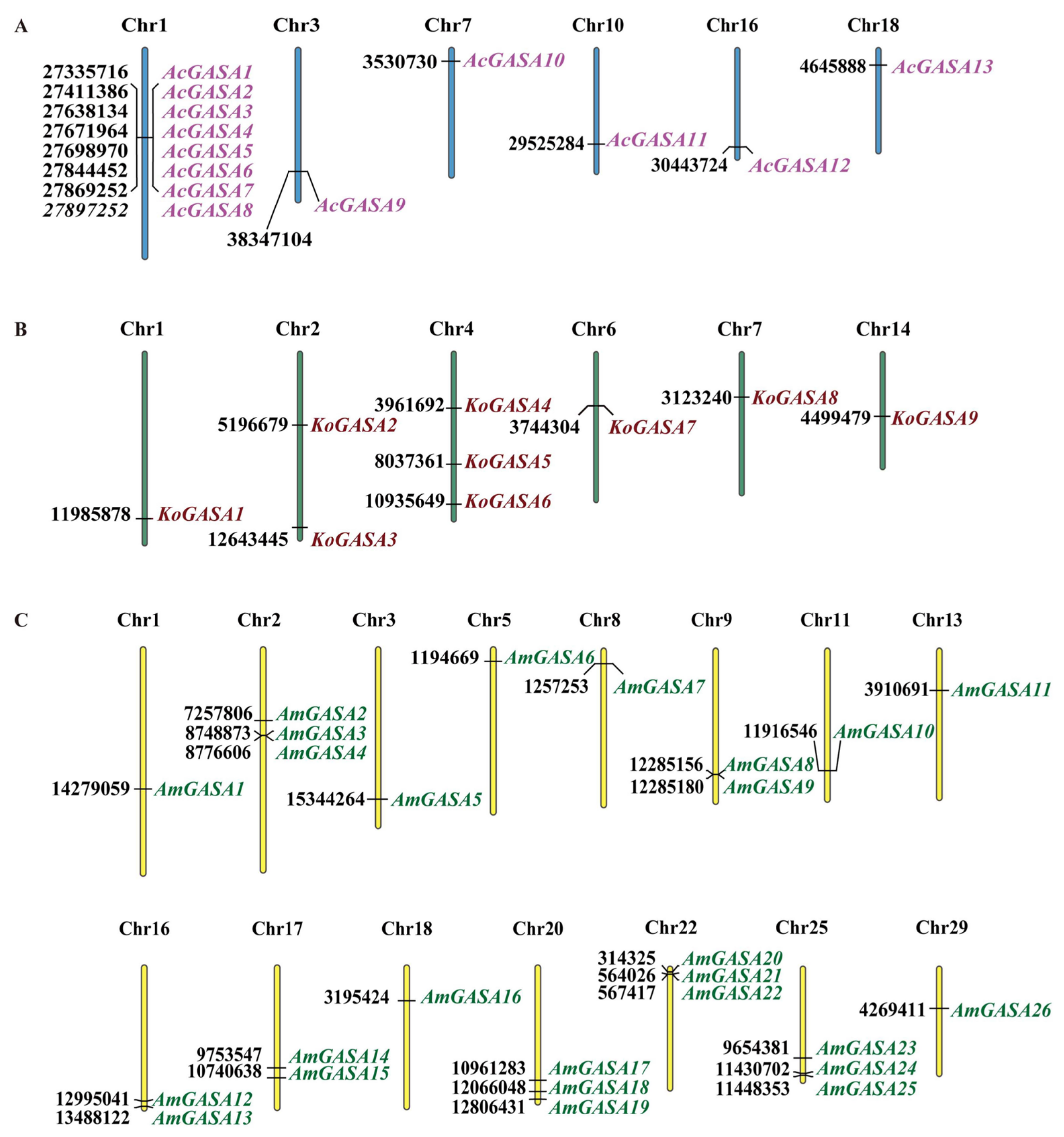
3.1. Identification of Snakin/GASA Genes in Three Mangrove Plants and Chromosomal Location
3.2. Physicochemical Properties Prediction
3.3. Subcellular Localization Prediction
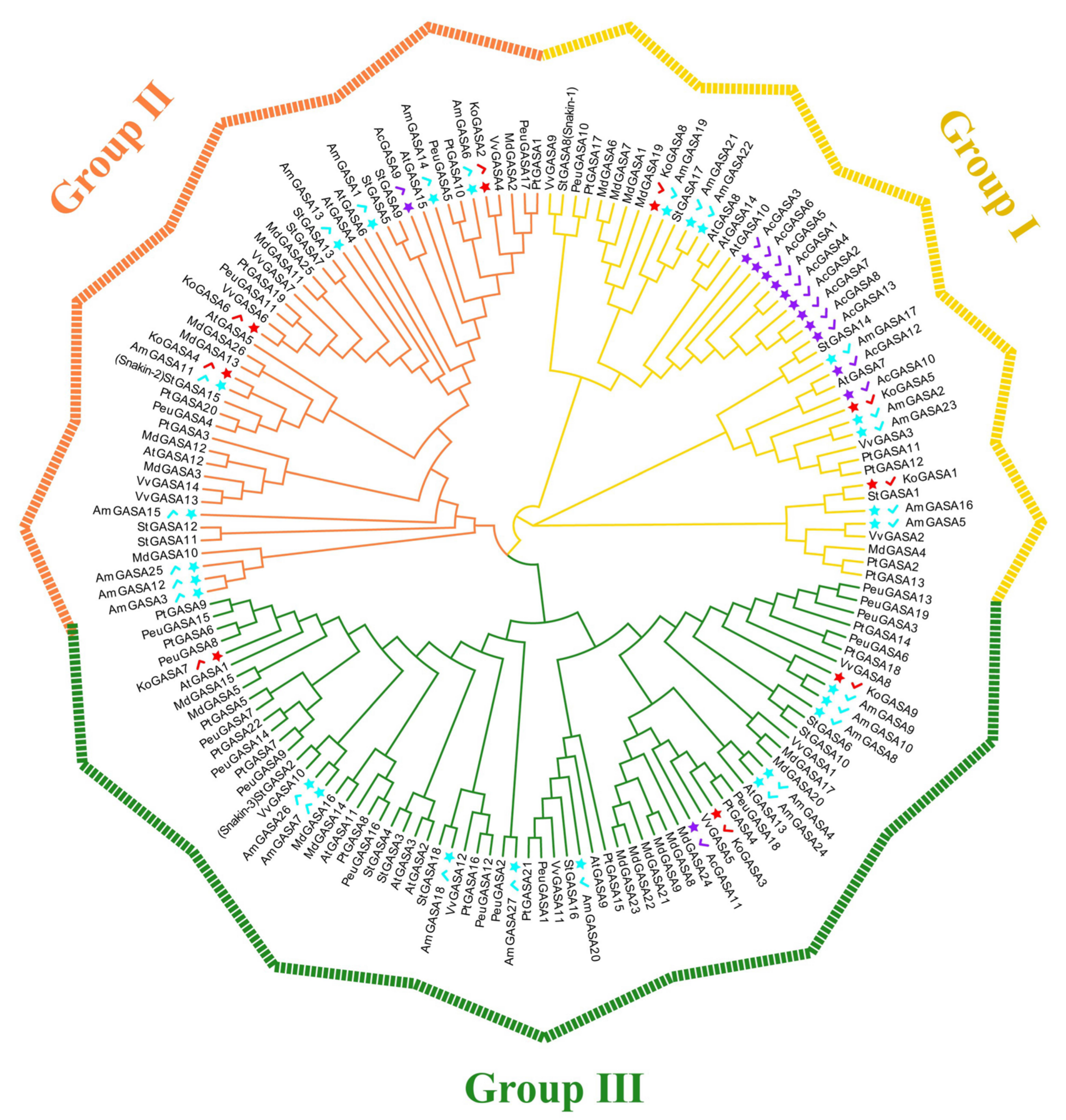
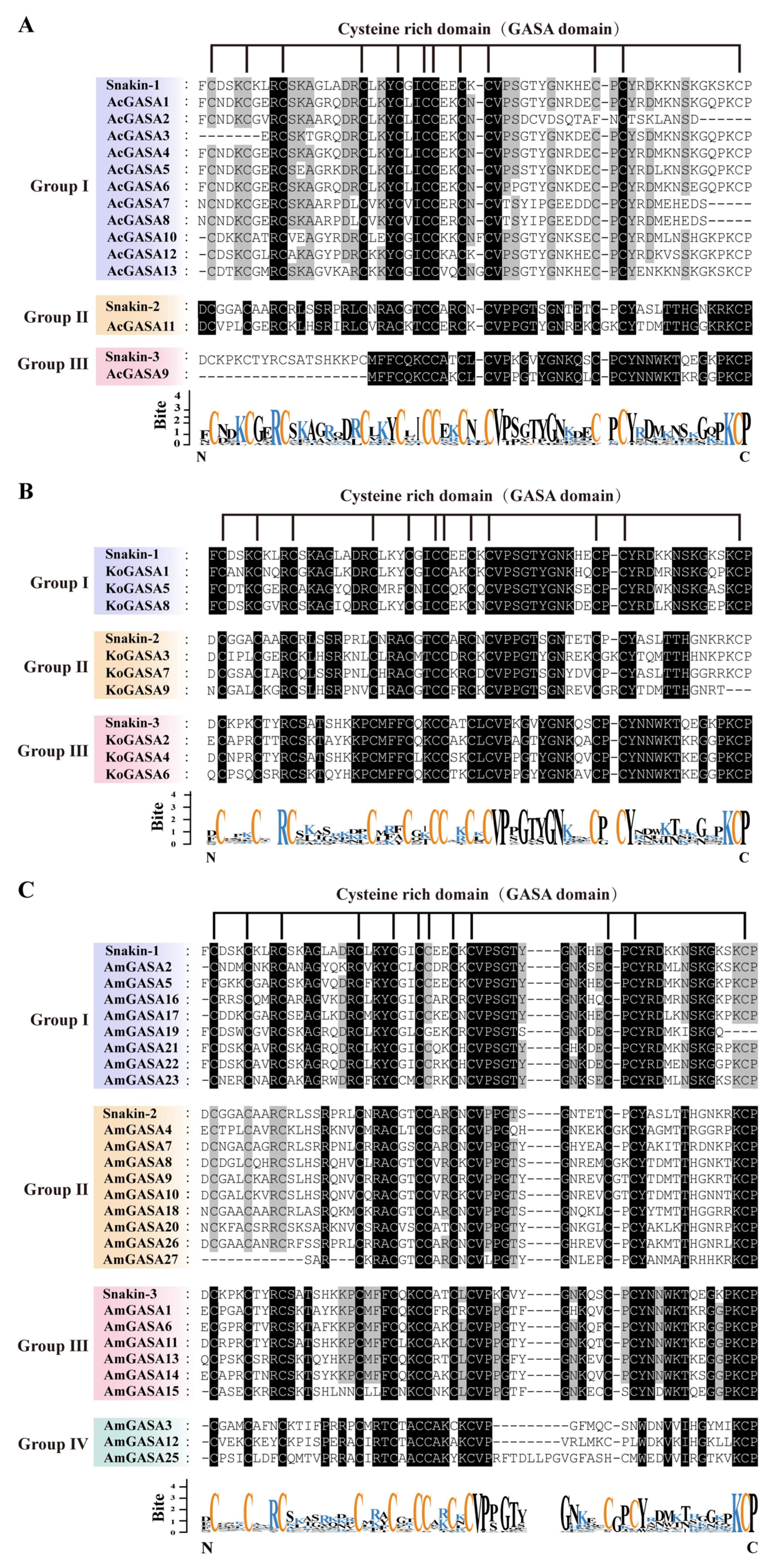
3.4. Phylogeny, and Protein Sequence Analysis of Snakin/GASA Family Members in Three Mangrove Species
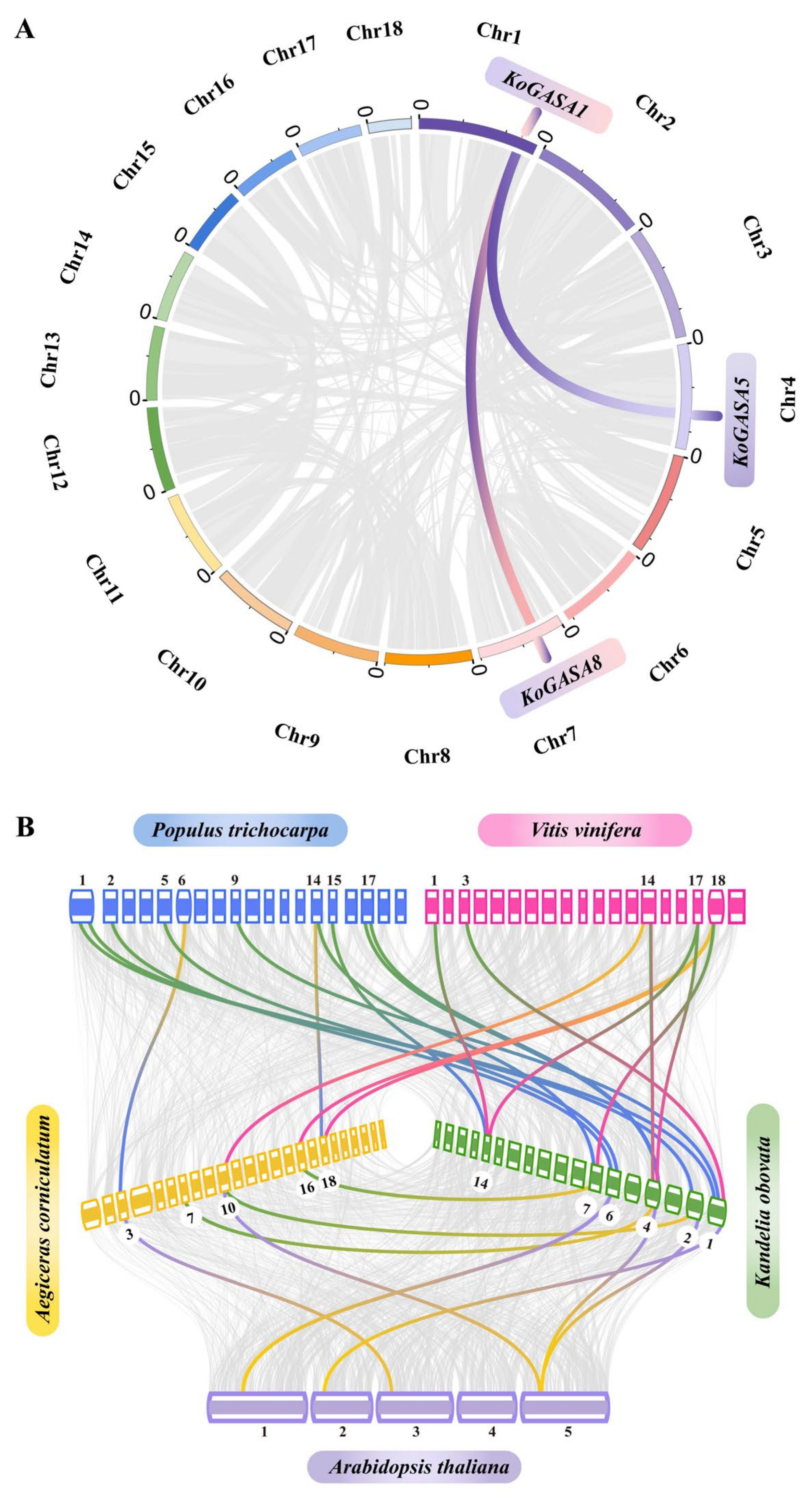
3.5. Collinearity Analysis among Snakin/GASA Family Members
3.6. Gene Structure and Motif Identification among Snakin/GASA Family Members
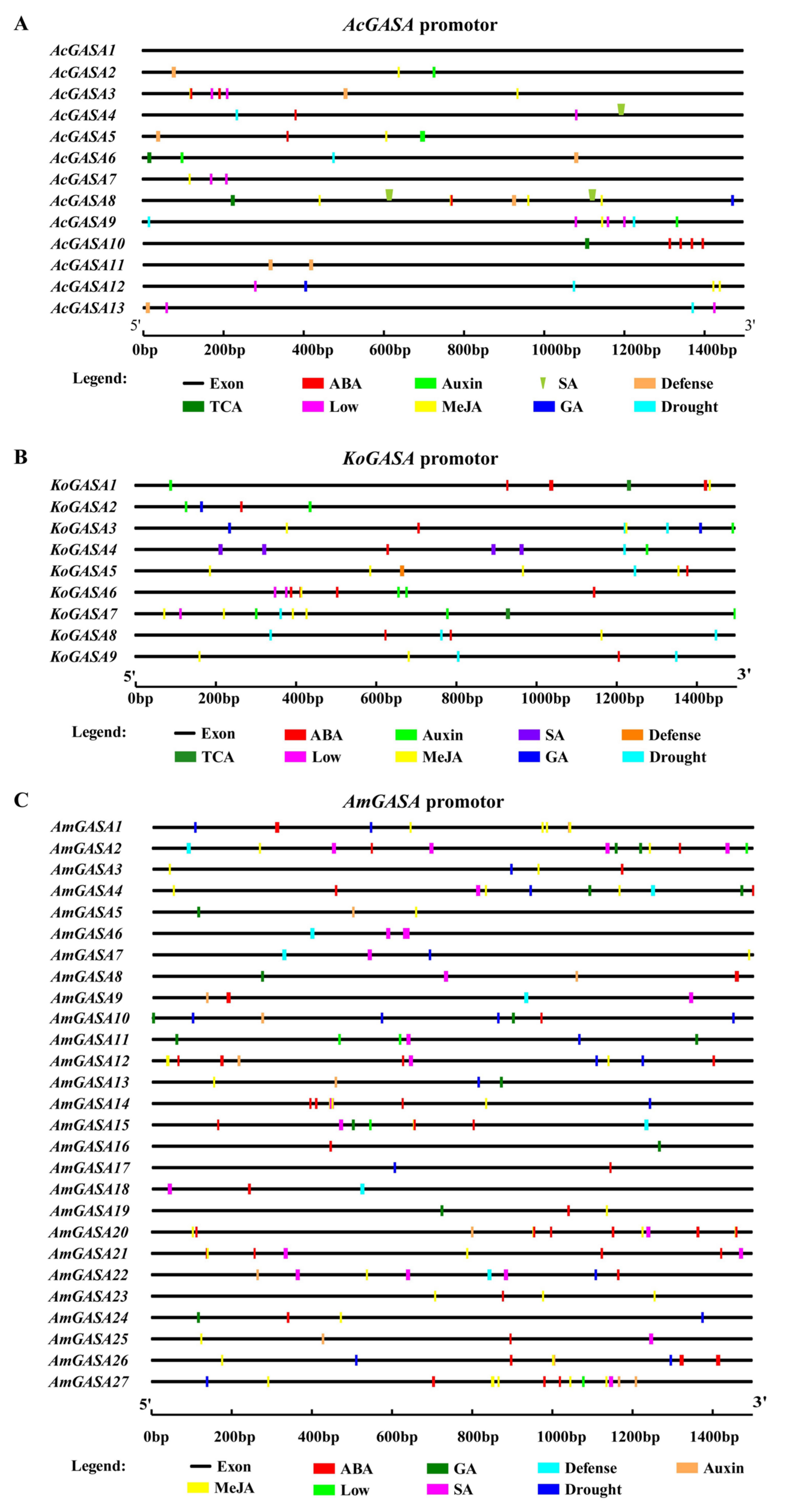
3.7. Gene upstream Element Analysis
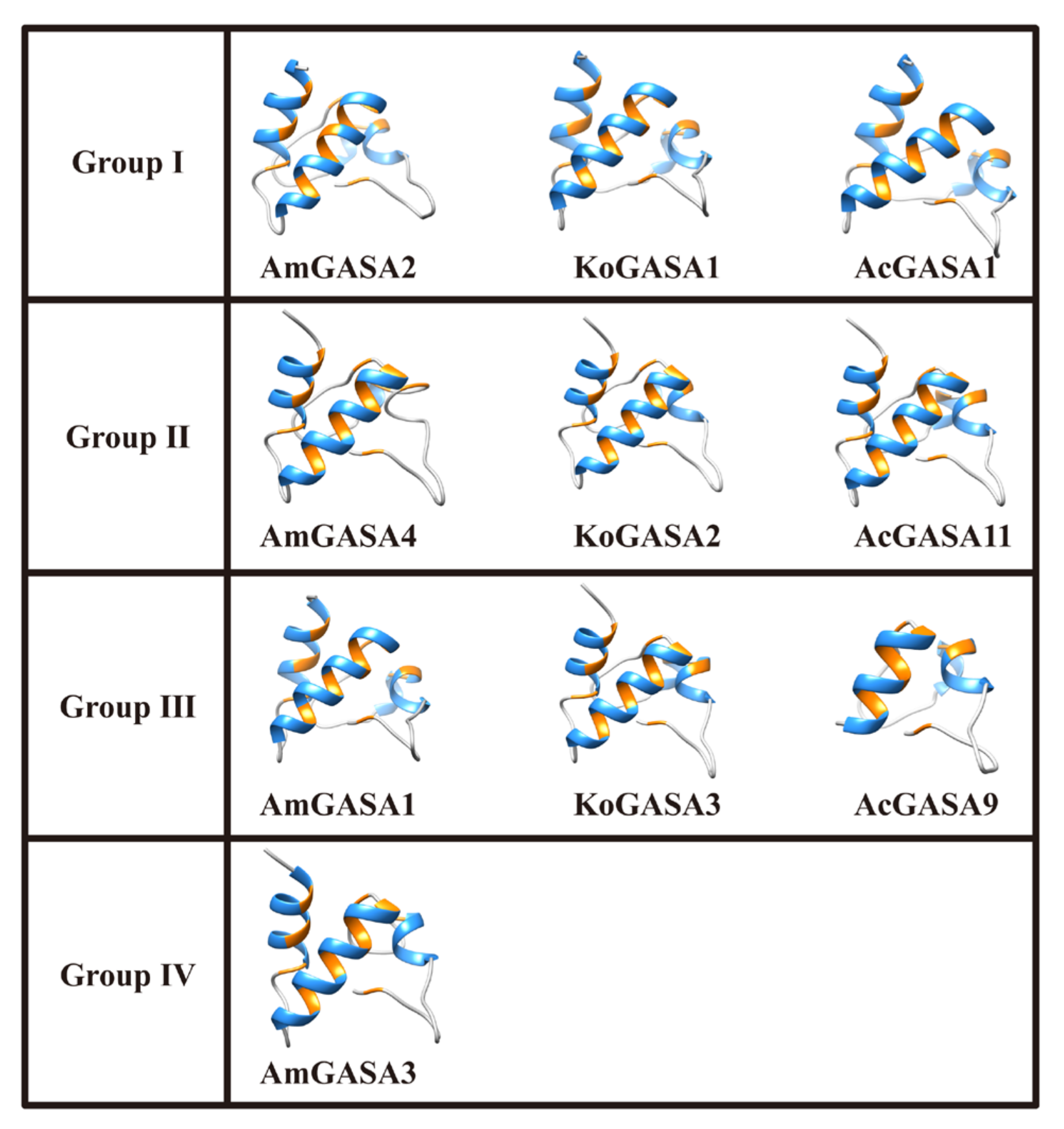
3.8. Protein Structure Prediction
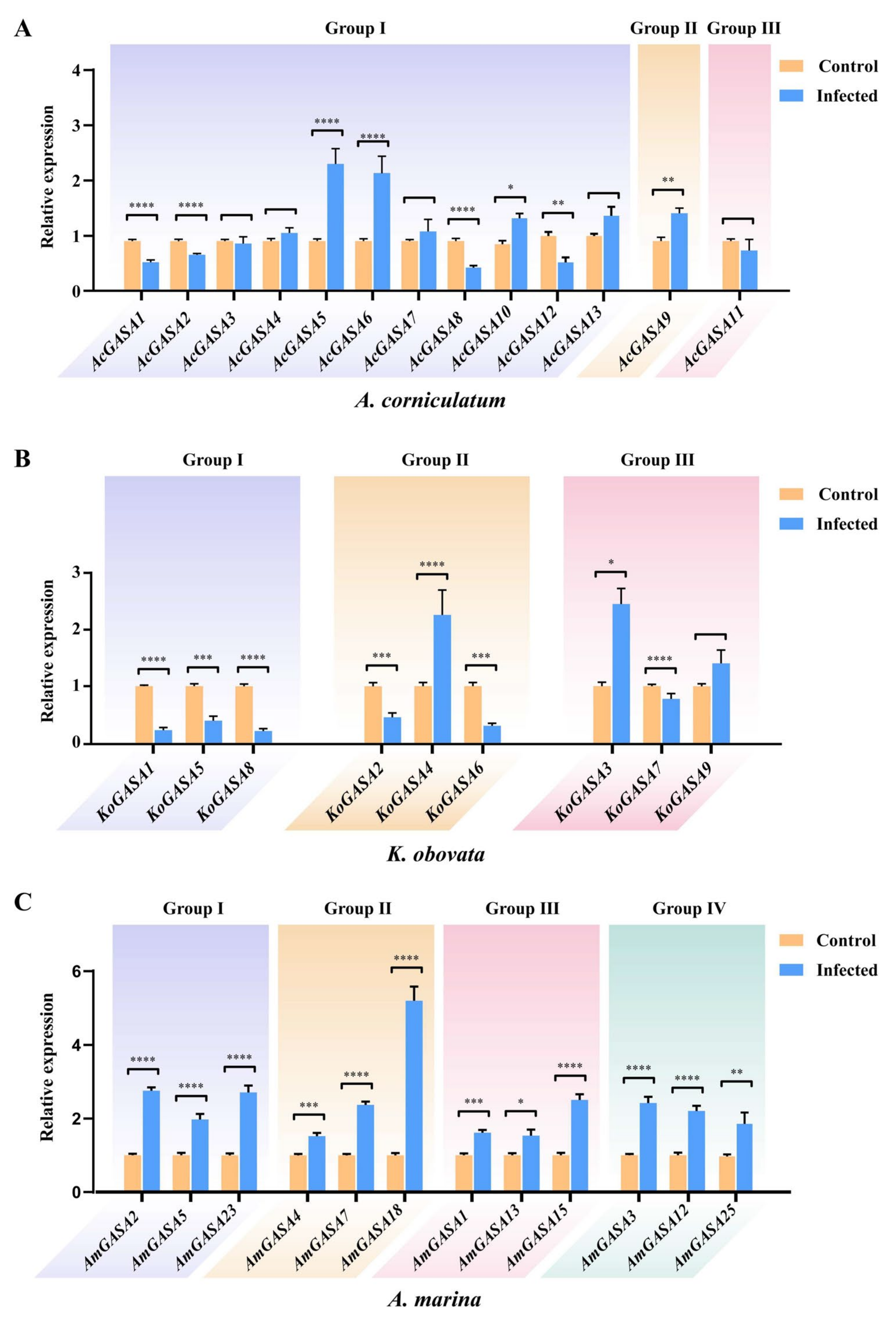
3.9. Responses to Pathogenic Microbial Threats
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, Y.; Pruitt, R.N.; Nürnberger, T.; Wang, Y. Evasion of Plant Immunity by Microbial Pathogens. Nat. Rev. Microbiol. 2022, 20, 449–464. [Google Scholar] [CrossRef] [PubMed]
- Varma, S.; Meena, A.K. Chapter-2 Integrated Defense Response of Plant against Pathogen. In Integrated Defense Response of Plant against Pathogen; Wiley: Hoboken, NJ, USA, 2020. [Google Scholar]
- Nishad, R.; Ahmed, T.; Rahman, V.J.; Kareem, A. Modulation of Plant Defense System in Response to Microbial Interactions. Front. Microbiol. 2020, 11, 1298. [Google Scholar] [CrossRef]
- Bukhat, S.; Imran, A.; Javaid, S.; Shahid, M.; Majeed, A.; Naqqash, T. Communication of Plants with Microbial World: Exploring the Regulatory Networks for PGPR Mediated Defense Signaling. Microbiol. Res. 2020, 238, 126486. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Hu, S.; Jian, W.; Xie, C.; Yang, X. Plant Antimicrobial Peptides: Structures, Functions, and Applications. Bot. Stud. 2021, 62, 1–15. [Google Scholar] [CrossRef]
- Moravej, H.; Moravej, Z.; Yazdanparast, M.; Heiat, M.; Mirhosseini, A.; Moosazadeh Moghaddam, M.; Mirnejad, R. Antimicrobial Peptides: Features, Action, and Their Resistance Mechanisms in Bacteria. Microbial. Drug Resist. 2018, 24, 747–767. [Google Scholar] [CrossRef] [PubMed]
- Mwangi, J.; Hao, X.; Lai, R.; Zhang, Z.-Y. Antimicrobial Peptides: New Hope in the War against Multidrug Resistance. Zool Res. 2019, 40, 488. [Google Scholar] [CrossRef]
- Li, W.; Tailhades, J.; O’Brien-Simpson, N.M.; Separovic, F.; Otvos, L.; Hossain, M.A.; Wade, J.D. Proline-Rich Antimicrobial Peptides: Potential Therapeutics against Antibiotic-Resistant Bacteria. Amino Acids 2014, 46, 2287–2294. [Google Scholar] [CrossRef]
- Li, W.; Separovic, F.; O’Brien-Simpson, N.M.; Wade, J.D. Chemically Modified and Conjugated Antimicrobial Peptides against Superbugs. Chem. Soc. Rev. 2021, 50, 4932–4973. [Google Scholar] [CrossRef]
- Su, T.; Han, M.; Cao, D.; Xu, M. Molecular and Biological Properties of Snakins: The Foremost Cysteine-Rich Plant Host Defense Peptides. J. Fungi 2020, 6, 220. [Google Scholar] [CrossRef]
- Koehbach, J.; Craik, D.J. The Vast Structural Diversity of Antimicrobial Peptides. Trends Pharmacol. Sci. 2019, 40, 517–528. [Google Scholar] [CrossRef]
- Campos, M.L.; de Souza, C.M.; de Oliveira, K.B.S.; Dias, S.C.; Franco, O.L. The Role of Antimicrobial Peptides in Plant Immunity. J. Exp. Bot. 2018, 69, 4997–5011. [Google Scholar] [CrossRef] [PubMed]
- Nahirñak, V.; Rivarola, M.; Gonzalez de Urreta, M.; Paniego, N.; Hopp, H.E.; Almasia, N.I.; Vazquez-Rovere, C. Genome-Wide Analysis of the Snakin/GASA Gene Family in Solanum tuberosum Cv. Kennebec. Am. J. Potato Res. 2016, 93, 172–188. [Google Scholar] [CrossRef]
- Segura, A.; Moreno, M.; Madueño, F.; Molina, A.; García-Olmedo, F. Snakin-1, a Peptide from Potato That Is Active against Plant Pathogens. Mol. Plant-Microbe Interact. 1999, 12, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Aubert, D.; Chevillard, M.; Dorne, A.-M.; Arlaud, G.; Herzog, M. Expression Patterns of GASA Genes in Arabidopsis thaliana: The GASA4 Gene is up-Regulated by Gibberellins in Meristematic Regions. Plant Mol. Biol. 1998, 36, 871–883. [Google Scholar] [CrossRef]
- Qu, J.; Kang, S.G.; Hah, C.; Jang, J.-C. Molecular and Cellular Characterization of GA-Stimulated Transcripts GASA4 and GASA6 in Arabidopsis thaliana. Plant Sci. 2016, 246, 1–10. [Google Scholar] [CrossRef]
- Qiu, B.; Zhang, Y.; Wang, Q.; Wang, Z.; Chen, H.; Cui, X.; Liu, D. Panax notoginseng Snakin Gene Increases Resistance to Fusarium solani in Transgenic Tobacco. Ind. Crops Prod. 2020, 157, 112902. [Google Scholar] [CrossRef]
- Ahmad, M.Z.; Sana, A.; Jamil, A.; Nasir, J.A.; Ahmed, S.; Hameed, M.U. A Genome-Wide Approach to the Comprehensive Analysis of GASA Gene Family in Glycine max. Plant Mol. Biol. 2019, 100, 607–620. [Google Scholar] [CrossRef]
- Ahmad, B.; Yao, J.; Zhang, S.; Li, X.; Zhang, X.; Yadav, V.; Wang, X. Genome-Wide Characterization and Expression Profiling of GASA Genes during Different Stages of Seed Development in Grapevine (Vitis vinifera L.) Predict Their Involvement in Seed Development. Int. J. Mol. Sci. 2020, 21, 1088. [Google Scholar] [CrossRef]
- Lanzotti, V.; Romano, A.; Lanzuise, S.; Bonanomi, G.; Scala, F. Antifungal Saponins from Bulbs of White Onion, Allium cepa L. Phytochemistry 2012, 74, 133–139. [Google Scholar] [CrossRef]
- Han, S.; Jiao, Z.; Niu, M.-X.; Yu, X.; Huang, M.; Liu, C.; Wang, H.-L.; Zhou, Y.; Mao, W.; Wang, X. Genome-Wide Comprehensive Analysis of the GASA Gene Family in Populus. Int. J. Mol. Sci. 2021, 22, 12336. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, X. One New Kind of Phytohormonal Signaling Integrator: Up-and-Coming GASA Family Genes. Plant Signal Behav. 2017, 12, e1226453. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Liu, W.; Li, X.; Li, H. Overexpression of GmGASA32 Promoted Soybean Height by Interacting with GmCDC25. Plant Signal Behav. 2021, 16, 1855017. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Decuadro, S.; Barraco-Vega, M.; Dans, P.D.; Pandolfi, V.; Benko-Iseppon, A.M.; Cecchetto, G. Antimicrobial and Structural Insights of a New Snakin-like Peptide Isolated from Peltophorum dubium (Fabaceae). Amino Acids 2018, 50, 1245–1259. [Google Scholar] [CrossRef]
- Hu, M.-J.; Sun, W.-H.; Tsai, W.-C.; Xiang, S.; Lai, X.-K.; Chen, D.-Q.; Liu, X.-D.; Wang, Y.-F.; Le, Y.-X.; Chen, S.-M.; et al. Chromosome-Scale Assembly of the Kandelia obovata Genome. Hortic. Res. 2020, 7, 75. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Guo, Z.; Ding, Q.; Zhao, Z.; Shen, Z.; Wei, M.; Gao, C.; Zhang, L.; Li, H.; Zhang, S.; et al. Chromosome-Level Assembly of the Mangrove Plant Aegiceras corniculatum Genome Generated through Illumina, PacBio and Hi-C Sequencing Technologies. Mol. Ecol. Resour. 2021, 21, 1593–1607. [Google Scholar] [CrossRef]
- Natarajan, P.; Murugesan, A.K.; Govindan, G.; Gopalakrishnan, A.; Kumar, R.; Duraisamy, P.; Balaji, R.; Shyamli, P.S.; Parida, A.K.; Parani, M. A Reference-Grade Genome Identifies Salt-Tolerance Genes from the Salt-Secreting Mangrove Species Avicennia marina. Commun. Biol. 2021, 4, 851. [Google Scholar] [CrossRef]
- Voorrips, R. MapChart: Software for the Graphical Presentation of Linkage Maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Nicholas, K.B. Genedoc: A Tool for Editing and Annoting Multiple Sequence Alignments. 1997. Available online: http://wwwpscedu/biomed/genedoc (accessed on 16 February 2023).
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Peng, Y.-L.; Wang, Y.-S.; Cheng, H.; Sun, C.-C.; Wu, P.; Wang, L.-Y.; Fei, J. Characterization and Expression Analysis of Three CBF/DREB1 Transcriptional Factor Genes from Mangrove Avicennia marina. Aquat. Toxicol. 2013, 140–141, 68–76. [Google Scholar] [CrossRef]
- Peng, Y.-L.; Wang, Y.-S.; Fei, J.; Sun, C.-C. Isolation and Expression Analysis of Two Novel C-Repeat Binding Factor (CBF) Genes Involved in Plant Growth and Abiotic Stress Response in Mangrove Kandelia obovata. Ecotoxicology 2020, 29, 718–725. [Google Scholar] [CrossRef]
- Peng, Y.-L.; Wang, Y.-S.; Gu, J.-D. Identification of Suitable Reference Genes in Mangrove Aegiceras corniculatum under Abiotic Stresses. Ecotoxicology 2015, 24, 1714–1721. [Google Scholar] [CrossRef]
- Rubinovich, L.; Weiss, D. The Arabidopsis Cysteine-rich Protein GASA4 Promotes GA Responses and Exhibits Redox Activity in Bacteria and in Planta. Plant J. 2010, 64, 1018–1027. [Google Scholar] [CrossRef]
- Lei, P.; Wei, X.; Gao, R.; Huo, F.; Nie, X.; Tong, W.; Song, W. Genome-Wide Identification of PYL Gene Family in Wheat: Evolution, Expression and 3D Structure Analysis. Genomics 2021, 113, 854–866. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, X.; Wang, Z. APD3: The Antimicrobial Peptide Database as a Tool for Research and Education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [PubMed]
- Seyfi, R.; Kahaki, F.A.; Ebrahimi, T.; Montazersaheb, S.; Eyvazi, S.; Babaeipour, V.; Tarhriz, V. Antimicrobial Peptides (AMPs): Roles, Functions and Mechanism of Action. Int. J. Pept. Res. Ther. 2020, 26, 1451–1463. [Google Scholar] [CrossRef]
- Fan, S.; Zhang, D.; Zhang, L.; Gao, C.; Xin, M.; Tahir, M.M.; Li, Y.; Ma, J.; Han, M. Comprehensive Analysis of GASA Family Members in the Malus domestica Genome: Identification, Characterization, and Their Expressions in Response to Apple Flower Induction. BMC Genom. 2017, 18, 827. [Google Scholar] [CrossRef]
- Porto, W.F.; Franco, O.L. Theoretical Structural Insights into the Snakin/GASA Family. Peptides 2013, 44, 163–167. [Google Scholar] [CrossRef]
- Mendu, V.; Singh, K.; Upadhyay, S.K. Insight into the Roles of Proline-Rich Extensin-like Receptor Protein Kinases of Bread Wheat (Triticum aestivum L.). Life 2022, 12, 941. [Google Scholar] [CrossRef]
- Sharma, H.; Sharma, A.; Rajput, R.; Sidhu, S.; Dhillon, H.; Verma, P.C.; Pandey, A.; Upadhyay, S.K. Molecular Characterization, Evolutionary Analysis, and Expression Profiling of BOR Genes in Important Cereals. Plants 2022, 11, 911. [Google Scholar] [CrossRef]
- Faraji, S.; Mehmood, F.; Malik, H.M.T.; Ahmed, I.; Heidari, P.; Poczai, P. The GASA Gene Family in Cacao (Theobroma cacao, Malvaceae): Genome Wide Identification and Expression Analysis. Agronomy 2021, 11, 1425. [Google Scholar]
- Boonpa, K.; Tantong, S.; Weerawanich, K.; Panpetch, P.; Pringsulaka, O.; Yingchutrakul, Y.; Roytrakul, S.; Sirikantaramas, S. Heterologous Expression and Antimicrobial Activity of OsGASR3 from Rice (Oryza sativa L.). J. Plant Physiol. 2018, 224, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Roxrud, I.; Lid, S.E.; Fletcher, J.C.; Schmidt, E.D.L.; Opsahl-Sorteberg, H.-G. GASA4, One of the 14-Member Arabidopsis GASA Family of Small Polypeptides, Regulates Flowering and Seed Development. Plant Cell Physiol. 2007, 48, 471–483. [Google Scholar] [CrossRef]
- An, B.; Wang, Q.; Zhang, X.; Zhang, B.; Luo, H.; He, C. Comprehensive Transcriptional and Functional Analyses of HbGASA Genes Reveal Their Roles in Fungal Pathogen Resistance in Hevea brasiliensis. Tree Genet. Genomes 2018, 14, 1–13. [Google Scholar] [CrossRef]
- Hernández-Martínez, M.A.; Suárez-Rodríguez, L.M.; López-Meza, J.E.; Ochoa-Zarzosa, A.; Salgado-Garciglia, R.; Fernández-Pavia, S.P.; López-Gómez, R. Antifungal Activity of Avocado Seed Recombinant GASA/Snakin PaSn. Antibiotics 2022, 11, 1558. [Google Scholar] [CrossRef]
- Ahmad, B.; Azeem, F.; Ali, M.A.; Nawaz, M.A.; Nadeem, H.; Abbas, A.; Batool, R.; Atif, R.M.; Ijaz, U.; Nieves-Cordones, M. Genome-Wide Identification and Expression Analysis of Two Component System Genes in Cicer arietinum. Genomics 2020, 112, 1371–1383. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wu, Y.; Liu, Y.; Yang, L.; Dong, R.; Jiang, L.; Liu, P.; Liu, G.; Wang, Z.; Luo, L. Genome-Wide Analysis of Tandem Duplicated Genes and Their Contribution to Stress Resistance in Pigeonpea (Cajanus cajan). Genomics 2021, 113, 728–735. [Google Scholar] [CrossRef]
- Faraji, S.; Filiz, E.; Kazemitabar, S.K.; Vannozzi, A.; Palumbo, F.; Barcaccia, G.; Heidari, P. The AP2/ERF Gene Family in Triticum durum: Genome-Wide Identification and Expression Analysis under Drought and Salinity Stresses. Genes 2020, 11, 1464. [Google Scholar] [CrossRef]
- Li, S.; Xing, Y.; Valdes, P.J.; Huang, Y.; Su, T.; Farnsworth, A.; Lunt, D.J.; Tang, H.; Kennedy, A.T.; Zhou, Z. Oligocene Climate Signals and Forcings in Eurasia Revealed by Plant Macrofossil and Modelling Results. Gondwana Res. 2018, 61, 115–127. [Google Scholar] [CrossRef]
- Sun, S.; Wang, H.; Yu, H.; Zhong, C.; Zhang, X.; Peng, J.; Wang, X. GASA14 Regulates Leaf Expansion and Abiotic Stress Resistance by Modulating Reactive Oxygen Species Accumulation. J. Exp. Bot. 2013, 64, 1637–1647. [Google Scholar] [CrossRef]
- Tripathi, D.; Zhang, T.; Koo, A.J.; Stacey, G.; Tanaka, K. Extracellular ATP Acts on Jasmonate Signaling to Reinforce Plant Defense. Plant Physiol. 2018, 176, 511–523. [Google Scholar] [CrossRef]
- Meena, R.K.; Jangra, S.; Wadhwa, Z.; Wati, M.; Wati, L. Role of Plant Volatiles in Defense and Communication. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 300–313. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, X. Salicylic Acid: Biosynthesis, Perception, and Contributions to Plant Immunity. Curr. Opin. Plant Biol. 2019, 50, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Wang, Y.; Li, X.; Zhang, Y. Biosynthesis and Regulation of Salicylic Acid and N-Hydroxypipecolic Acid in Plant Immunity. Mol. Plant 2020, 13, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Ding, Y.; Cai, B.; Qin, X.; Wu, J.; Yuan, M.; Wan, S.; Zhao, Y.; Xin, X.-F. Bacterial Effectors Manipulate Plant Abscisic Acid Signaling for Creation of an Aqueous Apoplast. Cell Host Microbe 2022, 30, 518–529. [Google Scholar] [CrossRef]
- Li, X.; He, S.; Gao, Y.; Xing, S.; Ren, H.; Yang, H.; Gao, Y.; Wang, J.; Li, N.; Duan, J. Ceratocystis and Related Genera Causing Wilt of Cunninghamia lanceolata Yunnan, China. For. Pathol. 2022, 52, e12744. [Google Scholar] [CrossRef]
- Kazemzadeh-Chakusary, M.; Mohammadi, H.; Sohrabi, M. Occurrence and Pathogenicity of Jattaea algeriensis (Calosphaeriales, Calosphaeriaceae) and Biscogniauxia mediterranea on Forest Trees in Guilan Province (Iran). J. Phytopathol. 2021, 169, 129–138. [Google Scholar] [CrossRef]
- Brown, A.A.; Lawrence, D.P.; Baumgartner, K. Role of Basidiomycete Fungi in the Grapevine Trunk Disease Esca. Plant Pathol. 2020, 69, 205–220. [Google Scholar] [CrossRef]
- Darqui, F.S.; Radonic, L.M.; Trotz, P.M.; López, N.; Vázquez Rovere, C.; Hopp, H.E.; López Bilbao, M. Potato Snakin-1 Gene Enhances Tolerance to Rhizoctonia solani and Sclerotinia sclerotiorum in Transgenic Lettuce Plants. J. Biotechnol. 2018, 283, 62–69. [Google Scholar] [CrossRef]
- Berrocal-Lobo, M.; Segura, A.; Moreno, M.; López, G.; García-Olmedo, F.; Molina, A. Snakin-2, an Antimicrobial Peptide from Potato Whose Gene Is Locally Induced by Wounding and Responds to Pathogen Infection. Plant Physiol. 2002, 128, 951–961. [Google Scholar] [CrossRef]
- Mao, Z.; Zheng, J.; Wang, Y.; Chen, G.; Yang, Y.; Feng, D.; Xie, B. The New CaSn Gene Belonging to the Snakin Family Induces Resistance against Root-Knot Nematode Infection in Pepper. Phytoparasitica 2011, 39, 151–164. [Google Scholar] [CrossRef]

| Name | Size (aa) | pI | Cys Number | Arg + Lys Number | Instability Index | Aliphatic Index | GRAVY 1 | SignalP 2 |
|---|---|---|---|---|---|---|---|---|
| AcGASA1 | 346 | 8.52 | 21 | 46 | 31.01 | 74.65 | −0.215 | No |
| AcGASA2 | 105 | 7.94 | 13 | 12 | 53.3 | 66 | −0.07 | Yes |
| AcGASA3 | 149 | 8.52 | 10 | 16 | 35.18 | 70 | −0.295 | No |
| AcGASA4 | 195 | 9.59 | 14 | 37 | 47.51 | 85.54 | −0.401 | No |
| AcGASA5 | 89 | 8.39 | 14 | 13 | 35.19 | 58.09 | −0.215 | Yes |
| AcGASA6 | 421 | 5.86 | 20 | 44 | 43.13 | 76.53 | −0.308 | No |
| AcGASA7 | 346 | 8.52 | 21 | 46 | 31.01 | 74.65 | −0.215 | No |
| AcGASA8 | 387 | 8.28 | 22 | 47 | 36.84 | 67.03 | −0.357 | No |
| AcGASA9 | 41 | 9.2 | 8 | 7 | 39.43 | 28.54 | −0.459 | Yes |
| AcGASA10 | 97 | 8.86 | 12 | 13 | 24.48 | 63.4 | −0.005 | Yes |
| AcGASA11 | 154 | 9.54 | 12 | 25 | 47.83 | 65.19 | −0.301 | Yes |
| AcGASA12 | 88 | 9.14 | 12 | 15 | 37.79 | 74.32 | −0.074 | Yes |
| AcGASA13 | 71 | 9.06 | 12 | 13 | 25.8 | 30.14 | −0.586 | No |
| KoGASA1 | 88 | 9.22 | 13 | 14 | 27.06 | 58.86 | −0.115 | Yes |
| KoGASA2 | 156 | 9.21 | 15 | 17 | 60.48 | 78.21 | 0.085 | No |
| KoGASA3 | 187 | 9.68 | 13 | 27 | 62.17 | 62.09 | −0.34 | Yes |
| KoGASA4 | 94 | 9.08 | 14 | 13 | 29.43 | 67.45 | −0.014 | Yes |
| KoGASA5 | 96 | 8.76 | 12 | 12 | 44.85 | 45.83 | −0.248 | Yes |
| KoGASA6 | 107 | 9.42 | 12 | 15 | 49.69 | 47.38 | −0.308 | Yes |
| KoGASA7 | 100 | 8.79 | 12 | 12 | 49.54 | 68.4 | −0.082 | Yes |
| KoGASA8 | 88 | 8.46 | 12 | 12 | 25.46 | 70.8 | 0.043 | Yes |
| KoGASA9 | 186 | 8.22 | 13 | 22 | 33.36 | 58.23 | −0.533 | No |
| AmGASA1 | 111 | 9.33 | 14 | 17 | 37.8 | 50.99 | −0.214 | Yes |
| AmGASA2 | 96 | 8.92 | 13 | 13 | 45.97 | 67.19 | 0.006 | Yes |
| AmGASA3 | 116 | 8.53 | 11 | 11 | 69.38 | 59.83 | −0.097 | No |
| AmGASA4 | 159 | 9.83 | 13 | 25 | 87.11 | 55.16 | −0.457 | Yes |
| AmGASA5 | 88 | 9.02 | 12 | 14 | 27.62 | 65.34 | −0.051 | Yes |
| AmGASA6 | 114 | 9.24 | 13 | 14 | 62.34 | 53.07 | −0.235 | Yes |
| AmGASA7 | 104 | 9.34 | 12 | 15 | 60.22 | 64.9 | −0.154 | Yes |
| AmGASA8 | 103 | 8.79 | 12 | 12 | 38.72 | 52.47 | −0.378 | No |
| AmGASA9 | 105 | 9.06 | 12 | 15 | 40.01 | 78.86 | −0.030 | Yes |
| AmGASA10 | 107 | 8.45 | 13 | 12 | 33.71 | 77.48 | 0.121 | Yes |
| AmGASA11 | 98 | 9.28 | 12 | 15 | 36.42 | 54.8 | −0.207 | Yes |
| AmGASA12 | 249 | 9.72 | 10 | 23 | 78.37 | 75.98 | −0.086 | Yes |
| AmGASA13 | 106 | 9.52 | 12 | 16 | 39.7 | 47.83 | −0.284 | Yes |
| AmGASA14 | 111 | 9.67 | 12 | 18 | 34.84 | 54.59 | −0.307 | Yes |
| AmGASA15 | 113 | 8.17 | 15 | 12 | 80.39 | 60.44 | −0.251 | Yes |
| AmGASA16 | 89 | 9.62 | 12 | 11 | 32.96 | 70.11 | −0.085 | Yes |
| AmGASA17 | 88 | 8.74 | 12 | 13 | 30.24 | 63.18 | −0.23 | Yes |
| AmGASA18 | 110 | 9.35 | 12 | 15 | 42.5 | 66.55 | −0.03 | Yes |
| AmGASA19 | 307 | 5.39 | 10 | 44 | 30.99 | 64.85 | −0.523 | Yes |
| AmGASA20 | 108 | 9.67 | 12 | 19 | 38.1 | 76.67 | 0.024 | Yes |
| AmGASA21 | 89 | 8.88 | 12 | 15 | 45.91 | 59.21 | −0.307 | Yes |
| AmGASA22 | 87 | 8.76 | 12 | 13 | 36.24 | 66.21 | −0.093 | Yes |
| AmGASA23 | 96 | 9.15 | 13 | 14 | 44.89 | 63.02 | 0.050 | Yes |
| AmGASA24 | 156 | 9.34 | 13 | 20 | 74.42 | 52.5 | −0.374 | Yes |
| AmGASA25 | 173 | 7.08 | 10 | 18 | 41.44 | 66.65 | −0.125 | Yes |
| AmGASA26 | 100 | 9.18 | 12 | 14 | 36.35 | 74.2 | 0.065 | Yes |
| AmGASA27 | 109 | 8.96 | 11 | 13 | 44.47 | 80.55 | −0.049 | Yes |
| Duplicated Gene Pairs | Ka | Ks | Ka/Ks | Duplicated Type | Time (MYE) |
|---|---|---|---|---|---|
| AcGASA7&AcGASA1 | 0.89 | 0.80 | 1.11 | Tandem | 29.67 |
| AcGASA8&AcGASA5 | 1.06 | 0.82 | 1.30 | Tandem | 35.37 |
| AcGASA8&AcGASA4 | 1.01 | 0.96 | 1.06 | Tandem | 33.74 |
| AcGASA5&AcGASA6 | 1.00 | 1.00 | 1.00 | Tandem | 33.36 |
| AcGASA5&AcGASA3 | 1.05 | 0.86 | 1.22 | Tandem | 34.92 |
| AcGASA5&AcGASA9 | 0.99 | 1.02 | 0.97 | Tandem | 33.13 |
| AcGASA6&AcGASA2 | 1.03 | 0.87 | 1.18 | Tandem | 34.48 |
| AcGASA6&AcGASA4 | 1.01 | 0.96 | 1.05 | Tandem | 33.65 |
| AcGASA2&AcGASA3 | 1.05 | 0.85 | 1.24 | Tandem | 35.10 |
| AcGASA4&AcGASA3 | 0.97 | 1.09 | 0.89 | Tandem | 32.38 |
| KoGASA1&KoGASA5 | 0.74 | 1.82 | 0.61 | Segmental | 60.75 |
| KoGASA1&KoGASA8 | 1.02 | 0.96 | 0.32 | Segmental | 31.89 |
| Relationship | Members | Location 1 | Members | Location 1 |
|---|---|---|---|---|
| A. thaliana and | AtGASA14 | 3 | AcGASA9 | 3 |
| A. corniculatum | AtGASA13 | 5 | AcGASA11 | 10 |
| A. thaliana and | AtGASA1 | 1 | KoGASA7 | 6 |
| K. obovata | AtGASA7 | 2 | KoGASA1 | 1 |
| AtGASA13 | 5 | KoGASA3 | 2 | |
| AtGASA4 | 5 | KoGASA6 | 4 | |
| AcGASA11 | 10 | KoGASA3 | 4 | |
| AcGASA12 | 16 | KoGASA8 | 7 | |
| AcGASA10 | 7 | KoGASA5 | 4 | |
| A. corniculatum | AcGASA11 | 10 | VvGASA5 | 14 |
| and V. vinifera | AcGASA12 | 16 | VvGASA9 | 18 |
| AcGASA13 | 18 | VvGASA9 | 18 | |
| A. corniculatum | AcGASA13 | 18 | PtGASA16 | 14 |
| and P. trichocarpa | AcGASA9 | 3 | PtGASA10 | 6 |
| K. obovata and | KoGASA1 | 1 | PtGASA2 | 1 |
| P. trichocarpa | KoGASA1 | 1 | PtGASA13 | 9 |
| KoGASA9 | 14 | PtGASA17 | 15 | |
| KoGASA3 | 2 | PtGASA4 | 1 | |
| KoGASA6 | 4 | PtGASA18 | 17 | |
| KoGASA4 | 4 | PtGASA19 | 17 | |
| KoGASA7 | 6 | PtGASA6 | 2 | |
| KoGASA7 | 6 | PtGASA9 | 5 | |
| KoGASA8 | 7 | PtGASA16 | 14 | |
| K. obovata and | KoGASA1 | 1 | VvGASA2 | 3 |
| V. vinifera | KoGASA9 | 14 | VvGASA1 | 1 |
| KoGASA9 | 14 | VvGASA8 | 17 | |
| KoGASA3 | 2 | VvGASA5 | 14 | |
| KoGASA6 | 4 | VvGASA6 | 14 | |
| KoGASA6 | 4 | VvGASA7 | 17 | |
| KoGASA8 | 7 | VvGASA9 | 18 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shang, C.; Ye, T.; Zhou, Q.; Chen, P.; Li, X.; Li, W.; Chen, S.; Hu, Z.; Zhang, W. Genome-Wide Identification and Bioinformatics Analyses of Host Defense Peptides Snakin/GASA in Mangrove Plants. Genes 2023, 14, 923. https://doi.org/10.3390/genes14040923
Shang C, Ye T, Zhou Q, Chen P, Li X, Li W, Chen S, Hu Z, Zhang W. Genome-Wide Identification and Bioinformatics Analyses of Host Defense Peptides Snakin/GASA in Mangrove Plants. Genes. 2023; 14(4):923. https://doi.org/10.3390/genes14040923
Chicago/Turabian StyleShang, Chenjing, Ting Ye, Qiao Zhou, Pengyu Chen, Xiangyu Li, Wenyi Li, Si Chen, Zhangli Hu, and Wei Zhang. 2023. "Genome-Wide Identification and Bioinformatics Analyses of Host Defense Peptides Snakin/GASA in Mangrove Plants" Genes 14, no. 4: 923. https://doi.org/10.3390/genes14040923
APA StyleShang, C., Ye, T., Zhou, Q., Chen, P., Li, X., Li, W., Chen, S., Hu, Z., & Zhang, W. (2023). Genome-Wide Identification and Bioinformatics Analyses of Host Defense Peptides Snakin/GASA in Mangrove Plants. Genes, 14(4), 923. https://doi.org/10.3390/genes14040923









