The Interplay between T Cells and Cancer: The Basis of Immunotherapy
Abstract
:1. Introduction
1.1. T Cell Receptor Structure and T Cell Signaling
1.2. Aberrant T Cell Signaling Associated with Various Malignancies
2. Mechanisms Responsible for the Current Challenges Faced in Immunotherapy
2.1. Exhaustion and Dysfunction of T Cells
2.1.1. Elevated Immune Checkpoint Expression Correlates with Poor Patient Outcomes and Reduced Responses to Cancer Immunotherapy
2.1.2. Changes in the Transcriptional and Epigenetic Landscapes of Dysfunctional T Cells and the Implications for T Cell Immunotherapy
2.2. Cancer-Cell-Intrinsic Characteristics Hinder Cancer Immunosurveillance and Limit the Efficacy of T-Cell-Based Cancer Immunotherapies
2.2.1. Molecular Alterations in Tumor Cells
Key Signaling Pathways Altered in Tumor Cells
Alterations of Proto-Oncogenes and Tumor Suppressors
Common Epigenetic Alterations Identified in Cancer Cells
2.2.2. Immunosuppressive Tumor Microenvironment
Metabolic Competition in the TME
Nutrients and Metabolites
TME-Derived Metabolites
Tumor-Associated Immune and Stromal Cells
- Tumor-Associated Macrophages (TAMs)
- Cancer-Associated Fibroblasts (CAFs)
- Tumor-Associated Neutrophils (TANs)
3. Exploring the Therapeutic Landscape for Cancer Treatment
3.1. Targeted Therapies: Small-Molecule Drugs, Therapeutic Antibodies, Oncolytic Viruses, and Gene Editing
3.2. Significant Emerging Immunotherapies: Cancer Vaccines, Engineered T Cells, and Immune Checkpoint Inhibitors
3.3. Challenges and Emerging Strategies in Cancer Immunotherapy
3.3.1. Combating Antigen Escape in CAR T Cell Therapy
Multiple Antigen-Targeting Strategies in CAR T Design
Combination of Single-Antigen Specific CAR T Therapies
Tandem CARs
Combination of CAR T Cells and BiTEs
Modular CAR T Designs
3.3.2. Addressing Additional Challenges: T Cell Trafficking and Persistence
3.3.3. Combinatorial Strategies for CAR T Therapy
3.3.4. Multidimensional Omics Data Analyses in the Advancement of T Cell Immunotherapy
3.3.5. Advanced Methods for Monitoring CAR T Cell Potency in Patients: Flow Cytometry, CyTOF, and ImmunoPET
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Waldman, A.D.; Fritz, J.M.; Lenardo, M.J. A guide to cancer immunotherapy: From T cell basic science to clinical practice. Nat. Rev. Immunol. 2020, 20, 651–668. [Google Scholar] [CrossRef] [PubMed]
- Smith-Garvin, J.E.; Koretzky, G.A.; Jordan, M.S. T cell activation. Annu. Rev. Immunol. 2009, 27, 591–619. [Google Scholar] [CrossRef] [PubMed]
- Tsimberidou, A.M.; Van Morris, K.; Vo, H.H.; Eck, S.; Lin, Y.F.; Rivas, J.M.; Andersson, B.S. T-cell receptor-based therapy: An innovative therapeutic approach for solid tumors. J. Hematol. Oncol. 2021, 14, 102. [Google Scholar] [CrossRef] [PubMed]
- Sterner, R.C.; Sterner, R.M. CAR-T cell therapy: Current limitations and potential strategies. Blood Cancer J. 2021, 11, 69. [Google Scholar] [CrossRef]
- Mullard, A. FDA approval of Immunocore’s first-in-class TCR therapeutic broadens depth of the T cell engager platform. Nat. Rev. Drug Discov. 2022, 21, 170. [Google Scholar] [CrossRef]
- Neelapu, S.S.; Locke, F.L.; Bartlett, N.L.; Lekakis, L.J.; Miklos, D.B.; Jacobson, C.A.; Braunschweig, I.; Oluwole, O.O.; Siddiqi, T.; Lin, Y.; et al. Axicabtagene Ciloleucel CAR T-Cell Therapy in Refractory Large B-Cell Lymphoma. N. Engl. J. Med. 2017, 377, 2531–2544. [Google Scholar] [CrossRef]
- Maude, S.L.; Laetsch, T.W.; Buechner, J.; Rives, S.; Boyer, M.; Bittencourt, H.; Bader, P.; Verneris, M.R.; Stefanski, H.E.; Myers, G.D.; et al. Tisagenlecleucel in Children and Young Adults with B-Cell Lymphoblastic Leukemia. N. Engl. J. Med. 2018, 378, 439–448. [Google Scholar] [CrossRef]
- Schuster, S.J.; Svoboda, J.; Chong, E.A.; Nasta, S.D.; Mato, A.R.; Anak, O.; Brogdon, J.L.; Pruteanu-Malinici, I.; Bhoj, V.; Landsburg, D.; et al. Chimeric Antigen Receptor T Cells in Refractory B-Cell Lymphomas. N. Engl. J. Med. 2017, 377, 2545–2554. [Google Scholar] [CrossRef]
- Brand, A.; Singer, K.; Koehl, G.E.; Kolitzus, M.; Schoenhammer, G.; Thiel, A.; Matos, C.; Bruss, C.; Klobuch, S.; Peter, K.; et al. LDHA-Associated Lactic Acid Production Blunts Tumor Immunosurveillance by T and NK Cells. Cell Metab. 2016, 24, 657–671. [Google Scholar] [CrossRef] [Green Version]
- Gorchakov, A.A.; Kulemzin, S.V.; Kochneva, G.V.; Taranin, A.V. Challenges and Prospects of Chimeric Antigen Receptor T-cell Therapy for Metastatic Prostate Cancer. Eur. Urol. 2020, 77, 299–308. [Google Scholar] [CrossRef]
- Crespo, J.; Sun, H.; Welling, T.H.; Tian, Z.; Zou, W. T cell anergy, exhaustion, senescence, and stemness in the tumor microenvironment. Curr. Opin. Immunol. 2013, 25, 214–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurachi, M. CD8+ T cell exhaustion. Semin. Immunopathol. 2019, 41, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Thommen, D.S.; Schumacher, T.N. T Cell Dysfunction in Cancer. Cancer Cell 2018, 33, 547–562. [Google Scholar] [CrossRef] [Green Version]
- Straathof, K.; Flutter, B.; Wallace, R.; Jain, N.; Loka, T.; Depani, S.; Wright, G.; Thomas, S.; Cheung, G.W.; Gileadi, T.; et al. Antitumor activity without on-target off-tumor toxicity of GD2-chimeric antigen receptor T cells in patients with neuroblastoma. Sci. Transl. Med. 2020, 12, eabd6169. [Google Scholar] [CrossRef] [PubMed]
- Ernst, M.; Oeser, A.; Besiroglu, B.; Caro-Valenzuela, J.; Abd El Aziz, M.; Monsef, I.; Borchmann, P.; Estcourt, L.J.; Skoetz, N.; Goldkuhle, M. Chimeric antigen receptor (CAR) T-cell therapy for people with relapsed or refractory diffuse large B-cell lymphoma. Cochrane Database Syst. Rev. 2021, 9, CD013365. [Google Scholar] [CrossRef] [PubMed]
- Kochenderfer, J.N.; Rosenberg, S.A. Treating B-cell cancer with T cells expressing anti-CD19 chimeric antigen receptors. Nat. Rev. Clin. Oncol. 2013, 10, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, R.O.; Evbuomwan, M.O.; Pittaluga, S.; Rose, J.J.; Raffeld, M.; Yang, S.; Gress, R.E.; Hakim, F.T.; Kochenderfer, J.N. B-cell maturation antigen is a promising target for adoptive T-cell therapy of multiple myeloma. Clin. Cancer Res. 2013, 19, 2048–2060. [Google Scholar] [CrossRef] [Green Version]
- Kochenderfer, J.N.; Dudley, M.E.; Feldman, S.A.; Wilson, W.H.; Spaner, D.E.; Maric, I.; Stetler-Stevenson, M.; Phan, G.Q.; Hughes, M.S.; Sherry, R.M.; et al. B-cell depletion and remissions of malignancy along with cytokine-associated toxicity in a clinical trial of anti-CD19 chimeric-antigen-receptor-transduced T cells. Blood 2012, 119, 2709–2720. [Google Scholar] [CrossRef]
- Frey, N.V.; Shaw, P.A.; Hexner, E.O.; Pequignot, E.; Gill, S.; Luger, S.M.; Mangan, J.K.; Loren, A.W.; Perl, A.E.; Maude, S.L.; et al. Optimizing Chimeric Antigen Receptor T-Cell Therapy for Adults With Acute Lymphoblastic Leukemia. J. Clin. Oncol. 2020, 38, 415–422. [Google Scholar] [CrossRef]
- Hassan, R.; Cohen, S.J.; Phillips, M.; Pastan, I.; Sharon, E.; Kelly, R.J.; Schweizer, C.; Weil, S.; Laheru, D. Phase I clinical trial of the chimeric anti-mesothelin monoclonal antibody MORAb-009 in patients with mesothelin-expressing cancers. Clin. Cancer Res. 2010, 16, 6132–6138. [Google Scholar] [CrossRef] [Green Version]
- Hassan, R.; Bullock, S.; Premkumar, A.; Kreitman, R.J.; Kindler, H.; Willingham, M.C.; Pastan, I. Phase I study of SS1P, a recombinant anti-mesothelin immunotoxin given as a bolus I.V. infusion to patients with mesothelin-expressing mesothelioma, ovarian, and pancreatic cancers. Clin. Cancer Res. 2007, 13, 5144–5149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dudley, M.E.; Wunderlich, J.; Nishimura, M.I.; Yu, D.; Yang, J.C.; Topalian, S.L.; Schwartzentruber, D.J.; Hwu, P.; Marincola, F.M.; Sherry, R.; et al. Adoptive transfer of cloned melanoma-reactive T lymphocytes for the treatment of patients with metastatic melanoma. J. Immunother. 2001, 24, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Dudley, M.E.; Wunderlich, J.R.; Yang, J.C.; Hwu, P.; Schwartzentruber, D.J.; Topalian, S.L.; Sherry, R.M.; Marincola, F.M.; Leitman, S.F.; Seipp, C.A.; et al. A phase I study of nonmyeloablative chemotherapy and adoptive transfer of autologous tumor antigen-specific T lymphocytes in patients with metastatic melanoma. J. Immunother. 2002, 25, 243–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maude, S.L.; Frey, N.; Shaw, P.A.; Aplenc, R.; Barrett, D.M.; Bunin, N.J.; Chew, A.; Gonzalez, V.E.; Zheng, Z.; Lacey, S.F.; et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med. 2014, 371, 1507–1517. [Google Scholar] [CrossRef] [Green Version]
- Van Bruggen, J.A.C.; Martens, A.W.J.; Fraietta, J.A.; Hofland, T.; Tonino, S.H.; Eldering, E.; Levin, M.D.; Siska, P.J.; Endstra, S.; Rathmell, J.C.; et al. Chronic lymphocytic leukemia cells impair mitochondrial fitness in CD8(+) T cells and impede CAR T-cell efficacy. Blood 2019, 134, 44–58. [Google Scholar] [CrossRef]
- Rosenberg, S.A. Raising the bar: The curative potential of human cancer immunotherapy. Sci. Transl. Med. 2012, 4, 127ps8. [Google Scholar] [CrossRef]
- Hinrichs, C.S.; Rosenberg, S.A. Exploiting the curative potential of adoptive T-cell therapy for cancer. Immunol. Rev. 2014, 257, 56–71. [Google Scholar] [CrossRef] [Green Version]
- Chaturvedi, A.K.; Engels, E.A.; Pfeiffer, R.M.; Hernandez, B.Y.; Xiao, W.; Kim, E.; Jiang, B.; Goodman, M.T.; Sibug-Saber, M.; Cozen, W.; et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J. Clin. Oncol. 2011, 29, 4294–4301. [Google Scholar] [CrossRef]
- Gyurdieva, A.; Zajic, S.; Chang, Y.F.; Houseman, E.A.; Zhong, S.; Kim, J.; Nathenson, M.; Faitg, T.; Woessner, M.; Turner, D.C.; et al. Biomarker correlates with response to NY-ESO-1 TCR T cells in patients with synovial sarcoma. Nat. Commun. 2022, 13, 5296. [Google Scholar] [CrossRef]
- D’Angelo, S.P.; Melchiori, L.; Merchant, M.S.; Bernstein, D.; Glod, J.; Kaplan, R.; Grupp, S.; Tap, W.D.; Chagin, K.; Binder, G.K.; et al. Antitumor Activity Associated with Prolonged Persistence of Adoptively Transferred NY-ESO-1 (c259)T Cells in Synovial Sarcoma. Cancer Discov. 2018, 8, 944–957. [Google Scholar] [CrossRef] [Green Version]
- Ramachandran, I.; Lowther, D.E.; Dryer-Minnerly, R.; Wang, R.; Fayngerts, S.; Nunez, D.; Betts, G.; Bath, N.; Tipping, A.J.; Melchiori, L.; et al. Systemic and local immunity following adoptive transfer of NY-ESO-1 SPEAR T cells in synovial sarcoma. J. Immunother. Cancer 2019, 7, 276. [Google Scholar] [CrossRef] [PubMed]
- Abate-Daga, D.; Hanada, K.; Davis, J.L.; Yang, J.C.; Rosenberg, S.A.; Morgan, R.A. Expression profiling of TCR-engineered T cells demonstrates overexpression of multiple inhibitory receptors in persisting lymphocytes. Blood 2013, 122, 1399–1410. [Google Scholar] [CrossRef] [Green Version]
- Kawakami, Y.; Eliyahu, S.; Delgado, C.H.; Robbins, P.F.; Rivoltini, L.; Topalian, S.L.; Miki, T.; Rosenberg, S.A. Cloning of the gene coding for a shared human melanoma antigen recognized by autologous T cells infiltrating into tumor. Proc. Natl. Acad. Sci. USA 1994, 91, 3515–3519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawakami, Y.; Eliyahu, S.; Delgado, C.H.; Robbins, P.F.; Sakaguchi, K.; Appella, E.; Yannelli, J.R.; Adema, G.J.; Miki, T.; Rosenberg, S.A. Identification of a human melanoma antigen recognized by tumor-infiltrating lymphocytes associated with in vivo tumor rejection. Proc. Natl. Acad. Sci. USA 1994, 91, 6458–6462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawakami, Y.; Eliyahu, S.; Sakaguchi, K.; Robbins, P.F.; Rivoltini, L.; Yannelli, J.R.; Appella, E.; Rosenberg, S.A. Identification of the immunodominant peptides of the MART-1 human melanoma antigen recognized by the majority of HLA-A2-restricted tumor infiltrating lymphocytes. J. Exp. Med. 1994, 180, 347–352. [Google Scholar] [CrossRef] [Green Version]
- Allison, J.P.; McIntyre, B.W.; Bloch, D. Tumor-specific antigen of murine T-lymphoma defined with monoclonal antibody. J. Immunol. 1982, 129, 2293–2300. [Google Scholar] [CrossRef] [PubMed]
- Fabbi, M.; Acuto, O.; Smart, J.E.; Reinherz, E.L. Homology of Ti alpha-subunit of a T-cell antigen-MHC receptor with immunoglobulin. Nature 1984, 312, 269–271. [Google Scholar] [CrossRef]
- Hedrick, S.M.; Cohen, D.I.; Nielsen, E.A.; Davis, M.M. Isolation of cDNA clones encoding T cell-specific membrane-associated proteins. Nature 1984, 308, 149–153. [Google Scholar] [CrossRef]
- Morath, A.; Schamel, W.W. alphabeta and gammadelta T cell receptors: Similar but different. J. Leukoc. Biol. 2020, 107, 1045–1055. [Google Scholar] [CrossRef]
- Call, M.E.; Pyrdol, J.; Wiedmann, M.; Wucherpfennig, K.W. The organizing principle in the formation of the T cell receptor-CD3 complex. Cell 2002, 111, 967–979. [Google Scholar] [CrossRef] [Green Version]
- Dushek, O.; Goyette, J.; van der Merwe, P.A. Non-catalytic tyrosine-phosphorylated receptors. Immunol. Rev. 2012, 250, 258–276. [Google Scholar] [CrossRef] [PubMed]
- Huse, M. The T-cell-receptor signaling network. J. Cell Sci. 2009, 122, 1269–1273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Kadlecek, T.A.; Au-Yeung, B.B.; Goodfellow, H.E.; Hsu, L.Y.; Freedman, T.S.; Weiss, A. ZAP-70: An essential kinase in T-cell signaling. Cold Spring Harb. Perspect. Biol. 2010, 2, a002279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, A.C.; Irving, B.A.; Fraser, J.D.; Weiss, A. The zeta chain is associated with a tyrosine kinase and upon T-cell antigen receptor stimulation associates with ZAP-70, a 70-kDa tyrosine phosphoprotein. Proc. Natl. Acad. Sci. USA 1991, 88, 9166–9170. [Google Scholar] [CrossRef] [Green Version]
- Balagopalan, L.; Coussens, N.P.; Sherman, E.; Samelson, L.E.; Sommers, C.L. The LAT story: A tale of cooperativity, coordination, and choreography. Cold Spring Harb. Perspect. Biol. 2010, 2, a005512. [Google Scholar] [CrossRef] [Green Version]
- Bunnell, S.C.; Diehn, M.; Yaffe, M.B.; Findell, P.R.; Cantley, L.C.; Berg, L.J. Biochemical interactions integrating Itk with the T cell receptor-initiated signaling cascade. J. Biol. Chem. 2000, 275, 2219–2230. [Google Scholar] [CrossRef] [Green Version]
- Martinez, G.J.; Pereira, R.M.; Aijo, T.; Kim, E.Y.; Marangoni, F.; Pipkin, M.E.; Togher, S.; Heissmeyer, V.; Zhang, Y.C.; Crotty, S.; et al. The transcription factor NFAT promotes exhaustion of activated CD8(+) T cells. Immunity 2015, 42, 265–278. [Google Scholar] [CrossRef] [Green Version]
- Shayan, G.; Srivastava, R.; Li, J.; Schmitt, N.; Kane, L.P.; Ferris, R.L. Adaptive resistance to anti-PD1 therapy by Tim-3 upregulation is mediated by the PI3K-Akt pathway in head and neck cancer. OncoImmunology 2017, 6, e1261779. [Google Scholar] [CrossRef] [Green Version]
- Jeon, H.-S.; Jin, G.; Kang, H.-G.; Choi, Y.Y.; Lee, W.K.; Choi, J.E.; Bae, E.Y.; Yoo, S.S.; Lee, S.Y.; Lee, E.B.; et al. A functional variant at 19q13.3, rs967591G>A, is associated with shorter survival of early-stage lung cancer. Clin. Cancer Res. 2013, 19, 4185–4195. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.Y.; Choi, J.E.; Jeon, H.-S.; Choi, Y.-Y.; Lee, W.K.; Lee, E.B.; Lee, H.C.; Kang, H.-G.; Yoo, S.S.; Lee, J.; et al. A Panel of Genetic Polymorphism for the Prediction of Prognosis in Patients with Early Stage Non-Small Cell Lung Cancer after Surgical Resection. PLoS ONE 2015, 10, e0140216. [Google Scholar] [CrossRef] [Green Version]
- Thommen, D.S.; Schreiner, J.; Müller, P.; Herzig, P.; Roller, A.; Belousov, A.; Umana, P.; Pisa, P.; Klein, C.; Bacac, M.; et al. Progression of Lung Cancer Is Associated with Increased Dysfunction of T Cells Defined by Coexpression of Multiple Inhibitory Receptors. Cancer Immunol. Res. 2015, 3, 1344–1355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, X.; Zhang, Y.; Zheng, L.; Zheng, C.; Song, J.; Zhang, Q.; Kang, B.; Liu, Z.; Jin, L.; Xing, R.; et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat. Med. 2018, 24, 978–985. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Xu, J.; Ni, J.; Gao, X.; Zhu, Z.; Dong, D.; Wang, X.; Shi, C.; Tao, X.; Dong, W.; et al. A functional insertion/deletion polymorphism in the proximal promoter of CD3G is associated with susceptibility for hepatocellular carcinoma in Chinese population. DNA Cell Biol. 2012, 31, 1480–1485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, C.; Zheng, L.; Yoo, J.-K.; Guo, H.; Zhang, Y.; Guo, X.; Kang, B.; Hu, R.; Huang, J.Y.; Zhang, Q.; et al. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell 2017, 169, 1342–1356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; He, Q.; Shen, H.; Xia, A.; Tian, W.; Yu, W.; Sun, B. TOX promotes the exhaustion of antitumor CD8+ T cells by preventing PD1 degradation in hepatocellular carcinoma. J. Hepatol. 2019, 71, 731–741. [Google Scholar] [CrossRef]
- Peña-Romero, A.C.; Orenes-Piñero, E. Dual Effect of Immune Cells within Tumour Microenvironment: Pro- and Anti-Tumour Effects and Their Triggers. Cancers 2022, 14, 1681. [Google Scholar] [CrossRef]
- Liang, C.; Huang, S.; Zhao, Y.; Chen, S.; Li, Y. TOX as a potential target for immunotherapy in lymphocytic malignancies. Biomarker Res. 2021, 9, 20. [Google Scholar] [CrossRef]
- Lu, X.; Yang, L.; Yao, D.; Wu, X.; Li, J.; Liu, X.; Deng, L.; Huang, C.; Wang, Y.; Li, D.; et al. Tumor antigen-specific CD8(+) T cells are negatively regulated by PD-1 and Tim-3 in human gastric cancer. Cell Immunol. 2017, 313, 43–51. [Google Scholar] [CrossRef]
- Tycko, B.; Smith, S.D.; Sklar, J. Chromosomal translocations joining LCK and TCRB loci in human T cell leukemia. J. Exp. Med. 1991, 174, 867–873. [Google Scholar] [CrossRef] [Green Version]
- Rassenti, L.Z.; Huynh, L.; Toy, T.L.; Chen, L.; Keating, M.J.; Gribben, J.G.; Neuberg, D.S.; Flinn, I.W.; Rai, K.R.; Byrd, J.C.; et al. ZAP-70 compared with immunoglobulin heavy-chain gene mutation status as a predictor of disease progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2004, 351, 893–901. [Google Scholar] [CrossRef]
- Ruiz, I.; Vicario, R.; Morancho, B.; Morales, C.; Arenas, E.; Herter, S.; Freimoser-Grundschober, A.; Somandin, J.; Sam, J.; Ast, O.; et al. p95HER2–T cell bispecific antibody for breast. Sci. Transl. Med. 2018, 10, eaat1445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zippelius, A.; Batard, P.; Rubio-Godoy, V.; Bioley, G.; Liénard, D.; Lejeune, F.; Rimoldi, D.; Guillaume, P.; Meidenbauer, N.; Mackensen, A.; et al. Effector Function of Human Tumor-Specific CD8 T Cells in Melanoma Lesions: A State of Local Functional Tolerance. Cancer Res. 2004, 64, 2865–2873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmadzadeh, M.; Johnson, L.A.; Heemskerk, B.; Wunderlich, J.R.; Dudley, M.E.; White, D.E.; Rosenberg, S.A. Tumor antigen–specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood 2009, 114, 1537–1544. [Google Scholar] [CrossRef] [PubMed]
- Baitsch, L.; Baumgaertner, P.; Devêvre, E.; Raghav, S.K.; Legat, A.; Barba, L.; Wieckowski, S.; Bouzourene, H.; Deplancke, B.; Romero, P.; et al. Exhaustion of tumor-specific CD8⁺ T cells in metastases from melanoma patients. J. Clin. Investig. 2011, 121, 2350–2360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vallois, D.; Dobay, M.P.D.; Morin, R.D.; Lemonnier, F.; Missiaglia, E.; Juilland, M.; Iwaszkiewicz, J.; Fataccioli, V.; Bisig, B.; Roberti, A.; et al. Activating mutations in genes related to TCR signaling in angioimmunoblastic and other follicular helper T-cell-derived lymphomas. Blood 2016, 128, 1490–1502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaqué, J.P.; Gómez-López, G.; Monsálvez, V.; Varela, I.; Martínez, N.; Pérez, C.; Domínguez, O.; Graña, O.; Rodríguez-Peralto, J.L.; Rodríguez-Pinilla, S.M.; et al. PLCG1 mutations in cutaneous T-cell lymphomas. Blood 2014, 123, 2034–2043. [Google Scholar] [CrossRef]
- Hayes, T.K.; Neel, N.F.; Hu, C.; Gautam, P.; Chenard, M.; Long, B.; Aziz, M.; Kassner, M.; Bryant, K.L.; Pierobon, M.; et al. Long-Term ERK Inhibition in KRAS-Mutant Pancreatic Cancer Is Associated with MYC Degradation and Senescence-like Growth Suppression. Cancer Cell 2016, 29, 75–89. [Google Scholar] [CrossRef] [Green Version]
- Hobbs, G.A.; Wittinghofer, A.; Der, C.J. Selective Targeting of the KRAS G12C Mutant: Kicking KRAS When It’s Down. Cancer Cell 2016, 29, 251–253. [Google Scholar] [CrossRef] [Green Version]
- Ng, S.Y.; Brown, L.; Stevenson, K.; deSouza, T.; Aster, J.C.; Louissaint, A., Jr.; Weinstock, D.M. RhoA G17V is sufficient to induce autoimmunity and promotes T-cell lymphomagenesis in mice. Blood 2018, 132, 935–947. [Google Scholar] [CrossRef] [Green Version]
- Barreira, M.; Fabbiano, S.; Couceiro, J.R.; Torreira, E.; Martínez-Torrecuadrada, J.L.; Montoya, G.; Llorca, O.; Bustelo, X.R. The C-terminal SH3 domain contributes to the intramolecular inhibition of Vav family proteins. Sci. Signal. 2014, 7, ra35. [Google Scholar] [CrossRef] [Green Version]
- Shah, K.; Al-Haidari, A.; Sun, J.; Kazi, J.U. T cell receptor (TCR) signaling in health and disease. Sig. Transduct. Target Ther. 2021, 6, 412. [Google Scholar] [CrossRef] [PubMed]
- Linka, R.M.; Risse, S.L.; Bienemann, K.; Werner, M.; Linka, Y.; Krux, F.; Synaeve, C.; Deenen, R.; Ginzel, S.; Dvorsky, R.; et al. Loss-of-function mutations within the IL-2 inducible kinase ITK in patients with EBV-associated lymphoproliferative diseases. Leukemia 2012, 26, 963–971. [Google Scholar] [CrossRef]
- Robles-Valero, J.; Lorenzo-Martín, L.F.; Menacho-Márquez, M.; Fernández-Pisonero, I.; Abad, A.; Camós, M.; Toribio, M.L.; Espinosa, L.; Bigas, A.; Bustelo, X.R. A Paradoxical Tumor-Suppressor Role for the Rac1 Exchange Factor Vav1 in T Cell Acute Lymphoblastic Leukemia. Cancer Cell 2017, 32, 608–623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rohr, J.; Guo, S.; Huo, J.; Bouska, A.; Lachel, C.; Li, Y.; Simone, P.D.; Zhang, W.; Gong, Q.; Wang, C.; et al. Recurrent activating mutations of CD28 in peripheral T-cell lymphomas. Leukemia 2016, 30, 1062–1070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quivoron, C.; Couronne, L.; Della Valle, V.; Lopez, C.K.; Plo, I.; Wagner-Ballon, O.; Do Cruzeiro, M.; Delhommeau, F.; Arnulf, B.; Stern, M.H.; et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell 2011, 20, 25–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cairns, R.A.; Iqbal, J.; Lemonnier, F.; Kucuk, C.; de Leval, L.; Jais, J.-P.; Parrens, M.; Martin, A.; Xerri, L.; Brousset, P.; et al. IDH2 mutations are frequent in angioimmunoblastic T-cell lymphoma. Blood 2012, 119, 1901–1903. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; McKeithan, T.W.; Gong, Q.; Zhang, W.; Bouska, A.; Rosenwald, A.; Gascoyne, R.D.; Wu, X.; Wang, J.; Muhammad, Z.; et al. IDH2R172 mutations define a unique subgroup of patients with angioimmunoblastic T-cell lymphoma. Blood 2015, 126, 1741–1752. [Google Scholar] [CrossRef] [Green Version]
- Palomero, T.; Couronné, L.; Khiabanian, H.; Kim, M.-Y.; Ambesi-Impiombato, A.; Perez-Garcia, A.; Carpenter, Z.; Abate, F.; Allegretta, M.; Haydu, J.E.; et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat. Genet. 2014, 46, 166–170. [Google Scholar] [CrossRef]
- Lee, S.H.; Kim, J.S.; Kim, J.; Kim, S.J.; Kim, W.S.; Lee, S.; Ko, Y.H.; Yoo, H.Y. A highly recurrent novel missense mutation in CD28 among angioimmunoblastic T-cell lymphoma patients. Haematologica 2015, 100, e505–e507. [Google Scholar] [CrossRef] [Green Version]
- Mizoguchi, H.; O’Shea, J.J.; Longo, D.L.; Loeffler, C.M.; McVicar, D.W.; Ochoa, A.C. Alterations in signal transduction molecules in T lymphocytes from tumor-bearing mice. Science 1992, 258, 1795–1798. [Google Scholar] [CrossRef]
- Matsuzaki, J.; Gnjatic, S.; Mhawech-Fauceglia, P.; Beck, A.; Miller, A.; Tsuji, T.; Eppolito, C.; Qian, F.; Lele, S.; Shrikant, P.; et al. Tumor-infiltrating NY-ESO-1-specific CD8+ T cells are negatively regulated by LAG-3 and PD-1 in human ovarian cancer. Proc. Natl. Acad. Sci. USA 2010, 107, 7875–7880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, E.; Zhang, Y.; Zhu, G.; Hao, J.; Persson, X.-M.T.; Egilmez, N.K.; Suttles, J.; Li, B. Deficiency of AMPK in CD8+ T cells suppresses their anti-tumor function by inducing protein phosphatase-mediated cell death. Oncotarget 2019, 6, 7944–7958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- June, C.H.; O’Connor, R.S.; Kawalekar, O.U.; Ghassemi, S.; Milone, M.C. CAR T cell immunotherapy for human cancer. Science 2018, 359, 1361–1365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wherry, E.J. T cell exhaustion. Nat. Immunol. 2011, 12, 492–499. [Google Scholar] [CrossRef] [PubMed]
- Chow, A.; Perica, K.; Klebanoff, C.A.; Wolchok, J.D. Clinical implications of T cell exhaustion for cancer immunotherapy. Nat. Rev. Clin. Oncol. 2022, 19, 775–790. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Zhang, Y.; Hu, Y.; Mei, H. T Cell Exhaustion and CAR-T Immunotherapy in Hematological Malignancies. Biomed. Res. Int. 2021, 2021, 6616391. [Google Scholar] [CrossRef] [PubMed]
- Alsaab, H.O.; Sau, S.; Alzhrani, R.; Tatiparti, K.; Bhise, K.; Kashaw, S.K.; Iyer, A.K. PD-1 and PD-L1 Checkpoint Signaling Inhibition for Cancer Immunotherapy: Mechanism, Combinations, and Clinical Outcome. Front. Pharmacol. 2017, 8, 561. [Google Scholar] [CrossRef] [Green Version]
- Blackburn, S.D.; Shin, H.; Haining, W.N.; Zou, T.; Workman, C.J.; Polley, A.; Betts, M.R.; Freeman, G.J.; Vignali, D.A.A.; Wherry, E.J. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat. Immunol. 2009, 10, 29–37. [Google Scholar] [CrossRef] [Green Version]
- Sakuishi, K.; Apetoh, L.; Sullivan, J.M.; Blazar, B.R.; Kuchroo, V.K.; Anderson, A.C. Targeting Tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity. J. Exp. Med. 2010, 207, 2187–2194. [Google Scholar] [CrossRef]
- Woo, S.-R.; Turnis, M.E.; Goldberg, M.V.; Bankoti, J.; Selby, M.; Nirschl, C.J.; Bettini, M.L.; Gravano, D.M.; Vogel, P.; Liu, C.L.; et al. Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T-cell function to promote tumoral immune escape. Cancer Res. 2012, 72, 917–927. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Wu, K.; Tao, K.; Chen, L.; Zheng, Q.; Lu, X.; Liu, J.; Shi, L.; Liu, C.; Wang, G.; et al. Tim-3/galectin-9 signaling pathway mediates T-cell dysfunction and predicts poor prognosis in patients with hepatitis B virus-associated hepatocellular carcinoma. Hepatology 2012, 56, 1342–1351. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Flies, D.B. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat. Rev. Immunol. 2013, 13, 227–242. [Google Scholar] [CrossRef]
- Kuchroo, V.K.; Anderson, A.C.; Petrovas, C. Coinhibitory receptors and CD8 T cell exhaustion in chronic infections. Curr. Opin. HIV AIDS 2014, 9, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Wherry, E.J.; Kurachi, M. Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 2015, 15, 486–499. [Google Scholar] [CrossRef] [PubMed]
- Tirosh, I.; Izar, B.; Prakadan, S.M.; Wadsworth, M.H.; Treacy, D.; Trombetta, J.J.; Rotem, A.; Rodman, C.; Lian, C.; Murphy, G.; et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016, 352, 189–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Leun, A.M.; Thommen, D.S.; Schumacher, T.N. CD8+ T cell states in human cancer: Insights from single-cell analysis. Nat. Rev. Cancer 2020, 20, 218–232. [Google Scholar] [CrossRef]
- Chauvin, J.-M.; Zarour, H.M. TIGIT in cancer immunotherapy. J. Immunother. Cancer 2020, 8, e000957. [Google Scholar] [CrossRef]
- Farber, D.L. Form and function for T cells in health and disease. Nat. Rev. Immunol. 2020, 20, 83–84. [Google Scholar] [CrossRef]
- Schnell, A.; Bod, L.; Madi, A.; Kuchroo, V.K. The yin and yang of co-inhibitory receptors: Toward anti-tumor immunity without autoimmunity. Cell Res. 2020, 30, 285–299. [Google Scholar] [CrossRef] [Green Version]
- Zebley, C.C.; Brown, C.; Mi, T.; Fan, Y.; Alli, S.; Boi, S.; Galletti, G.; Lugli, E.; Langfitt, D.; Metais, J.Y.; et al. CD19-CAR T cells undergo exhaustion DNA methylation programming in patients with acute lymphoblastic leukemia. Cell Rep. 2021, 37, 110079. [Google Scholar] [CrossRef]
- Kansy, B.A.; Concha-Benavente, F.; Srivastava, R.M.; Jie, H.B.; Shayan, G.; Lei, Y.; Moskovitz, J.; Moy, J.; Li, J.; Brandau, S.; et al. PD-1 Status in CD8(+) T Cells Associates with Survival and Anti-PD-1 Therapeutic Outcomes in Head and Neck Cancer. Cancer Res. 2017, 77, 6353–6364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, W.; Dimitri, A.; Wang, W.; Jung, I.Y.; Ott, C.J.; Fasolino, M.; Wang, Y.; Kulikovskaya, I.; Gupta, M.; Yoder, T.; et al. BET bromodomain protein inhibition reverses chimeric antigen receptor extinction and reinvigorates exhausted T cells in chronic lymphocytic leukemia. J. Clin. Investig. 2021, 131, e145459. [Google Scholar] [CrossRef] [PubMed]
- Fraietta, J.A.; Lacey, S.F.; Orlando, E.J.; Pruteanu-Malinici, I.; Gohil, M.; Lundh, S.; Boesteanu, A.C.; Wang, Y.; O’Connor, R.S.; Hwang, W.T.; et al. Determinants of response and resistance to CD19 chimeric antigen receptor (CAR) T cell therapy of chronic lymphocytic leukemia. Nat. Med. 2018, 24, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Deng, Q.; Han, G.; Puebla-Osorio, N.; Ma, M.C.J.; Strati, P.; Chasen, B.; Dai, E.; Dang, M.; Jain, N.; Yang, H.; et al. Characteristics of anti-CD19 CAR T cell infusion products associated with efficacy and toxicity in patients with large B cell lymphomas. Nat. Med. 2020, 26, 1878–1887. [Google Scholar] [CrossRef] [PubMed]
- Vahteristo, P.; Bartkova, J.; Eerola, H.; Syrjakoski, K.; Ojala, S.; Kilpivaara, O.; Tamminen, A.; Kononen, J.; Aittomaki, K.; Heikkila, P.; et al. A CHEK2 genetic variant contributing to a substantial fraction of familial breast cancer. Am. J. Hum. Genet. 2002, 71, 432–438. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.; Guan, Q.; Wang, Y.; Zhao, Z.J.; Zhou, G.W. SHP-1 suppresses cancer cell growth by promoting degradation of JAK kinases. J. Cell Biochem. 2003, 90, 1026–1037. [Google Scholar] [CrossRef]
- Wu, C.; Sun, M.; Liu, L.; Zhou, G.W. The function of the protein tyrosine phosphatase SHP-1 in cancer. Gene 2003, 306, 1–12. [Google Scholar] [CrossRef]
- Zhang, T.; Guo, W.; Yang, Y.; Liu, W.; Guo, L.; Gu, Y.; Shu, Y.; Wang, L.; Wu, X.; Hua, Z.; et al. Loss of SHP-2 activity in CD4+ T cells promotes melanoma progression and metastasis. Sci. Rep. 2013, 3, 2845. [Google Scholar] [CrossRef] [PubMed]
- Philip, M.; Fairchild, L.; Sun, L.; Horste, E.L.; Camara, S.; Shakiba, M.; Scott, A.C.; Viale, A.; Lauer, P.; Merghoub, T.; et al. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature 2017, 545, 452–456. [Google Scholar] [CrossRef] [Green Version]
- Mognol, G.P.; Spreafico, R.; Wong, V.; Scott-Browne, J.P.; Togher, S.; Hoffmann, A.; Hogan, P.G.; Rao, A.; Trifari, S. Exhaustion-associated regulatory regions in CD8+ tumor-infiltrating T cells. Proc. Natl. Acad. Sci. USA 2017, 114, E2776–E2785. [Google Scholar] [CrossRef] [Green Version]
- Soto-Nieves, N.; Puga, I.; Abe, B.T.; Bandyopadhyay, S.; Baine, I.; Rao, A.; Macian, F. Transcriptional complexes formed by NFAT dimers regulate the induction of T cell tolerance. J. Exp. Med. 2009, 206, 867–876. [Google Scholar] [CrossRef] [Green Version]
- Harant, H.; Lindley, I.J.D. Negative cross-talk between the human orphan nuclear receptor Nur77/NAK-1/TR3 and nuclear factor-κB. Nucleic Acids Res. 2004, 32, 5280–5290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Wang, Y.; Lu, H.; Li, J.; Yan, X.; Xiao, M.; Hao, J.; Alekseev, A.; Khong, H.; Chen, T.; et al. Genome-wide analysis identifies NR4A1 as a key mediator of T cell dysfunction. Nature 2019, 567, 525–529. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Cheng, S.; Luo, N.; Gao, R.; Yu, K.; Kang, B.; Wang, L.; Zhang, Q.; Fang, Q.; Zhang, L.; et al. Distinct epigenetic features of tumor-reactive CD8+ T cells in colorectal cancer patients revealed by genome-wide DNA methylation analysis. Genome Biol. 2019, 21, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; López-Moyado, I.F.; Seo, H.; Lio, C.-W.J.; Hempleman, L.J.; Sekiya, T.; Yoshimura, A.; Scott-Browne, J.P.; Rao, A. NR4A transcription factors limit CAR T cell function in solid tumours. Nature 2019, 567, 530–534. [Google Scholar] [CrossRef]
- Odagiu, L.; May, J.; Boulet, S.; Baldwin, T.A.; Labrecque, N. Role of the Orphan Nuclear Receptor NR4A Family in T-Cell Biology. Front. Endocrinol. 2020, 11, 624122. [Google Scholar] [CrossRef]
- Sen, D.R.; Kaminski, J.; Barnitz, R.A.; Kurachi, M.; Gerdemann, U.; Yates, K.B.; Tsao, H.W.; Godec, J.; LaFleur, M.W.; Brown, F.D.; et al. The epigenetic landscape of T cell exhaustion. Science 2016, 354, 1165–1169. [Google Scholar] [CrossRef] [Green Version]
- Scott, A.C.; Dündar, F.; Zumbo, P.; Chandran, S.S.; Klebanoff, C.A.; Shakiba, M.; Trivedi, P.; Menocal, L.; Appleby, H.; Camara, S.; et al. TOX is a critical regulator of tumour-specific T cell differentiation. Nature 2019, 571, 270–274. [Google Scholar] [CrossRef]
- Bordon, Y. TOX for tired T cells. Nat. Rev. Immunol. 2019, 19, 476. [Google Scholar] [CrossRef]
- Khan, O.; Giles, J.R.; McDonald, S.; Manne, S.; Ngiow, S.F.; Patel, K.P.; Werner, M.T.; Huang, A.C.; Alexander, K.A.; Wu, J.E.; et al. TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature 2019, 571, 211–218. [Google Scholar] [CrossRef]
- Blank, C.U.; Haining, W.N.; Held, W.; Hogan, P.G.; Kallies, A.; Lugli, E.; Lynn, R.C.; Philip, M.; Rao, A.; Restifo, N.P.; et al. Defining ‘T cell exhaustion’. Nat. Rev. Immunol. 2019, 19, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.; Chen, J.; Gonzalez-Avalos, E.; Samaniego-Castruita, D.; Das, A.; Wang, Y.H.; Lopez-Moyado, I.F.; Georges, R.O.; Zhang, W.; Onodera, A.; et al. TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8(+) T cell exhaustion. Proc. Natl. Acad. Sci. USA 2019, 116, 12410–12415. [Google Scholar] [CrossRef] [Green Version]
- Szabo, S.J.; Kim, S.T.; Costa, G.L.; Zhang, X.; Fathman, C.G.; Glimcher, L.H. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 2000, 100, 655–669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gacerez, A.T.; Sentman, C.L. T-bet promotes potent antitumor activity of CD4(+) CAR T cells. Cancer Gene Ther. 2018, 25, 117–128. [Google Scholar] [CrossRef] [Green Version]
- Tosolini, M.; Kirilovsky, A.; Mlecnik, B.; Fredriksen, T.; Mauger, S.; Bindea, G.; Berger, A.; Bruneval, P.; Fridman, W.H.; Pages, F.; et al. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res. 2011, 71, 1263–1271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haabeth, O.A.; Lorvik, K.B.; Hammarstrom, C.; Donaldson, I.M.; Haraldsen, G.; Bogen, B.; Corthay, A. Inflammation driven by tumour-specific Th1 cells protects against B-cell cancer. Nat. Commun. 2011, 2, 240. [Google Scholar] [CrossRef] [Green Version]
- Kallies, A.; Good-Jacobson, K.L. Transcription Factor T-bet Orchestrates Lineage Development and Function in the Immune System. Trends Immunol. 2017, 38, 287–297. [Google Scholar] [CrossRef]
- Li, J.; Shayan, G.; Avery, L.; Jie, H.-B.; Gildener-Leapman, N.; Schmitt, N.; Lu, B.F.; Kane, L.P.; Ferris, R.L. Tumor-infiltrating Tim-3+ T cells proliferate avidly except when PD-1 is co-expressed: Evidence for intracellular cross talk. OncoImmunology 2016, 5, e1200778. [Google Scholar] [CrossRef] [Green Version]
- Kao, C.; Oestreich, K.J.; Paley, M.A.; Crawford, A.; Angelosanto, J.M.; Ali, M.A.; Intlekofer, A.M.; Boss, J.M.; Reiner, S.L.; Weinmann, A.S.; et al. Transcription factor T-bet represses expression of the inhibitory receptor PD-1 and sustains virus-specific CD8+ T cell responses during chronic infection. Nat. Immunol. 2011, 12, 663–671. [Google Scholar] [CrossRef] [Green Version]
- Wherry, E.J.; Ha, S.-J.; Kaech, S.M.; Haining, W.N.; Sarkar, S.; Kalia, V.; Subramaniam, S.; Blattman, J.N.; Barber, D.L.; Ahmed, R. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity 2007, 27, 670–684. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; He, Y.; Hao, J.; Ni, L.; Dong, C. High Levels of Eomes Promote Exhaustion of Anti-tumor CD8(+) T Cells. Front. Immunol. 2018, 9, 2981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Man, K.; Gabriel, S.S.; Liao, Y.; Gloury, R.; Preston, S.; Henstridge, D.C.; Pellegrini, M.; Zehn, D.; Berberich-Siebelt, F.; Febbraio, M.A.; et al. Transcription Factor IRF4 Promotes CD8(+) T Cell Exhaustion and Limits the Development of Memory-like T Cells during Chronic Infection. Immunity 2017, 47, 1129–1141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, H.; Gonzalez-Avalos, E.; Zhang, W.; Ramchandani, P.; Yang, C.; Lio, C.J.; Rao, A.; Hogan, P.G. BATF and IRF4 cooperate to counter exhaustion in tumor-infiltrating CAR T cells. Nat. Immunol. 2021, 22, 983–995. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Yue, T.-T.; Yang, C.-L.; Wang, F.-X.; Luo, J.-H.; Rong, S.-J.; Zhang, M.; Guo, Y.; Xiong, F.; Wang, C.-Y. The MAPK dual specific phosphatase (DUSP) proteins: A versatile wrestler in T cell functionality. Int. Immunopharmacol. 2021, 98, 107906. [Google Scholar] [CrossRef]
- Lu, D.; Liu, L.; Sun, Y.Z.; Song, J.; Yin, Q.; Zhang, G.Z.; Qi, F.; Hu, Z.X.; Yang, Z.L.; Zhou, Z.; et al. The phosphatase PAC1 acts as a T cell suppressor and attenuates host antitumor immunity. Nat. Immunol. 2020, 21, 287–297. [Google Scholar] [CrossRef]
- Lissbrant, I.F.; Stattin, P.; Wikstrom, P.; Damber, J.E.; Egevad, L.; Bergh, A. Tumor associated macrophages in human prostate cancer: Relation to clinicopathological variables and survival. Int. J. Oncol. 2000, 17, 445–451. [Google Scholar] [CrossRef]
- Wiede, F.; Lu, K.H.; Du, X.; Liang, S.; Hochheiser, K.; Dodd, G.T.; Goh, P.K.; Kearney, C.; Meyran, D.; Beavis, P.A.; et al. PTPN2 phosphatase deletion in T cells promotes anti-tumour immunity and CAR T-cell efficacy in solid tumours. EMBO J. 2020, 39, e103637. [Google Scholar] [CrossRef]
- Ten Hoeve, J.; de Jesus Ibarra-Sanchez, M.; Fu, Y.; Zhu, W.; Tremblay, M.; David, M.; Shuai, K. Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol. Cell. Biol. 2002, 22, 5662–5668. [Google Scholar] [CrossRef] [Green Version]
- Simoncic, P.D.; Lee-Loy, A.; Barber, D.L.; Tremblay, M.L.; McGlade, C.J. The T cell protein tyrosine phosphatase is a negative regulator of janus family kinases 1 and 3. Curr. Biol. 2002, 12, 446–453. [Google Scholar] [CrossRef] [Green Version]
- Wiede, F.; Dudakov, J.A.; Lu, K.H.; Dodd, G.T.; Butt, T.; Godfrey, D.I.; Strasser, A.; Boyd, R.L.; Tiganis, T. PTPN2 regulates T cell lineage commitment and alphabeta versus gammadelta specification. J. Exp. Med. 2017, 214, 2733–2758. [Google Scholar] [CrossRef]
- LaFleur, M.W.; Nguyen, T.H.; Coxe, M.A.; Miller, B.C.; Yates, K.B.; Gillis, J.E.; Sen, D.R.; Gaudiano, E.F.; Al Abosy, R.; Freeman, G.J.; et al. PTPN2 regulates the generation of exhausted CD8(+) T cell subpopulations and restrains tumor immunity. Nat. Immunol. 2019, 20, 1335–1347. [Google Scholar] [CrossRef] [PubMed]
- Van Vliet, C.; Bukczynska, P.E.; Puryer, M.A.; Sadek, C.M.; Shields, B.J.; Tremblay, M.L.; Tiganis, T. Selective regulation of tumor necrosis factor-induced Erk signaling by Src family kinases and the T cell protein tyrosine phosphatase. Nat. Immunol. 2005, 6, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Sakata-Yanagimoto, M.; Enami, T.; Yoshida, K.; Shiraishi, Y.; Ishii, R.; Miyake, Y.; Muto, H.; Tsuyama, N.; Sato-Otsubo, A.; Okuno, Y.; et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat. Genet. 2014, 46, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Odejide, O.; Weigert, O.; Lane, A.A.; Toscano, D.; Lunning, M.A.; Kopp, N.; Kim, S.; van Bodegom, D.; Bolla, S.; Schatz, J.H.; et al. A targeted mutational landscape of angioimmunoblastic T-cell lymphoma. Blood 2014, 123, 1293–1296. [Google Scholar] [CrossRef]
- Feng, Y.; Li, X.; Cassady, K.; Zou, Z.; Zhang, X. TET2 Function in Hematopoietic Malignancies, Immune Regulation, and DNA Repair. Front. Oncol. 2019, 9, 210. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; van der Leun, A.; Yofe, I.; Lubling, Y.; Solodkin, D.G.; van Akkooi, A.; van den Braber, M.; Rozeman, L.; Haanen, J.; Blank, C.; et al. Dysfunctional CD8+ T cells form a proliferative, dynamically regulated compartment within human melanoma. Cell 2019, 176, 775–789. [Google Scholar] [CrossRef]
- Shin, H.; Blackburn, S.D.; Intlekofer, A.M.; Kao, C.; Angelosanto, J.M.; Reiner, S.L.; Wherry, E.J. A role for the transcriptional repressor Blimp-1 in CD8(+) T cell exhaustion during chronic viral infection. Immunity 2009, 31, 309–320. [Google Scholar] [CrossRef] [Green Version]
- Frenzel, A.; Grespi, F.; Chmelewskij, W.; Villunger, A. Bcl2 family proteins in carcinogenesis and the treatment of cancer. Apoptosis 2009, 14, 584–596. [Google Scholar] [CrossRef] [Green Version]
- Ren, R. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat. Rev. Cancer 2005, 5, 172–183. [Google Scholar] [CrossRef]
- Druker, B.J.; Tamura, S.; Buchdunger, E.; Ohno, S.; Segal, G.M.; Fanning, S.; Zimmermann, J.; Lydon, N.B. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat. Med. 1996, 2, 561–566. [Google Scholar] [CrossRef]
- Wan, P.T.; Garnett, M.J.; Roe, S.M.; Lee, S.; Niculescu-Duvaz, D.; Good, V.M.; Jones, C.M.; Marshall, C.J.; Springer, C.J.; Barford, D.; et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 2004, 116, 855–867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Leary, B.; Finn, R.S.; Turner, N.C. Treating cancer with selective CDK4/6 inhibitors. Nat. Rev. Clin. Oncol. 2016, 13, 417–430. [Google Scholar] [CrossRef]
- Vliagoftis, H.; Worobec, A.S.; Metcalfe, D.D. The protooncogene c-kit and c-kit ligand in human disease. J. Allergy Clin. Immunol. 1997, 100, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, R.I.; Gee, J.M.; Harper, M.E. EGFR and cancer prognosis. Eur. J. Cancer 2001, 37 (Suppl. 4), S9–S15. [Google Scholar] [CrossRef] [PubMed]
- Doherty, L.; Gigas, D.C.; Kesari, S.; Drappatz, J.; Kim, R.; Zimmerman, J.; Ostrowsky, L.; Wen, P.Y. Pilot study of the combination of EGFR and mTOR inhibitors in recurrent malignant gliomas. Neurology 2006, 67, 156–158. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.M.; Dominguez, G.; Gonzalez-Sancho, J.M.; Garcia, J.M.; Silva, J.; Garcia-Andrade, C.; Navarro, A.; Munoz, A.; Bonilla, F. Expression of thyroid hormone receptor/erbA genes is altered in human breast cancer. Oncogene 2002, 21, 4307–4316. [Google Scholar] [CrossRef] [Green Version]
- Amano, R.; Namekata, M.; Horiuchi, M.; Saso, M.; Yanagisawa, T.; Tanaka, Y.; Ghani, F.I.; Yamamoto, M.; Sakamoto, T. Specific inhibition of FGF5-induced cell proliferation by RNA aptamers. Sci. Rep. 2021, 11, 2976. [Google Scholar] [CrossRef]
- Hanada, K.; Yewdell, J.W.; Yang, J.C. Immune recognition of a human renal cancer antigen through post-translational protein splicing. Nature 2004, 427, 252–256. [Google Scholar] [CrossRef]
- El-Gamal, M.I.; Anbar, H.S.; Yoo, K.H.; Oh, C.H. FMS Kinase Inhibitors: Current Status and Future Prospects. Med. Res. Rev. 2013, 33, 599–636. [Google Scholar] [CrossRef]
- Goustin, A.S.; Leof, E.B.; Shipley, G.D.; Moses, H.L. Growth factors and cancer. Cancer Res. 1986, 46, 1015–1029. [Google Scholar]
- Ruiz i Altaba, A. Gli proteins and Hedgehog signaling: Development and cancer. Trends Genet. 1999, 15, 418–425. [Google Scholar] [CrossRef] [PubMed]
- Yarden, Y. Biology of HER2 and its importance in breast cancer. Oncology 2001, 61 (Suppl. 2), 1–13. [Google Scholar] [CrossRef]
- Nishihira, T.; Hashimoto, Y.; Katayama, M.; Mori, S.; Kuroki, T. Molecular and cellular features of esophageal cancer cells. J. Cancer Res. Clin. Oncol. 1993, 119, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Nateri, A.S.; Spencer-Dene, B.; Behrens, A. Interaction of phosphorylated c-Jun with TCF4 regulates intestinal cancer development. Nature 2005, 437, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Wade, M.; Li, Y.C.; Wahl, G.M. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat. Rev. Cancer 2013, 13, 83–96. [Google Scholar] [CrossRef] [Green Version]
- Natali, P.G.; Prat, M.; Nicotra, M.R.; Bigotti, A.; Olivero, M.; Comoglio, P.M.; Di Renzo, M.F. Overexpression of the met/HGF receptor in renal cell carcinomas. Int. J. Cancer 1996, 69, 212–217. [Google Scholar] [CrossRef]
- Dang, C.V. MYC on the path to cancer. Cell 2012, 149, 22–35. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Balmain, A.; Counter, C.M. A model for RAS mutation patterns in cancers: Finding the sweet spot. Nat. Rev. Cancer 2018, 18, 767–777. [Google Scholar] [CrossRef]
- Yunis, J.J.; Boot, A.J.; Mayer, M.G.; Bos, J.L. Mechanisms of ras mutation in myelodysplastic syndrome. Oncogene 1989, 4, 609–614. [Google Scholar]
- Riely, G.J.; Marks, J.; Pao, W. KRAS mutations in non-small cell lung cancer. Proc. Am. Thorac. Soc. 2009, 6, 201–205. [Google Scholar] [CrossRef]
- Locker, G.Y.; Hamilton, S.; Harris, J.; Jessup, J.M.; Kemeny, N.; Macdonald, J.S.; Somerfield, M.R.; Hayes, D.F.; Bast, R.C., Jr.; Asco. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J. Clin. Oncol. 2006, 24, 5313–5327. [Google Scholar] [CrossRef] [PubMed]
- Arighi, E.; Borrello, M.G.; Sariola, H. RET tyrosine kinase signaling in development and cancer. Cytokine Growth Factor Rev. 2005, 16, 441–467. [Google Scholar] [CrossRef] [PubMed]
- Johnsson, A.; Heldin, C.H.; Wasteson, A.; Westermark, B.; Deuel, T.F.; Huang, J.S.; Seeburg, P.H.; Gray, A.; Ullrich, A.; Scrace, G.; et al. The c-sis gene encodes a precursor of the B chain of platelet-derived growth factor. EMBO J. 1984, 3, 921–928. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.; Kharbanda, S.; Kufe, D. Association of the DF3/MUC1 breast cancer antigen with Grb2 and the Sos/Ras exchange protein. Cancer Res. 1995, 55, 4000–4003. [Google Scholar] [PubMed]
- Wheeler, D.L.; Iida, M.; Dunn, E.F. The role of Src in solid tumors. Oncologist 2009, 14, 667–678. [Google Scholar] [CrossRef]
- Galway, M.E.; Masucci, J.D.; Lloyd, A.M.; Walbot, V.; Davis, R.W.; Schiefelbein, J.W. The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev. Biol. 1994, 166, 740–754. [Google Scholar] [CrossRef]
- Ferrara, N.; Gerber, H.P.; LeCouter, J. The biology of VEGF and its receptors. Nat. Med. 2003, 9, 669–676. [Google Scholar] [CrossRef]
- LeCouter, J.; Moritz, D.R.; Li, B.; Phillips, G.L.; Liang, X.H.; Gerber, H.P.; Hillan, K.J.; Ferrara, N. Angiogenesis-independent endothelial protection of liver: Role of VEGFR-1. Science 2003, 299, 890–893. [Google Scholar] [CrossRef]
- Sitohy, B.; Nagy, J.A.; Dvorak, H.F. Anti-VEGF/VEGFR therapy for cancer: Reassessing the target. Cancer Res. 2012, 72, 1909–1914. [Google Scholar] [CrossRef] [Green Version]
- Cooper, G.M. The Cell: A Molecular Approach, 2nd ed.; Sinauer Associates Inc.: Sunderland, MA, USA, 2000; p. 689. [Google Scholar]
- Wang, L.H.; Wu, C.F.; Rajasekaran, N.; Shin, Y.K. Loss of Tumor Suppressor Gene Function in Human Cancer: An Overview. Cell. Physiol. Biochem. 2018, 51, 2647–2693. [Google Scholar] [CrossRef]
- Nik-Zainal, S.; Alexandrov, L.B.; Wedge, D.C.; Van Loo, P.; Greenman, C.D.; Raine, K.; Jones, D.; Hinton, J.; Marshall, J.; Stebbings, L.A.; et al. Mutational processes molding the genomes of 21 breast cancers. Cell 2012, 149, 979–993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martincorena, I.; Campbell, P.J. Somatic mutation in cancer and normal cells. Science 2015, 349, 1483–1489. [Google Scholar] [CrossRef]
- Jayaprakash, P.; Ai, M.; Liu, A.; Budhani, P.; Bartkowiak, T.; Sheng, J.; Ager, C.; Nicholas, C.; Jaiswal, A.R.; Sun, Y.; et al. Targeted hypoxia reduction restores T cell infiltration and sensitizes prostate cancer to immunotherapy. J. Clin. Investig. 2018, 128, 5137–5149. [Google Scholar] [CrossRef] [PubMed]
- Gerald, W.L.; Gramling, T.S.; Sens, D.A.; Garvin, A.J. Expression of the 11p13 Wilms’ tumor gene, WT1, correlates with histologic category of Wilms’ tumor. Am. J. Pathol. 1992, 140, 1031–1037. [Google Scholar]
- De Carvalho, D.D.; You, J.S.; Jones, P.A. DNA methylation and cellular reprogramming. Trends Cell Biol. 2010, 20, 609–617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanai, Y.; Ushijima, S.; Nakanishi, Y.; Sakamoto, M.; Hirohashi, S. Mutation of the DNA methyltransferase (DNMT) 1 gene in human colorectal cancers. Cancer Lett. 2003, 192, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Strawn, E.; Basir, Z.; Halverson, G.; Guo, S.W. Aberrant expression of deoxyribonucleic acid methyltransferases DNMT1, DNMT3A, and DNMT3B in women with endometriosis. Fertil. Steril. 2007, 87, 24–32. [Google Scholar] [CrossRef]
- Ley, T.J.; Ding, L.; Walter, M.J.; McLellan, M.D.; Lamprecht, T.; Larson, D.E.; Kandoth, C.; Payton, J.E.; Baty, J.; Welch, J.; et al. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 2010, 363, 2424–2433. [Google Scholar] [CrossRef] [Green Version]
- Yamashita, Y.; Yuan, J.; Suetake, I.; Suzuki, H.; Ishikawa, Y.; Choi, Y.L.; Ueno, T.; Soda, M.; Hamada, T.; Haruta, H.; et al. Array-based genomic resequencing of human leukemia. Oncogene 2010, 29, 3723–3731. [Google Scholar] [CrossRef] [Green Version]
- Yan, X.J.; Xu, J.; Gu, Z.H.; Pan, C.M.; Lu, G.; Shen, Y.; Shi, J.Y.; Zhu, Y.M.; Tang, L.; Zhang, X.W.; et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat. Genet. 2011, 43, 309–315. [Google Scholar] [CrossRef]
- Wijmenga, C.; Hansen, R.S.; Gimelli, G.; Bjorck, E.J.; Davies, E.G.; Valentine, D.; Belohradsky, B.H.; van Dongen, J.J.; Smeets, D.F.; van den Heuvel, L.P.; et al. Genetic variation in ICF syndrome: Evidence for genetic heterogeneity. Hum. Mutat. 2000, 16, 509–517. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.M.; Ohno, K.; Fukudome, T.; Tsujino, A.; Brengman, J.M.; De Vivo, D.C.; Packer, R.J.; Engel, A.G. Congenital myasthenic syndrome caused by low-expressor fast-channel AChR delta subunit mutation. Neurology 2002, 59, 1881–1888. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, M.E.; Abdel-Wahab, O.; Lu, C.; Ward, P.S.; Patel, J.; Shih, A.; Li, Y.; Bhagwat, N.; Vasanthakumar, A.; Fernandez, H.F.; et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 2010, 18, 553–567. [Google Scholar] [CrossRef] [Green Version]
- Lu, C.; Ward, P.S.; Kapoor, G.S.; Rohle, D.; Turcan, S.; Abdel-Wahab, O.; Edwards, C.R.; Khanin, R.; Figueroa, M.E.; Melnick, A.; et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 2012, 483, 474–478. [Google Scholar] [CrossRef] [Green Version]
- Turcan, S.; Rohle, D.; Goenka, A.; Walsh, L.A.; Fang, F.; Yilmaz, E.; Campos, C.; Fabius, A.W.; Lu, C.; Ward, P.S.; et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 2012, 483, 479–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sansom, O.J.; Maddison, K.; Clarke, A.R. Mechanisms of disease: Methyl-binding domain proteins as potential therapeutic targets in cancer. Nat. Clin. Pract. Oncol. 2007, 4, 305–315. [Google Scholar] [CrossRef]
- Gui, Y.; Guo, G.; Huang, Y.; Hu, X.; Tang, A.; Gao, S.; Wu, R.; Chen, C.; Li, X.; Zhou, L.; et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat. Genet. 2011, 43, 875–878. [Google Scholar] [CrossRef]
- Morin, R.D.; Mendez-Lago, M.; Mungall, A.J.; Goya, R.; Mungall, K.L.; Corbett, R.D.; Johnson, N.A.; Severson, T.M.; Chiu, R.; Field, M.; et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 2011, 476, 298–303. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.; Li, J.; Song, L. Bmi-1, stem cells and cancer. Acta Biochim. Biophys. Sin. 2009, 41, 527–534. [Google Scholar] [CrossRef] [Green Version]
- Lukacs, R.U.; Memarzadeh, S.; Wu, H.; Witte, O.N. Bmi-1 is a crucial regulator of prostate stem cell self-renewal and malignant transformation. Cell Stem Cell 2010, 7, 682–693. [Google Scholar] [CrossRef] [Green Version]
- Miremadi, A.; Oestergaard, M.Z.; Pharoah, P.D.; Caldas, C. Cancer genetics of epigenetic genes. Hum. Mol. Genet. 2007, 16, R28–R49. [Google Scholar] [CrossRef] [PubMed]
- Chase, A.; Cross, N.C. Aberrations of EZH2 in cancer. Clin. Cancer Res. 2011, 17, 2613–2618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsang, D.P.; Cheng, A.S. Epigenetic regulation of signaling pathways in cancer: Role of the histone methyltransferase EZH2. J. Gastroenterol. Hepatol. 2011, 26, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Varier, R.A.; Timmers, H.T. Histone lysine methylation and demethylation pathways in cancer. Biochim. Biophys. Acta 2011, 1815, 75–89. [Google Scholar] [CrossRef]
- Ropero, S.; Fraga, M.F.; Ballestar, E.; Hamelin, R.; Yamamoto, H.; Boix-Chornet, M.; Caballero, R.; Alaminos, M.; Setien, F.; Paz, M.F.; et al. A truncating mutation of HDAC2 in human cancers confers resistance to histone deacetylase inhibition. Nat. Genet. 2006, 38, 566–569. [Google Scholar] [CrossRef]
- Rotili, D.; Mai, A. Targeting Histone Demethylases: A New Avenue for the Fight against Cancer. Genes Cancer 2011, 2, 663–679. [Google Scholar] [CrossRef]
- Jones, S.; Wang, T.L.; Shih Ie, M.; Mao, T.L.; Nakayama, K.; Roden, R.; Glas, R.; Slamon, D.; Diaz, L.A., Jr.; Vogelstein, B.; et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 2010, 330, 228–231. [Google Scholar] [CrossRef] [Green Version]
- Guan, B.; Mao, T.L.; Panuganti, P.K.; Kuhn, E.; Kurman, R.J.; Maeda, D.; Chen, E.; Jeng, Y.M.; Wang, T.L.; Shih Ie, M. Mutation and loss of expression of ARID1A in uterine low-grade endometrioid carcinoma. Am. J. Surg. Pathol. 2011, 35, 625–632. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Zhao, H.; Zhang, X.; Wood, L.D.; Anders, R.A.; Choti, M.A.; Pawlik, T.M.; Daniel, H.D.; Kannangai, R.; Offerhaus, G.J.; et al. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat. Genet. 2011, 43, 828–829. [Google Scholar] [CrossRef] [Green Version]
- Drost, J.; Mantovani, F.; Tocco, F.; Elkon, R.; Comel, A.; Holstege, H.; Kerkhoven, R.; Jonkers, J.; Voorhoeve, P.M.; Agami, R.; et al. BRD7 is a candidate tumour suppressor gene required for p53 function. Nat. Cell Biol. 2010, 12, 380–389. [Google Scholar] [CrossRef]
- Sun, A.; Tawfik, O.; Gayed, B.; Thrasher, J.B.; Hoestje, S.; Li, C.; Li, B. Aberrant expression of SWI/SNF catalytic subunits BRG1/BRM is associated with tumor development and increased invasiveness in prostate cancers. Prostate 2007, 67, 203–213. [Google Scholar] [CrossRef] [PubMed]
- De Zwaan, S.E.; Haass, N.K. Genetics of basal cell carcinoma. Australas. J. Dermatol. 2010, 51, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Bagchi, A.; Papazoglu, C.; Wu, Y.; Capurso, D.; Brodt, M.; Francis, D.; Bredel, M.; Vogel, H.; Mills, A.A. CHD5 is a tumor suppressor at human 1p36. Cell 2007, 128, 459–475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.S.; Chung, N.G.; Kang, M.R.; Yoo, N.J.; Lee, S.H. Genetic and expressional alterations of CHD genes in gastric and colorectal cancers. Histopathology 2011, 58, 660–668. [Google Scholar] [CrossRef]
- Wang, J.; Chen, H.; Fu, S.; Xu, Z.M.; Sun, K.L.; Fu, W.N. The involvement of CHD5 hypermethylation in laryngeal squamous cell carcinoma. Oral. Oncol. 2011, 47, 601–608. [Google Scholar] [CrossRef]
- Wessels, K.; Bohnhorst, B.; Luhmer, I.; Morlot, S.; Bohring, A.; Jonasson, J.; Epplen, J.T.; Gadzicki, D.; Glaser, S.; Gohring, G.; et al. Novel CHD7 mutations contributing to the mutation spectrum in patients with CHARGE syndrome. Eur. J. Med. Genet. 2010, 53, 280–285. [Google Scholar] [CrossRef]
- Mattera, L.; Escaffit, F.; Pillaire, M.J.; Selves, J.; Tyteca, S.; Hoffmann, J.S.; Gourraud, P.A.; Chevillard-Briet, M.; Cazaux, C.; Trouche, D. The p400/Tip60 ratio is critical for colorectal cancer cell proliferation through DNA damage response pathways. Oncogene 2009, 28, 1506–1517. [Google Scholar] [CrossRef] [Green Version]
- Varela, I.; Tarpey, P.; Raine, K.; Huang, D.; Ong, C.K.; Stephens, P.; Davies, H.; Jones, D.; Lin, M.L.; Teague, J.; et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 2011, 469, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Wilson, B.G.; Roberts, C.W. SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer 2011, 11, 481–492. [Google Scholar] [CrossRef]
- Balakrishnan, A.; Bleeker, F.E.; Lamba, S.; Rodolfo, M.; Daniotti, M.; Scarpa, A.; van Tilborg, A.A.; Leenstra, S.; Zanon, C.; Bardelli, A. Novel somatic and germline mutations in cancer candidate genes in glioblastoma, melanoma, and pancreatic carcinoma. Cancer Res. 2007, 67, 3545–3550. [Google Scholar] [CrossRef] [Green Version]
- Morrison, D.K. MAP kinase pathways. Cold Spring Harb. Perspect. Biol. 2012, 4, a011254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hemmings, B.A.; Restuccia, D.F. PI3K-PKB/Akt pathway. Cold Spring Harb. Perspect. Biol. 2012, 4, a011189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogelstein, B.; Kinzler, K.W. Cancer genes and the pathways they control. Nat. Med. 2004, 10, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Vega, F.; Mina, M.; Armenia, J.; Chatila, W.K.; Luna, A.; La, K.C.; Dimitriadoy, S.; Liu, D.L.; Kantheti, H.S.; Saghafinia, S.; et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018, 173, 321–337. [Google Scholar] [CrossRef] [Green Version]
- Ciriello, G.; Miller, M.L.; Aksoy, B.A.; Senbabaoglu, Y.; Schultz, N.; Sander, C. Emerging landscape of oncogenic signatures across human cancers. Nat. Genet. 2013, 45, 1127–1133. [Google Scholar] [CrossRef] [Green Version]
- Hoadley, K.A.; Yau, C.; Hinoue, T.; Wolf, D.M.; Lazar, A.J.; Drill, E.; Shen, R.; Taylor, A.M.; Cherniack, A.D.; Thorsson, V.; et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell 2018, 173, 291–304. [Google Scholar] [CrossRef] [Green Version]
- Hoadley, K.A.; Yau, C.; Wolf, D.M.; Cherniack, A.D.; Tamborero, D.; Ng, S.; Leiserson, M.D.M.; Niu, B.; McLellan, M.D.; Uzunangelov, V.; et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 2014, 158, 929–944. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Yang, J. Functional mechanisms for human tumor suppressors. J. Cancer 2010, 1, 136–140. [Google Scholar] [CrossRef]
- Shortt, J.; Johnstone, R.W. Oncogenes in cell survival and cell death. Cold Spring Harb. Perspect. Biol. 2012, 4, a009829. [Google Scholar] [CrossRef] [Green Version]
- Prior, I.A.; Lewis, P.D.; Mattos, C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012, 72, 2457–2467. [Google Scholar] [CrossRef] [Green Version]
- Rodenhuis, S. Ras and human tumors. Semin. Cancer Biol. 1992, 3, 241–247. [Google Scholar] [PubMed]
- Brison, O. Gene amplification and tumor progression. Biochim. Biophys. Acta 1993, 1155, 25–41. [Google Scholar] [CrossRef] [PubMed]
- Schwab, M.; Alitalo, K.; Klempnauer, K.H.; Varmus, H.E.; Bishop, J.M.; Gilbert, F.; Brodeur, G.; Goldstein, M.; Trent, J. Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature 1983, 305, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Kruidering, M. N-MYC overexpression and neuroblastomas. Trends Pharmacol. Sci. 2002, 23, 452. [Google Scholar] [CrossRef] [PubMed]
- Zajac-Kaye, M. Myc oncogene: A key component in cell cycle regulation and its implication for lung cancer. Lung Cancer 2001, 34 (Suppl. 2), S43–S46. [Google Scholar] [CrossRef]
- Lane, D.P. p53, guardian of the genome. Nature 1992, 358, 15–16. [Google Scholar] [CrossRef]
- Olivier, M.; Hollstein, M.; Hainaut, P. TP53 mutations in human cancers: Origins, consequences, and clinical use. Cold Spring Harb. Perspect. Biol. 2010, 2, a001008. [Google Scholar] [CrossRef] [Green Version]
- Li, F.P.; Fraumeni, J.F., Jr.; Mulvihill, J.J.; Blattner, W.A.; Dreyfus, M.G.; Tucker, M.A.; Miller, R.W. A cancer family syndrome in twenty-four kindreds. Cancer Res. 1988, 48, 5358–5362. [Google Scholar]
- Malkin, D.; Li, F.P.; Strong, L.C.; Fraumeni, J.F., Jr.; Nelson, C.E.; Kim, D.H.; Kassel, J.; Gryka, M.A.; Bischoff, F.Z.; Tainsky, M.A.; et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science 1990, 250, 1233–1238. [Google Scholar] [CrossRef]
- You, J.S.; Jones, P.A. Cancer genetics and epigenetics: Two sides of the same coin? Cancer Cell 2012, 22, 9–20. [Google Scholar] [CrossRef] [Green Version]
- Baylin, S.B.; Jones, P.A. Epigenetic Determinants of Cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, a019505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatziapostolou, M.; Iliopoulos, D. Epigenetic aberrations during oncogenesis. Cell. Mol. Life Sci. 2011, 68, 1681–1702. [Google Scholar] [CrossRef] [PubMed]
- Baylin, S.B.; Jones, P.A. A decade of exploring the cancer epigenome—Biological and translational implications. Nat. Rev. Cancer 2011, 11, 726–734. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007, 8, 286–298. [Google Scholar] [CrossRef] [PubMed]
- Krivtsov, A.V.; Armstrong, S.A. MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer 2007, 7, 823–833. [Google Scholar] [CrossRef]
- Hawkins, R.D.; Hon, G.C.; Yang, C.; Antosiewicz-Bourget, J.E.; Lee, L.K.; Ngo, Q.M.; Klugman, S.; Ching, K.A.; Edsall, L.E.; Ye, Z.; et al. Dynamic chromatin states in human ES cells reveal potential regulatory sequences and genes involved in pluripotency. Cell Res. 2011, 21, 1393–1409. [Google Scholar] [CrossRef] [Green Version]
- Hon, G.C.; Hawkins, R.D.; Ren, B. Predictive chromatin signatures in the mammalian genome. Hum. Mol. Genet. 2009, 18, R195–R201. [Google Scholar] [CrossRef] [Green Version]
- Mills, A.A. Throwing the cancer switch: Reciprocal roles of polycomb and trithorax proteins. Nat. Rev. Cancer 2010, 10, 669–682. [Google Scholar] [CrossRef]
- Lin, H.; Wong, R.P.; Martinka, M.; Li, G. Loss of SNF5 expression correlates with poor patient survival in melanoma. Clin. Cancer Res. 2009, 15, 6404–6411. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zhang, Y.; Yang, Y.; Niu, M.; Sun, S.; Ji, H.; Ma, Y.; Yao, G.; Jiang, Y.; Shan, M.; et al. Frequent low expression of chromatin remodeling gene ARID1A in breast cancer and its clinical significance. Cancer Epidemiol. 2012, 36, 288–293. [Google Scholar] [CrossRef]
- MacIver, N.J.; Michalek, R.D.; Rathmell, J.C. Metabolic regulation of T lymphocytes. Annu. Rev. Immunol. 2013, 31, 259–283. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J. Energy metabolism of cancer: Glycolysis versus oxidative phosphorylation (Review). Oncol. Lett. 2012, 4, 1151–1157. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.H.; Qiu, J.; O’Sullivan, D.; Buck, M.D.; Noguchi, T.; Curtis, J.D.; Chen, Q.; Gindin, M.; Gubin, M.M.; van der Windt, G.J.; et al. Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell 2015, 162, 1229–1241. [Google Scholar] [CrossRef] [Green Version]
- Sugiura, A.; Rathmell, J.C. Metabolic Barriers to T Cell Function in Tumors. J. Immunol. 2018, 200, 400–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ottensmeier, C.H.; Perry, K.L.; Harden, E.L.; Stasakova, J.; Jenei, V.; Fleming, J.; Wood, O.; Woo, J.; Woelk, C.H.; Thomas, G.J.; et al. Upregulated Glucose Metabolism Correlates Inversely with CD8+ T-cell Infiltration and Survival in Squamous Cell Carcinoma. Cancer Res. 2016, 76, 4136–4148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis-Yadley, A.H.; Abbott, A.M.; Pimiento, J.M.; Chen, D.T.; Malafa, M.P. Increased Expression of the Glucose Transporter Type 1 Gene Is Associated With Worse Overall Survival in Resected Pancreatic Adenocarcinoma. Pancreas 2016, 45, 974–979. [Google Scholar] [CrossRef] [Green Version]
- Ho, P.C.; Bihuniak, J.D.; Macintyre, A.N.; Staron, M.; Liu, X.; Amezquita, R.; Tsui, Y.C.; Cui, G.; Micevic, G.; Perales, J.C.; et al. Phosphoenolpyruvate Is a Metabolic Checkpoint of Anti-tumor T Cell Responses. Cell 2015, 162, 1217–1228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, C.H.; Curtis, J.D.; Maggi, L.B., Jr.; Faubert, B.; Villarino, A.V.; O’Sullivan, D.; Huang, S.C.; van der Windt, G.J.; Blagih, J.; Qiu, J.; et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 2013, 153, 1239–1251. [Google Scholar] [CrossRef] [Green Version]
- Macintyre, A.N.; Gerriets, V.A.; Nichols, A.G.; Michalek, R.D.; Rudolph, M.C.; Deoliveira, D.; Anderson, S.M.; Abel, E.D.; Chen, B.J.; Hale, L.P.; et al. The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metab. 2014, 20, 61–72. [Google Scholar] [CrossRef] [Green Version]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef] [Green Version]
- Pavlova, N.N.; Zhu, J.; Thompson, C.B. The hallmarks of cancer metabolism: Still emerging. Cell Metab. 2022, 34, 355–377. [Google Scholar] [CrossRef] [PubMed]
- Andrejeva, G.; Rathmell, J.C. Similarities and Distinctions of Cancer and Immune Metabolism in Inflammation and Tumors. Cell Metab. 2017, 26, 49–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Hardie, R.A.; Hoy, A.J.; van Geldermalsen, M.; Gao, D.; Fazli, L.; Sadowski, M.C.; Balaban, S.; Schreuder, M.; Nagarajah, R.; et al. Targeting ASCT2-mediated glutamine uptake blocks prostate cancer growth and tumour development. J. Pathol. 2015, 236, 278–289. [Google Scholar] [CrossRef] [Green Version]
- Mocellin, S.; Bronte, V.; Nitti, D. Nitric oxide, a double edged sword in cancer biology: Searching for therapeutic opportunities. Med. Res. Rev. 2007, 27, 317–352. [Google Scholar] [CrossRef]
- Grzywa, T.M.; Sosnowska, A.; Matryba, P.; Rydzynska, Z.; Jasinski, M.; Nowis, D.; Golab, J. Myeloid Cell-Derived Arginase in Cancer Immune Response. Front. Immunol. 2020, 11, 938. [Google Scholar] [CrossRef] [PubMed]
- Geiger, R.; Rieckmann, J.C.; Wolf, T.; Basso, C.; Feng, Y.; Fuhrer, T.; Kogadeeva, M.; Picotti, P.; Meissner, F.; Mann, M.; et al. L-Arginine Modulates T Cell Metabolism and Enhances Survival and Anti-tumor Activity. Cell 2016, 167, 829–842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brody, J.R.; Costantino, C.L.; Berger, A.C.; Sato, T.; Lisanti, M.P.; Yeo, C.J.; Emmons, R.V.; Witkiewicz, A.K. Expression of indoleamine 2,3-dioxygenase in metastatic malignant melanoma recruits regulatory T cells to avoid immune detection and affects survival. Cell Cycle 2009, 8, 1930–1934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Witkiewicz, A.K.; Costantino, C.L.; Metz, R.; Muller, A.J.; Prendergast, G.C.; Yeo, C.J.; Brody, J.R. Genotyping and expression analysis of IDO2 in human pancreatic cancer: A novel, active target. J. Am. Coll. Surg. 2009, 208, 781–787; discussion 787–789. [Google Scholar] [CrossRef] [Green Version]
- Inaba, T.; Ino, K.; Kajiyama, H.; Yamamoto, E.; Shibata, K.; Nawa, A.; Nagasaka, T.; Akimoto, H.; Takikawa, O.; Kikkawa, F. Role of the immunosuppressive enzyme indoleamine 2,3-dioxygenase in the progression of ovarian carcinoma. Gynecol. Oncol. 2009, 115, 185–192. [Google Scholar] [CrossRef]
- Fallarino, F.; Grohmann, U.; You, S.; McGrath, B.C.; Cavener, D.R.; Vacca, C.; Orabona, C.; Bianchi, R.; Belladonna, M.L.; Volpi, C.; et al. The combined effects of tryptophan starvation and tryptophan catabolites down-regulate T cell receptor zeta-chain and induce a regulatory phenotype in naive T cells. J. Immunol. 2006, 176, 6752–6761. [Google Scholar] [CrossRef] [Green Version]
- Petrova, V.; Annicchiarico-Petruzzelli, M.; Melino, G.; Amelio, I. The hypoxic tumour microenvironment. Oncogenesis 2018, 7, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben-Shoshan, J.; Maysel-Auslender, S.; Mor, A.; Keren, G.; George, J. Hypoxia controls CD4+CD25+ regulatory T-cell homeostasis via hypoxia-inducible factor-1alpha. Eur. J. Immunol. 2008, 38, 2412–2418. [Google Scholar] [CrossRef] [PubMed]
- Clambey, E.T.; McNamee, E.N.; Westrich, J.A.; Glover, L.E.; Campbell, E.L.; Jedlicka, P.; de Zoeten, E.F.; Cambier, J.C.; Stenmark, K.R.; Colgan, S.P.; et al. Hypoxia-inducible factor-1 alpha-dependent induction of FoxP3 drives regulatory T-cell abundance and function during inflammatory hypoxia of the mucosa. Proc. Natl. Acad. Sci. USA 2012, 109, E2784–E2793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohta, A.; Gorelik, E.; Prasad, S.J.; Ronchese, F.; Lukashev, D.; Wong, M.K.; Huang, X.; Caldwell, S.; Liu, K.; Smith, P.; et al. A2A adenosine receptor protects tumors from antitumor T cells. Proc. Natl. Acad. Sci. USA 2006, 103, 13132–13137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ford, B.R.; Vignali, P.D.A.; Rittenhouse, N.L.; Scharping, N.E.; Peralta, R.; Lontos, K.; Frisch, A.T.; Delgoffe, G.M.; Poholek, A.C. Tumor microenvironmental signals reshape chromatin landscapes to limit the functional potential of exhausted T cells. Sci. Immunol. 2022, 7, eabj9123. [Google Scholar] [CrossRef]
- Vignali, P.D.A.; DePeaux, K.; Watson, M.J.; Ye, C.; Ford, B.R.; Lontos, K.; McGaa, N.K.; Scharping, N.E.; Menk, A.V.; Robson, S.C.; et al. Hypoxia drives CD39-dependent suppressor function in exhausted T cells to limit antitumor immunity. Nat. Immunol. 2023, 24, 267–279. [Google Scholar] [CrossRef]
- Facciabene, A.; Peng, X.; Hagemann, I.S.; Balint, K.; Barchetti, A.; Wang, L.P.; Gimotty, P.A.; Gilks, C.B.; Lal, P.; Zhang, L.; et al. Tumour hypoxia promotes tolerance and angiogenesis via CCL28 and T(reg) cells. Nature 2011, 475, 226–230. [Google Scholar] [CrossRef]
- Noman, M.Z.; Desantis, G.; Janji, B.; Hasmim, M.; Karray, S.; Dessen, P.; Bronte, V.; Chouaib, S. PD-L1 is a novel direct target of HIF-1 alpha., and its blockade under hypoxia enhanced MDSC-mediated T cell activation. J. Exp. Med. 2014, 211, 781–790. [Google Scholar] [CrossRef] [Green Version]
- Barsoum, I.B.; Smallwood, C.A.; Siemens, D.R.; Graham, C.H. A mechanism of hypoxia-mediated escape from adaptive immunity in cancer cells. Cancer Res. 2014, 74, 665–674. [Google Scholar] [CrossRef] [Green Version]
- Conforti, L. Potassium channels of T lymphocytes take center stage in the fight against cancer. J. Immunother. Cancer 2017, 5, 2. [Google Scholar] [CrossRef] [Green Version]
- Eil, R.; Vodnala, S.K.; Clever, D.; Klebanoff, C.A.; Sukumar, M.; Pan, J.H.; Palmer, D.C.; Gros, A.; Yamamoto, T.N.; Patel, S.J.; et al. Ionic immune suppression within the tumour microenvironment limits T cell effector function. Nature 2016, 537, 539–543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baixauli, F.; Villa, M.; Pearce, E.L. Potassium shapes antitumor immunity. Science 2019, 363, 1395–1396. [Google Scholar] [CrossRef] [PubMed]
- Vodnala, S.K.; Eil, R.; Kishton, R.J.; Sukumar, M.; Yamamoto, T.N.; Ha, N.H.; Lee, P.H.; Shin, M.; Patel, S.J.; Yu, Z.; et al. T cell stemness and dysfunction in tumors are triggered by a common mechanism. Science 2019, 363, eaau0135. [Google Scholar] [CrossRef] [PubMed]
- Chimote, A.A.; Balajthy, A.; Arnold, M.J.; Newton, H.S.; Hajdu, P.; Qualtieri, J.; Wise-Draper, T.; Conforti, L. A defect in KCa3.1 channel activity limits the ability of CD8(+) T cells from cancer patients to infiltrate an adenosine-rich microenvironment. Sci. Signal 2018, 11, eaaq1616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terrie, E.; Coronas, V.; Constantin, B. Role of the calcium toolkit in cancer stem cells. Cell Calcium 2019, 80, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Immler, R.; Simon, S.I.; Sperandio, M. Calcium signalling and related ion channels in neutrophil recruitment and function. Eur. J. Clin. Investig. 2018, 48 (Suppl. 2), e12964. [Google Scholar] [CrossRef]
- Bagur, R.; Hajnoczky, G. Intracellular Ca(2+) Sensing: Its Role in Calcium Homeostasis and Signaling. Mol. Cell 2017, 66, 780–788. [Google Scholar] [CrossRef] [Green Version]
- Bauer, C.A.; Kim, E.Y.; Marangoni, F.; Carrizosa, E.; Claudio, N.M.; Mempel, T.R. Dynamic Treg interactions with intratumoral APCs promote local CTL dysfunction. J. Clin. Investig. 2014, 124, 2425–2440. [Google Scholar] [CrossRef]
- Marangoni, F.; Murooka, T.T.; Manzo, T.; Kim, E.Y.; Carrizosa, E.; Elpek, N.M.; Mempel, T.R. The transcription factor NFAT exhibits signal memory during serial T cell interactions with antigen-presenting cells. Immunity 2013, 38, 237–249. [Google Scholar] [CrossRef] [Green Version]
- Stewart, T.A.; Yapa, K.T.; Monteith, G.R. Altered calcium signaling in cancer cells. Biochim. Biophys. Acta 2015, 1848, 2502–2511. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.D.; Bae, S.; Capece, T.; Nedelkovska, H.; de Rubio, R.G.; Smrcka, A.V.; Jun, C.D.; Jung, W.; Park, B.; Kim, T.I.; et al. Targeted calcium influx boosts cytotoxic T lymphocyte function in the tumour microenvironment. Nat. Commun. 2017, 8, 15365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yarosz, E.L.; Chang, C.H. The Role of Reactive Oxygen Species in Regulating T Cell-mediated Immunity and Disease. Immune Netw. 2018, 18, e14. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Song, M.; Zhang, B.; Zhang, Y. Reactive Oxygen Species Regulate T Cell Immune Response in the Tumor Microenvironment. Oxid. Med. Cell. Longev. 2016, 2016, 1580967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franchina, D.G.; Dostert, C.; Brenner, D. Reactive Oxygen Species: Involvement in T Cell Signaling and Metabolism. Trends Immunol. 2018, 39, 489–502. [Google Scholar] [CrossRef] [PubMed]
- Nagaraj, S.; Gupta, K.; Pisarev, V.; Kinarsky, L.; Sherman, S.; Kang, L.; Herber, D.L.; Schneck, J.; Gabrilovich, D.I. Altered recognition of antigen is a mechanism of CD8+ T cell tolerance in cancer. Nat. Med. 2007, 13, 828–835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kusmartsev, S.; Nefedova, Y.; Yoder, D.; Gabrilovich, D.I. Antigen-specific inhibition of CD8+ T cell response by immature myeloid cells in cancer is mediated by reactive oxygen species. J. Immunol. 2004, 172, 989–999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmielau, J.; Finn, O.J. Activated granulocytes and granulocyte-derived hydrogen peroxide are the underlying mechanism of suppression of t-cell function in advanced cancer patients. Cancer Res. 2001, 61, 4756–4760. [Google Scholar] [PubMed]
- Ma, X.; Bi, E.; Lu, Y.; Su, P.; Huang, C.; Liu, L.; Wang, Q.; Yang, M.; Kalady, M.F.; Qian, J.; et al. Cholesterol Induces CD8(+) T Cell Exhaustion in the Tumor Microenvironment. Cell Metab. 2019, 30, 143–156. [Google Scholar] [CrossRef]
- Fischer, K.; Hoffmann, P.; Voelkl, S.; Meidenbauer, N.; Ammer, J.; Edinger, M.; Gottfried, E.; Schwarz, S.; Rothe, G.; Hoves, S.; et al. Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood 2007, 109, 3812–3819. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.L.; Lucas, J.E.; Schroeder, T.; Mori, S.; Wu, J.; Nevins, J.; Dewhirst, M.; West, M.; Chi, J.T. The genomic analysis of lactic acidosis and acidosis response in human cancers. PLoS Genet. 2008, 4, e1000293. [Google Scholar] [CrossRef] [Green Version]
- Chi, H. Regulation and function of mTOR signalling in T cell fate decisions. Nat. Rev. Immunol. 2012, 12, 325–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angelin, A.; Gil-de-Gomez, L.; Dahiya, S.; Jiao, J.; Guo, L.; Levine, M.H.; Wang, Z.; Quinn, W.J., 3rd; Kopinski, P.K.; Wang, L.; et al. Foxp3 Reprograms T Cell Metabolism to Function in Low-Glucose, High-Lactate Environments. Cell Metab. 2017, 25, 1282–1293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laoui, D.; Movahedi, K.; Van Overmeire, E.; Van den Bossche, J.; Schouppe, E.; Mommer, C.; Nikolaou, A.; Morias, Y.; De Baetselier, P.; Van Ginderachter, J.A. Tumor-associated macrophages in breast cancer: Distinct subsets, distinct functions. Int. J. Dev. Biol. 2011, 55, 861–867. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Sozzani, S.; Locati, M.; Allavena, P.; Sica, A. Macrophage polarization: Tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002, 23, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; He, Y.; Sun, X.; Li, Q.; Wang, W.; Zhao, A.; Di, W. A high M1/M2 ratio of tumor-associated macrophages is associated with extended survival in ovarian cancer patients. J. Ovarian Res. 2014, 7, 19. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Xu, J.; Lan, H. Tumor-associated macrophages in tumor metastasis: Biological roles and clinical therapeutic applications. J. Hematol. Oncol. 2019, 12, 76. [Google Scholar] [CrossRef]
- Li, C.; Xu, X.; Wei, S.; Jiang, P.; Xue, L.; Wang, J.; Senior, C. Tumor-associated macrophages: Potential therapeutic strategies and future prospects in cancer. J. Immunother. Cancer 2021, 9, e001341. [Google Scholar] [CrossRef]
- Gazzaniga, S.; Bravo, A.I.; Guglielmotti, A.; van Rooijen, N.; Maschi, F.; Vecchi, A.; Mantovani, A.; Mordoh, J.; Wainstok, R. Targeting tumor-associated macrophages and inhibition of MCP-1 reduce angiogenesis and tumor growth in a human melanoma xenograft. J. Investig. Dermatol. 2007, 127, 2031–2041. [Google Scholar] [CrossRef] [Green Version]
- Grivennikov, S.I.; Wang, K.; Mucida, D.; Stewart, C.A.; Schnabl, B.; Jauch, D.; Taniguchi, K.; Yu, G.Y.; Osterreicher, C.H.; Hung, K.E.; et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 2012, 491, 254–258. [Google Scholar] [CrossRef] [Green Version]
- Kong, L.; Zhou, Y.; Bu, H.; Lv, T.; Shi, Y.; Yang, J. Deletion of interleukin-6 in monocytes/macrophages suppresses the initiation of hepatocellular carcinoma in mice. J. Exp. Clin. Cancer Res. 2016, 35, 131. [Google Scholar] [CrossRef] [Green Version]
- Fu, X.T.; Dai, Z.; Song, K.; Zhang, Z.J.; Zhou, Z.J.; Zhou, S.L.; Zhao, Y.M.; Xiao, Y.S.; Sun, Q.M.; Ding, Z.B.; et al. Macrophage-secreted IL-8 induces epithelial-mesenchymal transition in hepatocellular carcinoma cells by activating the JAK2/STAT3/Snail pathway. Int. J. Oncol. 2015, 46, 587–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Deng, J.; Rychahou, P.G.; Qiu, S.; Evers, B.M.; Zhou, B.P. Stabilization of snail by NF-kappaB is required for inflammation-induced cell migration and invasion. Cancer Cell 2009, 15, 416–428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawata, M.; Koinuma, D.; Ogami, T.; Umezawa, K.; Iwata, C.; Watabe, T.; Miyazono, K. TGF-beta-induced epithelial-mesenchymal transition of A549 lung adenocarcinoma cells is enhanced by pro-inflammatory cytokines derived from RAW 264.7 macrophage cells. J. Biochem. 2012, 151, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Kessenbrock, K.; Plaks, V.; Werb, Z. Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell 2010, 141, 52–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasiljeva, O.; Papazoglou, A.; Kruger, A.; Brodoefel, H.; Korovin, M.; Deussing, J.; Augustin, N.; Nielsen, B.S.; Almholt, K.; Bogyo, M.; et al. Tumor cell-derived and macrophage-derived cathepsin B promotes progression and lung metastasis of mammary cancer. Cancer Res. 2006, 66, 5242–5250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gocheva, V.; Wang, H.W.; Gadea, B.B.; Shree, T.; Hunter, K.E.; Garfall, A.L.; Berman, T.; Joyce, J.A. IL-4 induces cathepsin protease activity in tumor-associated macrophages to promote cancer growth and invasion. Genes Dev. 2010, 24, 241–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, E.Y.; Li, J.F.; Gnatovskiy, L.; Deng, Y.; Zhu, L.; Grzesik, D.A.; Qian, H.; Xue, X.N.; Pollard, J.W. Macrophages regulate the angiogenic switch in a mouse model of breast cancer. Cancer Res. 2006, 66, 11238–11246. [Google Scholar] [CrossRef] [Green Version]
- Lin, E.Y.; Pollard, J.W. Tumor-associated macrophages press the angiogenic switch in breast cancer. Cancer Res. 2007, 67, 5064–5066. [Google Scholar] [CrossRef] [Green Version]
- Bergers, G.; Brekken, R.; McMahon, G.; Vu, T.H.; Itoh, T.; Tamaki, K.; Tanzawa, K.; Thorpe, P.; Itohara, S.; Werb, Z.; et al. Matrix metalloproteinase-9 triggers the angiogenic switch during carcinogenesis. Nat. Cell Biol. 2000, 2, 737–744. [Google Scholar] [CrossRef]
- Giraudo, E.; Inoue, M.; Hanahan, D. An amino-bisphosphonate targets MMP-9-expressing macrophages and angiogenesis to impair cervical carcinogenesis. J. Clin. Investig. 2004, 114, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Leek, R.D.; Harris, A.L.; Lewis, C.E. Cytokine networks in solid human tumors: Regulation of angiogenesis. J. Leukoc. Biol. 1994, 56, 423–435. [Google Scholar] [CrossRef]
- Gil-Bernabe, A.M.; Ferjancic, S.; Tlalka, M.; Zhao, L.; Allen, P.D.; Im, J.H.; Watson, K.; Hill, S.A.; Amirkhosravi, A.; Francis, J.L.; et al. Recruitment of monocytes/macrophages by tissue factor-mediated coagulation is essential for metastatic cell survival and premetastatic niche establishment in mice. Blood 2012, 119, 3164–3175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Q.; Zhang, X.H.; Massague, J. Macrophage binding to receptor VCAM-1 transmits survival signals in breast cancer cells that invade the lungs. Cancer Cell 2011, 20, 538–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, X.; Mu, E.; Wei, Y.; Riethdorf, S.; Yang, Q.; Yuan, M.; Yan, J.; Hua, Y.; Tiede, B.J.; Lu, X.; et al. VCAM-1 promotes osteolytic expansion of indolent bone micrometastasis of breast cancer by engaging alpha4beta1-positive osteoclast progenitors. Cancer Cell 2011, 20, 701–714. [Google Scholar] [CrossRef] [Green Version]
- Koukourakis, M.I.; Giatromanolaki, A.; Kakolyris, S.; O’Byrne, K.J.; Apostolikas, N.; Skarlatos, J.; Gatter, K.C.; Harris, A.L. Different patterns of stromal and cancer cell thymidine phosphorylase reactivity in non-small-cell lung cancer: Impact on tumour neoangiogenesis and survival. Br. J. Cancer 1998, 77, 1696–1703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossi, M.L.; Jones, N.R.; Candy, E.; Nicoll, J.A.; Compton, J.S.; Hughes, J.T.; Esiri, M.M.; Moss, T.H.; Cruz-Sanchez, F.F.; Coakham, H.B. The mononuclear cell infiltrate compared with survival in high-grade astrocytomas. Acta Neuropathol. 1989, 78, 189–193. [Google Scholar] [CrossRef]
- Shimura, S.; Yang, G.; Ebara, S.; Wheeler, T.M.; Frolov, A.; Thompson, T.C. Reduced infiltration of tumor-associated macrophages in human prostate cancer: Association with cancer progression. Cancer Res. 2000, 60, 5857–5861. [Google Scholar]
- Yang, L.; Zhang, Y. Tumor-associated macrophages: From basic research to clinical application. J. Hematol. Oncol. 2017, 10, 58. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Li, Y.; Zhu, B. T-cell exhaustion in the tumor microenvironment. Cell Death Dis. 2015, 6, e1792. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Yang, L.; Yue, D.; Cao, L.; Li, L.; Wang, D.; Ping, Y.; Shen, Z.; Zheng, Y.; Wang, L.; et al. Macrophage-derived CCL22 promotes an immunosuppressive tumor microenvironment via IL-8 in malignant pleural effusion. Cancer Lett. 2019, 452, 244–253. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, S.; Zhang, B.; Qiao, L.; Zhang, Y.; Zhang, Y. T Cell Dysfunction and Exhaustion in Cancer. Front. Cell Dev. Biol. 2020, 8, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahai, E.; Astsaturov, I.; Cukierman, E.; DeNardo, D.G.; Egeblad, M.; Evans, R.M.; Fearon, D.; Greten, F.R.; Hingorani, S.R.; Hunter, T.; et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer 2020, 20, 174–186. [Google Scholar] [CrossRef] [Green Version]
- Ping, Q.; Yan, R.; Cheng, X.; Wang, W.; Zhong, Y.; Hou, Z.; Shi, Y.; Wang, C.; Li, R. Cancer-associated fibroblasts: Overview, progress, challenges, and directions. Cancer Gene Ther. 2021, 28, 984–999. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, E.V.; Pereira, B.A.; Lawrence, M.G.; Ma, X.; Rebello, R.J.; Chan, H.; Niranjan, B.; Wu, Y.; Ellem, S.; Guan, X.; et al. Proteomic Profiling of Human Prostate Cancer-associated Fibroblasts (CAF) Reveals LOXL2-dependent Regulation of the Tumor Microenvironment. Mol. Cell Proteom. 2019, 18, 1410–1427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, X.; Hou, Y.; Yang, G.; Wang, X.; Tang, S.; Du, Y.E.; Yang, L.; Yu, T.; Zhang, H.; Zhou, M.; et al. Stromal miR-200s contribute to breast cancer cell invasion through CAF activation and ECM remodeling. Cell Death Differ. 2016, 23, 132–145. [Google Scholar] [CrossRef] [Green Version]
- Mohammadi, H.; Sahai, E. Mechanisms and impact of altered tumour mechanics. Nat. Cell Biol. 2018, 20, 766–774. [Google Scholar] [CrossRef]
- Jain, R.K.; Martin, J.D.; Stylianopoulos, T. The role of mechanical forces in tumor growth and therapy. Annu. Rev. Biomed. Eng. 2014, 16, 321–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DuFort, C.C.; DelGiorno, K.E.; Hingorani, S.R. Mounting Pressure in the Microenvironment: Fluids, Solids, and Cells in Pancreatic Ductal Adenocarcinoma. Gastroenterology 2016, 150, 1545–1557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paszek, M.J.; Zahir, N.; Johnson, K.R.; Lakins, J.N.; Rozenberg, G.I.; Gefen, A.; Reinhart-King, C.A.; Margulies, S.S.; Dembo, M.; Boettiger, D.; et al. Tensional homeostasis and the malignant phenotype. Cancer Cell 2005, 8, 241–254. [Google Scholar] [CrossRef] [Green Version]
- Monteran, L.; Erez, N. The Dark Side of Fibroblasts: Cancer-Associated Fibroblasts as Mediators of Immunosuppression in the Tumor Microenvironment. Front. Immunol. 2019, 10, 1835. [Google Scholar] [CrossRef] [Green Version]
- Dumont, N.; Liu, B.; Defilippis, R.A.; Chang, H.; Rabban, J.T.; Karnezis, A.N.; Tjoe, J.A.; Marx, J.; Parvin, B.; Tlsty, T.D. Breast fibroblasts modulate early dissemination, tumorigenesis, and metastasis through alteration of extracellular matrix characteristics. Neoplasia 2013, 15, 249–262. [Google Scholar] [CrossRef] [Green Version]
- Madsen, C.D.; Pedersen, J.T.; Venning, F.A.; Singh, L.B.; Moeendarbary, E.; Charras, G.; Cox, T.R.; Sahai, E.; Erler, J.T. Hypoxia and loss of PHD2 inactivate stromal fibroblasts to decrease tumour stiffness and metastasis. EMBO Rep. 2015, 16, 1394–1408. [Google Scholar] [CrossRef] [PubMed]
- Calon, A.; Espinet, E.; Palomo-Ponce, S.; Tauriello, D.V.; Iglesias, M.; Cespedes, M.V.; Sevillano, M.; Nadal, C.; Jung, P.; Zhang, X.H.; et al. Dependency of colorectal cancer on a TGF-beta-driven program in stromal cells for metastasis initiation. Cancer Cell 2012, 22, 571–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaggioli, C.; Hooper, S.; Hidalgo-Carcedo, C.; Grosse, R.; Marshall, J.F.; Harrington, K.; Sahai, E. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat. Cell Biol. 2007, 9, 1392–1400. [Google Scholar] [CrossRef]
- Malanchi, I.; Santamaria-Martinez, A.; Susanto, E.; Peng, H.; Lehr, H.A.; Delaloye, J.F.; Huelsken, J. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature 2011, 481, 85–89. [Google Scholar] [CrossRef] [Green Version]
- Oskarsson, T.; Acharyya, S.; Zhang, X.H.; Vanharanta, S.; Tavazoie, S.F.; Morris, P.G.; Downey, R.J.; Manova-Todorova, K.; Brogi, E.; Massague, J. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat. Med. 2011, 17, 867–874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Wever, O.; Nguyen, Q.D.; Van Hoorde, L.; Bracke, M.; Bruyneel, E.; Gespach, C.; Mareel, M. Tenascin-C and SF/HGF produced by myofibroblasts in vitro provide convergent pro-invasive signals to human colon cancer cells through RhoA and Rac. FASEB J. 2004, 18, 1016–1018. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Gao, W.; Lytle, N.K.; Huang, P.; Yuan, X.; Dann, A.M.; Ridinger-Saison, M.; DelGiorno, K.E.; Antal, C.E.; Liang, G.; et al. Targeting LIF-mediated paracrine interaction for pancreatic cancer therapy and monitoring. Nature 2019, 569, 131–135. [Google Scholar] [CrossRef]
- Cazet, A.S.; Hui, M.N.; Elsworth, B.L.; Wu, S.Z.; Roden, D.; Chan, C.L.; Skhinas, J.N.; Collot, R.; Yang, J.; Harvey, K.; et al. Targeting stromal remodeling and cancer stem cell plasticity overcomes chemoresistance in triple negative breast cancer. Nat. Commun. 2018, 9, 2897. [Google Scholar] [CrossRef] [Green Version]
- Bruzzese, F.; Hagglof, C.; Leone, A.; Sjoberg, E.; Roca, M.S.; Kiflemariam, S.; Sjoblom, T.; Hammarsten, P.; Egevad, L.; Bergh, A.; et al. Local and systemic protumorigenic effects of cancer-associated fibroblast-derived GDF15. Cancer Res. 2014, 74, 3408–3417. [Google Scholar] [CrossRef] [Green Version]
- Olumi, A.F.; Grossfeld, G.D.; Hayward, S.W.; Carroll, P.R.; Tlsty, T.D.; Cunha, G.R. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res. 1999, 59, 5002–5011. [Google Scholar] [CrossRef] [PubMed]
- Bhome, R.; Goh, R.W.; Bullock, M.D.; Pillar, N.; Thirdborough, S.M.; Mellone, M.; Mirnezami, R.; Galea, D.; Veselkov, K.; Gu, Q.; et al. Exosomal microRNAs derived from colorectal cancer-associated fibroblasts: Role in driving cancer progression. Aging 2017, 9, 2666–2694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luga, V.; Zhang, L.; Viloria-Petit, A.M.; Ogunjimi, A.A.; Inanlou, M.R.; Chiu, E.; Buchanan, M.; Hosein, A.N.; Basik, M.; Wrana, J.L. Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell 2012, 151, 1542–1556. [Google Scholar] [CrossRef] [Green Version]
- Fearon, D.T. The carcinoma-associated fibroblast expressing fibroblast activation protein and escape from immune surveillance. Cancer Immunol. Res. 2014, 2, 187–193. [Google Scholar] [CrossRef] [Green Version]
- Elyada, E.; Bolisetty, M.; Laise, P.; Flynn, W.F.; Courtois, E.T.; Burkhart, R.A.; Teinor, J.A.; Belleau, P.; Biffi, G.; Lucito, M.S.; et al. Cross-Species Single-Cell Analysis of Pancreatic Ductal Adenocarcinoma Reveals Antigen-Presenting Cancer-Associated Fibroblasts. Cancer Discov. 2019, 9, 1102–1123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lakins, M.A.; Ghorani, E.; Munir, H.; Martins, C.P.; Shields, J.D. Cancer-associated fibroblasts induce antigen-specific deletion of CD8(+) T Cells to protect tumour cells. Nat. Commun. 2018, 9, 948. [Google Scholar] [CrossRef]
- Costa, A.; Kieffer, Y.; Scholer-Dahirel, A.; Pelon, F.; Bourachot, B.; Cardon, M.; Sirven, P.; Magagna, I.; Fuhrmann, L.; Bernard, C.; et al. Fibroblast Heterogeneity and Immunosuppressive Environment in Human Breast Cancer. Cancer Cell 2018, 33, 463–479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinchuk, I.V.; Saada, J.I.; Beswick, E.J.; Boya, G.; Qiu, S.M.; Mifflin, R.C.; Raju, G.S.; Reyes, V.E.; Powell, D.W. PD-1 ligand expression by human colonic myofibroblasts/fibroblasts regulates CD4+ T-cell activity. Gastroenterology 2008, 135, 1228–1237. [Google Scholar] [CrossRef] [Green Version]
- Fridlender, Z.G.; Albelda, S.M. Tumor-associated neutrophils: Friend or foe? Carcinogenesis 2012, 33, 949–955. [Google Scholar] [CrossRef] [Green Version]
- Fridlender, Z.G.; Sun, J.; Kim, S.; Kapoor, V.; Cheng, G.; Ling, L.; Worthen, G.S.; Albelda, S.M. Polarization of tumor-associated neutrophil phenotype by TGF-beta: “N1” versus “N2” TAN. Cancer Cell 2009, 16, 183–194. [Google Scholar] [CrossRef] [Green Version]
- Swierczak, A.; Mouchemore, K.A.; Hamilton, J.A.; Anderson, R.L. Neutrophils: Important contributors to tumor progression and metastasis. Cancer Metastasis Rev. 2015, 34, 735–751. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A. The yin-yang of tumor-associated neutrophils. Cancer Cell 2009, 16, 173–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uribe-Querol, E.; Rosales, C. Neutrophils in Cancer: Two Sides of the Same Coin. J. Immunol. Res. 2015, 2015, 983698. [Google Scholar] [CrossRef] [Green Version]
- Flavell, R.A.; Sanjabi, S.; Wrzesinski, S.H.; Licona-Limon, P. The polarization of immune cells in the tumour environment by TGFbeta. Nat. Rev. Immunol. 2010, 10, 554–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coffelt, S.B.; Wellenstein, M.D.; de Visser, K.E. Neutrophils in cancer: Neutral no more. Nat. Rev. Cancer 2016, 16, 431–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eruslanov, E.B.; Singhal, S.; Albelda, S.M. Mouse versus Human Neutrophils in Cancer: A Major Knowledge Gap. Trends Cancer 2017, 3, 149–160. [Google Scholar] [CrossRef] [Green Version]
- Taya, M.; Garcia-Hernandez, M.L.; Rangel-Moreno, J.; Minor, B.; Gibbons, E.; Hammes, S.R. Neutrophil elastase from myeloid cells promotes TSC2-null tumor growth. Endocr. Relat. Cancer 2020, 27, 261–274. [Google Scholar] [CrossRef]
- Lerman, I.; Garcia-Hernandez, M.L.; Rangel-Moreno, J.; Chiriboga, L.; Pan, C.; Nastiuk, K.L.; Krolewski, J.J.; Sen, A.; Hammes, S.R. Infiltrating Myeloid Cells Exert Protumorigenic Actions via Neutrophil Elastase. Mol. Cancer Res. 2017, 15, 1138–1152. [Google Scholar] [CrossRef] [Green Version]
- Liao, Z.; Chua, D.; Tan, N.S. Reactive oxygen species: A volatile driver of field cancerization and metastasis. Mol. Cancer 2019, 18, 65. [Google Scholar] [CrossRef]
- Wu, L.; Saxena, S.; Singh, R.K. Neutrophils in the Tumor Microenvironment. In Tumor Microenvironment; Advances in Experimental Medicine and Biology; Springer: Berlin/Heidelberg, Germany, 2020; Volume 1224, pp. 1–20. [Google Scholar] [CrossRef]
- Coussens, L.M.; Tinkle, C.L.; Hanahan, D.; Werb, Z. MMP-9 supplied by bone marrow-derived cells contributes to skin carcinogenesis. Cell 2000, 103, 481–490. [Google Scholar] [CrossRef] [Green Version]
- Masucci, M.T.; Minopoli, M.; Del Vecchio, S.; Carriero, M.V. The Emerging Role of Neutrophil Extracellular Traps (NETs) in Tumor Progression and Metastasis. Front. Immunol. 2020, 11, 1749. [Google Scholar] [CrossRef]
- Spicer, J.D.; McDonald, B.; Cools-Lartigue, J.J.; Chow, S.C.; Giannias, B.; Kubes, P.; Ferri, L.E. Neutrophils promote liver metastasis via Mac-1-mediated interactions with circulating tumor cells. Cancer Res. 2012, 72, 3919–3927. [Google Scholar] [CrossRef] [Green Version]
- Cools-Lartigue, J.; Spicer, J.; McDonald, B.; Gowing, S.; Chow, S.; Giannias, B.; Bourdeau, F.; Kubes, P.; Ferri, L. Neutrophil extracellular traps sequester circulating tumor cells and promote metastasis. J. Clin. Investig. 2013, 123, 3446–3458. [Google Scholar] [CrossRef] [PubMed]
- Szczerba, B.M.; Castro-Giner, F.; Vetter, M.; Krol, I.; Gkountela, S.; Landin, J.; Scheidmann, M.C.; Donato, C.; Scherrer, R.; Singer, J.; et al. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature 2019, 566, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zhang, X.H. Tumor-Associated Neutrophils and Macrophages-Heterogenous but Not Chaotic. Front. Immunol. 2020, 11, 553967. [Google Scholar] [CrossRef]
- Itatani, Y.; Yamamoto, T.; Zhong, C.; Molinolo, A.A.; Ruppel, J.; Hegde, P.; Taketo, M.M.; Ferrara, N. Suppressing neutrophil-dependent angiogenesis abrogates resistance to anti-VEGF antibody in a genetic model of colorectal cancer. Proc. Natl. Acad. Sci. USA 2020, 117, 21598–21608. [Google Scholar] [CrossRef] [PubMed]
- Grosse-Steffen, T.; Giese, T.; Giese, N.; Longerich, T.; Schirmacher, P.; Hansch, G.M.; Gaida, M.M. Epithelial-to-mesenchymal transition in pancreatic ductal adenocarcinoma and pancreatic tumor cell lines: The role of neutrophils and neutrophil-derived elastase. Clin. Dev. Immunol. 2012, 2012, 720768. [Google Scholar] [CrossRef]
- Li, S.; Cong, X.; Gao, H.; Lan, X.; Li, Z.; Wang, W.; Song, S.; Wang, Y.; Li, C.; Zhang, H.; et al. Tumor-associated neutrophils induce EMT by IL-17a to promote migration and invasion in gastric cancer cells. J. Exp. Clin. Cancer Res. 2019, 38, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, P.; Sun, Y.; Ma, L. ZEB1: At the crossroads of epithelial-mesenchymal transition, metastasis and therapy resistance. Cell Cycle 2015, 14, 481–487. [Google Scholar] [CrossRef] [Green Version]
- Mollinedo, F. Neutrophil Degranulation, Plasticity, and Cancer Metastasis. Trends Immunol. 2019, 40, 228–242. [Google Scholar] [CrossRef]
- Mishalian, I.; Bayuh, R.; Eruslanov, E.; Michaeli, J.; Levy, L.; Zolotarov, L.; Singhal, S.; Albelda, S.M.; Granot, Z.; Fridlender, Z.G. Neutrophils recruit regulatory T-cells into tumors via secretion of CCL17—A new mechanism of impaired antitumor immunity. Int. J. Cancer 2014, 135, 1178–1186. [Google Scholar] [CrossRef] [PubMed]
- Raftopoulou, S.; Valadez-Cosmes, P.; Mihalic, Z.N.; Schicho, R.; Kargl, J. Tumor-Mediated Neutrophil Polarization and Therapeutic Implications. Int. J. Mol. Sci. 2022, 23, 3218. [Google Scholar] [CrossRef] [PubMed]
- Bedard, P.L.; Hyman, D.M.; Davids, M.S.; Siu, L.L. Small molecules, big impact: 20 years of targeted therapy in oncology. Lancet 2020, 395, 1078–1088. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Li, Y.; Xiong, L.; Wang, W.; Wu, M.; Yuan, T.; Yang, W.; Tian, C.; Miao, Z.; Wang, T.; et al. Small molecules in targeted cancer therapy: Advances, challenges, and future perspectives. Signal. Transduct. Target Ther. 2021, 6, 201. [Google Scholar] [CrossRef]
- Ostrem, J.M.; Peters, U.; Sos, M.L.; Wells, J.A.; Shokat, K.M. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 2013, 503, 548–551. [Google Scholar] [CrossRef] [Green Version]
- Lito, P.; Solomon, M.; Li, L.S.; Hansen, R.; Rosen, N. Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science 2016, 351, 604–608. [Google Scholar] [CrossRef] [Green Version]
- Janes, M.R.; Zhang, J.; Li, L.S.; Hansen, R.; Peters, U.; Guo, X.; Chen, Y.; Babbar, A.; Firdaus, S.J.; Darjania, L.; et al. Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell 2018, 172, 578–589. [Google Scholar] [CrossRef] [Green Version]
- Zeng, M.; Lu, J.; Li, L.; Feru, F.; Quan, C.; Gero, T.W.; Ficarro, S.B.; Xiong, Y.; Ambrogio, C.; Paranal, R.M.; et al. Potent and Selective Covalent Quinazoline Inhibitors of KRAS G12C. Cell Chem. Biol. 2017, 24, 1005–1016. [Google Scholar] [CrossRef] [Green Version]
- Kohler, G.; Milstein, C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 1975, 256, 495–497. [Google Scholar] [CrossRef]
- Pietras, R.J.; Fendly, B.M.; Chazin, V.R.; Pegram, M.D.; Howell, S.B.; Slamon, D.J. Antibody to HER-2/neu receptor blocks DNA repair after cisplatin in human breast and ovarian cancer cells. Oncogene 1994, 9, 1829–1838. [Google Scholar]
- Izumi, Y.; Xu, L.; di Tomaso, E.; Fukumura, D.; Jain, R.K. Tumour biology: Herceptin acts as an anti-angiogenic cocktail. Nature 2002, 416, 279–280. [Google Scholar] [CrossRef]
- Harding, J.; Burtness, B. Cetuximab: An epidermal growth factor receptor chemeric human-murine monoclonal antibody. Drugs Today 2005, 41, 107–127. [Google Scholar] [CrossRef]
- Maier, L.A.; Xu, F.J.; Hester, S.; Boyer, C.M.; McKenzie, S.; Bruskin, A.M.; Argon, Y.; Bast, R.C., Jr. Requirements for the internalization of a murine monoclonal antibody directed against the HER-2/neu gene product c-erbB-2. Cancer Res. 1991, 51, 5361–5369. [Google Scholar] [PubMed]
- Redman, J.M.; Hill, E.M.; AlDeghaither, D.; Weiner, L.M. Mechanisms of action of therapeutic antibodies for cancer. Mol. Immunol. 2015, 67, 28–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukuhara, H.; Ino, Y.; Todo, T. Oncolytic virus therapy: A new era of cancer treatment at dawn. Cancer Sci. 2016, 107, 1373–1379. [Google Scholar] [CrossRef] [PubMed]
- Platanias, L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005, 5, 375–386. [Google Scholar] [CrossRef]
- Coffin, R. Interview with Robert Coffin, inventor of T-VEC: The first oncolytic immunotherapy approved for the treatment of cancer. Immunotherapy 2016, 8, 103–106. [Google Scholar] [CrossRef] [Green Version]
- Azangou-Khyavy, M.; Ghasemi, M.; Khanali, J.; Boroomand-Saboor, M.; Jamalkhah, M.; Soleimani, M.; Kiani, J. CRISPR/Cas: From Tumor Gene Editing to T Cell-Based Immunotherapy of Cancer. Front. Immunol. 2020, 11, 2062. [Google Scholar] [CrossRef]
- Chira, S.; Gulei, D.; Hajitou, A.; Berindan-Neagoe, I. Restoring the p53 ‘Guardian’ Phenotype in p53-Deficient Tumor Cells with CRISPR/Cas9. Trends Biotechnol. 2018, 36, 653–660. [Google Scholar] [CrossRef]
- Wang, H.; Sun, W. CRISPR-mediated targeting of HER2 inhibits cell proliferation through a dominant negative mutation. Cancer Lett. 2017, 385, 137–143. [Google Scholar] [CrossRef]
- Su, S.; Hu, B.; Shao, J.; Shen, B.; Du, J.; Du, Y.; Zhou, J.; Yu, L.; Zhang, L.; Chen, F.; et al. CRISPR-Cas9 mediated efficient PD-1 disruption on human primary T cells from cancer patients. Sci. Rep. 2016, 6, 20070. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.; Zou, Y.; Zhang, L.; Tang, J.; Niedermann, G.; Firat, E.; Huang, X.; Zhu, X. Nucleofection with Plasmid DNA for CRISPR/Cas9-Mediated Inactivation of Programmed Cell Death Protein 1 in CD133-Specific CAR T Cells. Hum. Gene Ther. 2019, 30, 446–458. [Google Scholar] [CrossRef]
- Dai, X.; Park, J.J.; Du, Y.; Kim, H.R.; Wang, G.; Errami, Y.; Chen, S. One-step generation of modular CAR-T cells with AAV-Cpf1. Nat. Methods 2019, 16, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Eyquem, J.; Mansilla-Soto, J.; Giavridis, T.; van der Stegen, S.J.; Hamieh, M.; Cunanan, K.M.; Odak, A.; Gonen, M.; Sadelain, M. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 2017, 543, 113–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caldwell, K.J.; Gottschalk, S.; Talleur, A.C. Allogeneic CAR Cell Therapy-More Than a Pipe Dream. Front. Immunol. 2020, 11, 618427. [Google Scholar] [CrossRef] [PubMed]
- Pavel-Dinu, M.; Wiebking, V.; Dejene, B.T.; Srifa, W.; Mantri, S.; Nicolas, C.E.; Lee, C.; Bao, G.; Kildebeck, E.J.; Punjya, N.; et al. Gene correction for SCID-X1 in long-term hematopoietic stem cells. Nat. Commun. 2019, 10, 1634. [Google Scholar] [CrossRef] [Green Version]
- Couzin-Frankel, J. Breakthrough of the year 2013. Cancer immunotherapy. Science 2013, 342, 1432–1433. [Google Scholar] [CrossRef] [Green Version]
- Schumacher, T.N.; Schreiber, R.D. Neoantigens in cancer immunotherapy. Science 2015, 348, 69–74. [Google Scholar] [CrossRef] [Green Version]
- Butterfield, L.H. Cancer vaccines. BMJ 2015, 350, h988. [Google Scholar] [CrossRef]
- Carreno, B.M.; Magrini, V.; Becker-Hapak, M.; Kaabinejadian, S.; Hundal, J.; Petti, A.A.; Ly, A.; Lie, W.R.; Hildebrand, W.H.; Mardis, E.R.; et al. Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science 2015, 348, 803–808. [Google Scholar] [CrossRef] [Green Version]
- Sahin, U.; Derhovanessian, E.; Miller, M.; Kloke, B.P.; Simon, P.; Lower, M.; Bukur, V.; Tadmor, A.D.; Luxemburger, U.; Schrors, B.; et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 2017, 547, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Ott, P.A.; Hu, Z.; Keskin, D.B.; Shukla, S.A.; Sun, J.; Bozym, D.J.; Zhang, W.; Luoma, A.; Giobbie-Hurder, A.; Peter, L.; et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 2017, 547, 217–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arbelaez, C.A.; Estrada, J.; Gessner, M.A.; Glaus, C.; Morales, A.B.; Mohn, D.; Phee, H.; Lipford, J.R.; Johnston, J.A. A nanoparticle vaccine that targets neoantigen peptides to lymphoid tissues elicits robust antitumor T cell responses. NPJ Vaccines 2020, 5, 106. [Google Scholar] [CrossRef] [PubMed]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug. Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef] [Green Version]
- Cafri, G.; Gartner, J.J.; Zaks, T.; Hopson, K.; Levin, N.; Paria, B.C.; Parkhurst, M.R.; Yossef, R.; Lowery, F.J.; Jafferji, M.S.; et al. mRNA vaccine-induced neoantigen-specific T cell immunity in patients with gastrointestinal cancer. J. Clin. Investig. 2020, 130, 5976–5988. [Google Scholar] [CrossRef]
- Wu, J.; Liu, T.; Rios, Z.; Mei, Q.; Lin, X.; Cao, S. Heat Shock Proteins and Cancer. Trends Pharmacol. Sci. 2017, 38, 226–256. [Google Scholar] [CrossRef]
- Ciocca, D.R.; Calderwood, S.K. Heat shock proteins in cancer: Diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones 2005, 10, 86–103. [Google Scholar] [CrossRef]
- Calderwood, S.K.; Gong, J. Heat Shock Proteins Promote Cancer: It’s a Protection Racket. Trends Biochem. Sci. 2016, 41, 311–323. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.F.; Ren, W.; Rollins, L.; Pittman, P.; Shah, M.; Shen, L.; Gu, Q.; Strube, R.; Hu, F.; Chen, S.Y. A broadly applicable, personalized heat shock protein-mediated oncolytic tumor vaccine. Cancer Res. 2003, 63, 7321–7329. [Google Scholar]
- Taha, E.A.; Ono, K.; Eguchi, T. Roles of Extracellular HSPs as Biomarkers in Immune Surveillance and Immune Evasion. Int. J. Mol. Sci. 2019, 20, 4588. [Google Scholar] [CrossRef] [Green Version]
- Murshid, A.; Gong, J.; Stevenson, M.A.; Calderwood, S.K. Heat shock proteins and cancer vaccines: Developments in the past decade and chaperoning in the decade to come. Expert Rev. Vaccines 2011, 10, 1553–1568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, J.K.; Xiong, X.; Ren, X.; Yang, J.M.; Song, J. Heat Shock Proteins in Cancer Immunotherapy. J. Oncol. 2019, 2019, 3267207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueda, G.; Tamura, Y.; Hirai, I.; Kamiguchi, K.; Ichimiya, S.; Torigoe, T.; Hiratsuka, H.; Sunakawa, H.; Sato, N. Tumor-derived heat shock protein 70-pulsed dendritic cells elicit tumor-specific cytotoxic T lymphocytes (CTLs) and tumor immunity. Cancer Sci. 2004, 95, 248–253. [Google Scholar] [CrossRef]
- Hoos, A.; Levey, D.L. Vaccination with heat shock protein-peptide complexes: From basic science to clinical applications. Expert. Rev. Vaccines 2003, 2, 369–379. [Google Scholar] [CrossRef]
- Blachere, N.E.; Udono, H.; Janetzki, S.; Li, Z.; Heike, M.; Srivastava, P.K. Heat shock protein vaccines against cancer. J. Immunother. Emphas. Tumor Immunol. 1993, 14, 352–356. [Google Scholar] [CrossRef]
- Liu, X.; Xu, Y.; Xiong, W.; Yin, B.; Huang, Y.; Chu, J.; Xing, C.; Qian, C.; Du, Y.; Duan, T.; et al. Development of a TCR-like antibody and chimeric antigen receptor against NY-ESO-1/HLA-A2 for cancer immunotherapy. J. Immunother. Cancer 2022, 10, e004035. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Ivica, N.A.; Jia, B.; Li, Y.; Dong, H.; Liang, Y.; Brown, D.; Romee, R.; Chen, J. CAR-T cells targeting a nucleophosmin neoepitope exhibit potent specific activity in mouse models of acute myeloid leukaemia. Nat. Biomed. Eng. 2021, 5, 399–413. [Google Scholar] [CrossRef]
- Hsiue, E.H.; Wright, K.M.; Douglass, J.; Hwang, M.S.; Mog, B.J.; Pearlman, A.H.; Paul, S.; DiNapoli, S.R.; Konig, M.F.; Wang, Q.; et al. Targeting a neoantigen derived from a common TP53 mutation. Science 2021, 371, eabc8697. [Google Scholar] [CrossRef]
- Douglass, J.; Hsiue, E.H.; Mog, B.J.; Hwang, M.S.; DiNapoli, S.R.; Pearlman, A.H.; Miller, M.S.; Wright, K.M.; Azurmendi, P.A.; Wang, Q.; et al. Bispecific antibodies targeting mutant RAS neoantigens. Sci. Immunol. 2021, 6, eabd5515. [Google Scholar] [CrossRef]
- Sergeeva, A.; He, H.; Ruisaard, K.; St John, L.; Alatrash, G.; Clise-Dwyer, K.; Li, D.; Patenia, R.; Hong, R.; Sukhumalchandra, P.; et al. Activity of 8F4, a T-cell receptor-like anti-PR1/HLA-A2 antibody, against primary human AML in vivo. Leukemia 2016, 30, 1475–1484. [Google Scholar] [CrossRef] [Green Version]
- Dao, T.; Yan, S.; Veomett, N.; Pankov, D.; Zhou, L.; Korontsvit, T.; Scott, A.; Whitten, J.; Maslak, P.; Casey, E.; et al. Targeting the intracellular WT1 oncogene product with a therapeutic human antibody. Sci. Transl. Med. 2013, 5, 176ra33. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Li, Y.M.; Zhou, J.J.; Zhou, Z.; Xu, Y.C.; Zhao, W.B.; Chen, S.Q. The Antitumor Activity of TCR-Mimic Antibody-Drug Conjugates (TCRm-ADCs) Targeting the Intracellular Wilms Tumor 1 (WT1) Oncoprotein. Int. J. Mol. Sci. 2019, 20, 3912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dao, T.; Mun, S.S.; Scott, A.C.; Jarvis, C.A.; Korontsvit, T.; Yang, Z.; Liu, L.; Klatt, M.G.; Guerreiro, M.; Selvakumar, A.; et al. Depleting T regulatory cells by targeting intracellular Foxp3 with a TCR mimic antibody. Oncoimmunology 2019, 8, 1570778. [Google Scholar] [CrossRef] [PubMed]
- Lowe, D.B.; Bivens, C.K.; Mobley, A.S.; Herrera, C.E.; McCormick, A.L.; Wichner, T.; Sabnani, M.K.; Wood, L.M.; Weidanz, J.A. TCR-like antibody drug conjugates mediate killing of tumor cells with low peptide/HLA targets. mAbs 2017, 9, 603–614. [Google Scholar] [CrossRef] [Green Version]
- Ma, Q.; Garber, H.R.; Lu, S.; He, H.; Tallis, E.; Ding, X.; Sergeeva, A.; Wood, M.S.; Dotti, G.; Salvado, B.; et al. A novel TCR-like CAR with specificity for PR1/HLA-A2 effectively targets myeloid leukemia in vitro when expressed in human adult peripheral blood and cord blood T cells. Cytotherapy 2016, 18, 985–994. [Google Scholar] [CrossRef] [Green Version]
- Dao, T.; Pankov, D.; Scott, A.; Korontsvit, T.; Zakhaleva, V.; Xu, Y.; Xiang, J.; Yan, S.; de Morais Guerreiro, M.D.; Veomett, N.; et al. Therapeutic bispecific T-cell engager antibody targeting the intracellular oncoprotein WT1. Nat. Biotechnol. 2015, 33, 1079–1086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veomett, N.; Dao, T.; Liu, H.; Xiang, J.; Pankov, D.; Dubrovsky, L.; Whitten, J.A.; Park, S.M.; Korontsvit, T.; Zakhaleva, V.; et al. Therapeutic efficacy of an Fc-enhanced TCR-like antibody to the intracellular WT1 oncoprotein. Clin. Cancer Res. 2014, 20, 4036–4046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubrovsky, L.; Pankov, D.; Brea, E.J.; Dao, T.; Scott, A.; Yan, S.; O’Reilly, R.J.; Liu, C.; Scheinberg, D.A. A TCR-mimic antibody to WT1 bypasses tyrosine kinase inhibitor resistance in human BCR-ABL+ leukemias. Blood 2014, 123, 3296–3304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribas, A.; Wolchok, J.D. Cancer immunotherapy using checkpoint blockade. Science 2018, 359, 1350–1355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majzner, R.G.; Mackall, C.L. Tumor Antigen Escape from CAR T-cell Therapy. Cancer Discov. 2018, 8, 1219–1226. [Google Scholar] [CrossRef] [Green Version]
- Kailayangiri, S.; Altvater, B.; Wiebel, M.; Jamitzky, S.; Rossig, C. Overcoming Heterogeneity of Antigen Expression for Effective CAR T Cell Targeting of Cancers. Cancers 2020, 12, 1075. [Google Scholar] [CrossRef]
- Anurathapan, U.; Chan, R.C.; Hindi, H.F.; Mucharla, R.; Bajgain, P.; Hayes, B.C.; Fisher, W.E.; Heslop, H.E.; Rooney, C.M.; Brenner, M.K.; et al. Kinetics of tumor destruction by chimeric antigen receptor-modified T cells. Mol. Ther. 2014, 22, 623–633. [Google Scholar] [CrossRef] [Green Version]
- Ramakrishna, S.; Highfill, S.L.; Walsh, Z.; Nguyen, S.M.; Lei, H.; Shern, J.F.; Qin, H.; Kraft, I.L.; Stetler-Stevenson, M.; Yuan, C.M.; et al. Modulation of Target Antigen Density Improves CAR T-cell Functionality and Persistence. Clin. Cancer Res. 2019, 25, 5329–5341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pont, M.J.; Hill, T.; Cole, G.O.; Abbott, J.J.; Kelliher, J.; Salter, A.I.; Hudecek, M.; Comstock, M.L.; Rajan, A.; Patel, B.K.R.; et al. gamma-Secretase inhibition increases efficacy of BCMA-specific chimeric antigen receptor T cells in multiple myeloma. Blood 2019, 134, 1585–1597. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Zuo, S.; Deng, B.; Xu, X.; Li, C.; Zheng, Q.; Ling, Z.; Song, W.; Xu, J.; Duan, J.; et al. Sequential CD19-22 CAR T therapy induces sustained remission in children with r/r B-ALL. Blood 2020, 135, 387–391. [Google Scholar] [CrossRef]
- Wang, N.; Hu, X.; Cao, W.; Li, C.; Xiao, Y.; Cao, Y.; Gu, C.; Zhang, S.; Chen, L.; Cheng, J.; et al. Efficacy and safety of CAR19/22 T-cell cocktail therapy in patients with refractory/relapsed B-cell malignancies. Blood 2020, 135, 17–27. [Google Scholar] [CrossRef]
- Dai, H.; Wu, Z.; Jia, H.; Tong, C.; Guo, Y.; Ti, D.; Han, X.; Liu, Y.; Zhang, W.; Wang, C.; et al. Bispecific CAR-T cells targeting both CD19 and CD22 for therapy of adults with relapsed or refractory B cell acute lymphoblastic leukemia. J. Hematol. Oncol. 2020, 13, 30. [Google Scholar] [CrossRef] [PubMed]
- Grada, Z.; Hegde, M.; Byrd, T.; Shaffer, D.R.; Ghazi, A.; Brawley, V.S.; Corder, A.; Schonfeld, K.; Koch, J.; Dotti, G.; et al. TanCAR: A Novel Bispecific Chimeric Antigen Receptor for Cancer Immunotherapy. Mol. Ther. Nucleic Acids 2013, 2, e105. [Google Scholar] [CrossRef]
- Hegde, M.; Mukherjee, M.; Grada, Z.; Pignata, A.; Landi, D.; Navai, S.A.; Wakefield, A.; Fousek, K.; Bielamowicz, K.; Chow, K.K.; et al. Tandem CAR T cells targeting HER2 and IL13Ralpha2 mitigate tumor antigen escape. J. Clin. Investig. 2016, 126, 3036–3052. [Google Scholar] [CrossRef] [Green Version]
- Schneider, D.; Xiong, Y.; Wu, D.; Nölle, V.; Schmitz, S.; Haso, W.; Kaiser, A.; Dropulic, B.; Orentas, R.J. A tandem CD19/CD20 CAR lentiviral vector drives on-target and off-target antigen modulation in leukemia cell lines. J. Immunother. Cancer 2017, 5, 42. [Google Scholar] [CrossRef] [PubMed]
- Huehls, A.M.; Coupet, T.A.; Sentman, C.L. Bispecific T-cell engagers for cancer immunotherapy. Immunol. Cell Biol. 2015, 93, 290–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwahori, K.; Kakarla, S.; Velasquez, M.P.; Yu, F.; Yi, Z.; Gerken, C.; Song, X.T.; Gottschalk, S. Engager T cells: A new class of antigen-specific T cells that redirect bystander T cells. Mol. Ther. 2015, 23, 171–178. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.G.; Marks, I.; Srinivasarao, M.; Kanduluru, A.K.; Mahalingam, S.M.; Liu, X.; Chu, H.; Low, P.S. Use of a Single CAR T Cell and Several Bispecific Adapters Facilitates Eradication of Multiple Antigenically Different Solid Tumors. Cancer Res. 2019, 79, 387–396. [Google Scholar] [CrossRef] [Green Version]
- Sutherland, A.R.; Owens, M.N.; Geyer, C.R. Modular Chimeric Antigen Receptor Systems for Universal CAR T Cell Retargeting. Int. J. Mol. Sci. 2020, 21, 7222. [Google Scholar] [CrossRef] [PubMed]
- Urbanska, K.; Lanitis, E.; Poussin, M.; Lynn, R.C.; Gavin, B.P.; Kelderman, S.; Yu, J.; Scholler, N.; Powell, D.J., Jr. A universal strategy for adoptive immunotherapy of cancer through use of a novel T-cell antigen receptor. Cancer Res. 2012, 72, 1844–1852. [Google Scholar] [CrossRef] [Green Version]
- Brown, C.E.; Aguilar, B.; Starr, R.; Yang, X.; Chang, W.C.; Weng, L.; Chang, B.; Sarkissian, A.; Brito, A.; Sanchez, J.F.; et al. Optimization of IL13Ralpha2-Targeted Chimeric Antigen Receptor T Cells for Improved Anti-tumor Efficacy against Glioblastoma. Mol. Ther. 2018, 26, 31–44. [Google Scholar] [CrossRef] [Green Version]
- Whilding, L.M.; Halim, L.; Draper, B.; Parente-Pereira, A.C.; Zabinski, T.; Davies, D.M.; Maher, J. CAR T-Cells Targeting the Integrin alphavbeta6 and Co-Expressing the Chemokine Receptor CXCR2 Demonstrate Enhanced Homing and Efficacy against Several Solid Malignancies. Cancers 2019, 11, 674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, L.; Tao, H.; Karachi, A.; Long, Y.; Hou, A.Y.; Na, M.; Dyson, K.A.; Grippin, A.J.; Deleyrolle, L.P.; Zhang, W.; et al. CXCR1- or CXCR2-modified CAR T cells co-opt IL-8 for maximal antitumor efficacy in solid tumors. Nat. Commun. 2019, 10, 4016. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Rui, W.; Zheng, H.; Huang, D.; Yu, F.; Zhang, Y.; Dong, J.; Zhao, X.; Lin, X. CXCR2-modified CAR-T cells have enhanced trafficking ability that improves treatment of hepatocellular carcinoma. Eur. J. Immunol. 2020, 50, 712–724. [Google Scholar] [CrossRef]
- Gregor, C.E.; Foeng, J.; Comerford, I.; McColl, S.R. Chemokine-Driven CD4(+) T Cell Homing: New Concepts and Recent Advances. Adv. Immunol. 2017, 135, 119–181. [Google Scholar] [CrossRef]
- Nagarsheth, N.; Wicha, M.S.; Zou, W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat. Rev. Immunol. 2017, 17, 559–572. [Google Scholar] [CrossRef] [Green Version]
- Guedan, S.; Posey, A.D., Jr.; Shaw, C.; Wing, A.; Da, T.; Patel, P.R.; McGettigan, S.E.; Casado-Medrano, V.; Kawalekar, O.U.; Uribe-Herranz, M.; et al. Enhancing CAR T cell persistence through ICOS and 4-1BB costimulation. JCI Insight 2018, 3, e96976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turtle, C.J.; Hanafi, L.A.; Berger, C.; Gooley, T.A.; Cherian, S.; Hudecek, M.; Sommermeyer, D.; Melville, K.; Pender, B.; Budiarto, T.M.; et al. CD19 CAR-T cells of defined CD4+:CD8+ composition in adult B cell ALL patients. J. Clin. Investig. 2016, 126, 2123–2138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jensen, M.C.; Popplewell, L.; Cooper, L.J.; DiGiusto, D.; Kalos, M.; Ostberg, J.R.; Forman, S.J. Antitransgene rejection responses contribute to attenuated persistence of adoptively transferred CD20/CD19-specific chimeric antigen receptor redirected T cells in humans. Biol. Blood Marrow Transplant. 2010, 16, 1245–1256. [Google Scholar] [CrossRef] [Green Version]
- Lamers, C.H.; Gratama, J.W.; Pouw, N.M.; Langeveld, S.C.; Krimpen, B.A.; Kraan, J.; Stoter, G.; Debets, R. Parallel detection of transduced T lymphocytes after immunogene therapy of renal cell cancer by flow cytometry and real-time polymerase chain reaction: Implications for loss of transgene expression. Hum. Gene Ther. 2005, 16, 1452–1462. [Google Scholar] [CrossRef] [PubMed]
- Van der Stegen, S.J.; Hamieh, M.; Sadelain, M. The pharmacology of second-generation chimeric antigen receptors. Nat. Rev. Drug Discov. 2015, 14, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Sadelain, M.; Brentjens, R.; Riviere, I. The basic principles of chimeric antigen receptor design. Cancer Discov. 2013, 3, 388–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Zhang, M.; Ramos, C.A.; Durett, A.; Liu, E.; Dakhova, O.; Liu, H.; Creighton, C.J.; Gee, A.P.; Heslop, H.E.; et al. Closely related T-memory stem cells correlate with in vivo expansion of CAR.CD19-T cells and are preserved by IL-7 and IL-15. Blood 2014, 123, 3750–3759. [Google Scholar] [CrossRef] [Green Version]
- Sallusto, F.; Geginat, J.; Lanzavecchia, A. Central memory and effector memory T cell subsets: Function, generation, and maintenance. Annu. Rev. Immunol. 2004, 22, 745–763. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Borresen-Dale, A.L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef] [Green Version]
- Lim, W.A.; June, C.H. The Principles of Engineering Immune Cells to Treat Cancer. Cell 2017, 168, 724–740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liot, S.; Balas, J.; Aubert, A.; Prigent, L.; Mercier-Gouy, P.; Verrier, B.; Bertolino, P.; Hennino, A.; Valcourt, U.; Lambert, E. Stroma Involvement in Pancreatic Ductal Adenocarcinoma: An Overview Focusing on Extracellular Matrix Proteins. Front. Immunol. 2021, 12, 612271. [Google Scholar] [CrossRef] [PubMed]
- Himes, B.T.; Geiger, P.A.; Ayasoufi, K.; Bhargav, A.G.; Brown, D.A.; Parney, I.F. Immunosuppression in Glioblastoma: Current Understanding and Therapeutic Implications. Front. Oncol. 2021, 11, 770561. [Google Scholar] [CrossRef]
- Walker, A.J.; Majzner, R.G.; Zhang, L.; Wanhainen, K.; Long, A.H.; Nguyen, S.M.; Lopomo, P.; Vigny, M.; Fry, T.J.; Orentas, R.J.; et al. Tumor Antigen and Receptor Densities Regulate Efficacy of a Chimeric Antigen Receptor Targeting Anaplastic Lymphoma Kinase. Mol. Ther. 2017, 25, 2189–2201. [Google Scholar] [CrossRef] [Green Version]
- Xiao, X.; Wang, Y.; Zou, Z.; Yang, Y.; Wang, X.; Xin, X.; Tu, S.; Li, Y. Combination strategies to optimize the efficacy of chimeric antigen receptor T cell therapy in haematological malignancies. Front. Immunol. 2022, 13, 954235. [Google Scholar] [CrossRef] [PubMed]
- Perica, K.; Flynn, J.; Curran, K.J.; Rivere, I.; Wang, X.; Senechal, B.; Halton, E.; Diamonte, C.; Pineda, J.; Bernal, Y.; et al. Impact of bridging chemotherapy on clinical outcome of CD19 CAR T therapy in adult acute lymphoblastic leukemia. Leukemia 2021, 35, 3268–3271. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, X.; Chen, D.; Yu, J. Radiotherapy combined with immunotherapy: The dawn of cancer treatment. Signal. Transduct. Target Ther. 2022, 7, 258. [Google Scholar] [CrossRef]
- Funk, C.R.; Wang, S.; Chen, K.Z.; Waller, A.; Sharma, A.; Edgar, C.L.; Gupta, V.A.; Chandrakasan, S.; Zoine, J.T.; Fedanov, A.; et al. PI3Kdelta/gamma inhibition promotes human CART cell epigenetic and metabolic reprogramming to enhance antitumor cytotoxicity. Blood 2022, 139, 523–537. [Google Scholar] [CrossRef]
- Zheng, W.; O’Hear, C.E.; Alli, R.; Basham, J.H.; Abdelsamed, H.A.; Palmer, L.E.; Jones, L.L.; Youngblood, B.; Geiger, T.L. PI3K orchestration of the in vivo persistence of chimeric antigen receptor-modified T cells. Leukemia 2018, 32, 1157–1167. [Google Scholar] [CrossRef]
- Kotla, V.; Goel, S.; Nischal, S.; Heuck, C.; Vivek, K.; Das, B.; Verma, A. Mechanism of action of lenalidomide in hematological malignancies. J. Hematol. Oncol. 2009, 2, 36. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Walter, M.; Urak, R.; Weng, L.; Huynh, C.; Lim, L.; Wong, C.W.; Chang, W.C.; Thomas, S.H.; Sanchez, J.F.; et al. Lenalidomide Enhances the Function of CS1 Chimeric Antigen Receptor-Redirected T Cells Against Multiple Myeloma. Clin. Cancer Res. 2018, 24, 106–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Works, M.; Soni, N.; Hauskins, C.; Sierra, C.; Baturevych, A.; Jones, J.C.; Curtis, W.; Carlson, P.; Johnstone, T.G.; Kugler, D.; et al. Anti-B-cell Maturation Antigen Chimeric Antigen Receptor T cell Function against Multiple Myeloma Is Enhanced in the Presence of Lenalidomide. Mol. Cancer Ther. 2019, 18, 2246–2257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chong, E.A.; Alanio, C.; Svoboda, J.; Nasta, S.D.; Landsburg, D.J.; Lacey, S.F.; Ruella, M.; Bhattacharyya, S.; Wherry, E.J.; Schuster, S.J. Pembrolizumab for B-cell lymphomas relapsing after or refractory to CD19-directed CAR T-cell therapy. Blood 2022, 139, 1026–1038. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Lu, W.; Sun, R.; Jin, X.; Cheng, L.; He, X.; Wang, L.; Yuan, T.; Lyu, C.; Zhao, M. Anti-CD19 Chimeric Antigen Receptor T Cells in Combination With Nivolumab Are Safe and Effective Against Relapsed/Refractory B-Cell Non-hodgkin Lymphoma. Front. Oncol. 2019, 9, 767. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Chen, Y.; Jing, Y.; Green, M.R.; Han, L. Advancing CAR T cell therapy through the use of multidimensional omics data. Nat. Rev. Clin. Oncol. 2023, 20, 211–228. [Google Scholar] [CrossRef]
- Dannenfelser, R.; Allen, G.M.; VanderSluis, B.; Koegel, A.K.; Levinson, S.; Stark, S.R.; Yao, V.; Tadych, A.; Troyanskaya, O.G.; Lim, W.A. Discriminatory Power of Combinatorial Antigen Recognition in Cancer T Cell Therapies. Cell Syst. 2020, 11, 215–228. [Google Scholar] [CrossRef]
- Hu, Z.; Yuan, J.; Long, M.; Jiang, J.; Zhang, Y.; Zhang, T.; Xu, M.; Fan, Y.; Tanyi, J.L.; Montone, K.T.; et al. The Cancer Surfaceome Atlas integrates genomic, functional and drug response data to identify actionable targets. Nat. Cancer 2021, 2, 1406–1422. [Google Scholar] [CrossRef]
- Olivier, M.; Asmis, R.; Hawkins, G.A.; Howard, T.D.; Cox, L.A. The Need for Multi-Omics Biomarker Signatures in Precision Medicine. Int. J. Mol. Sci. 2019, 20, 4781. [Google Scholar] [CrossRef] [Green Version]
- Perna, F.; Berman, S.H.; Soni, R.K.; Mansilla-Soto, J.; Eyquem, J.; Hamieh, M.; Hendrickson, R.C.; Brennan, C.W.; Sadelain, M. Integrating Proteomics and Transcriptomics for Systematic Combinatorial Chimeric Antigen Receptor Therapy of AML. Cancer Cell 2017, 32, 506–519. [Google Scholar] [CrossRef] [Green Version]
- Cho, J.H.; Collins, J.J.; Wong, W.W. Universal Chimeric Antigen Receptors for Multiplexed and Logical Control of T Cell Responses. Cell 2018, 173, 1426–1438. [Google Scholar] [CrossRef] [Green Version]
- Cho, J.H.; Okuma, A.; Sofjan, K.; Lee, S.; Collins, J.J.; Wong, W.W. Engineering advanced logic and distributed computing in human CAR immune cells. Nat. Commun. 2021, 12, 792. [Google Scholar] [CrossRef]
- Garcia-Prieto, C.A.; Villanueva, L.; Bueno-Costa, A.; Davalos, V.; Gonzalez-Navarro, E.A.; Juan, M.; Urbano-Ispizua, A.; Delgado, J.; Ortiz-Maldonado, V.; Del Bufalo, F.; et al. Epigenetic Profiling and Response to CD19 Chimeric Antigen Receptor T-Cell Therapy in B-Cell Malignancies. J. Natl. Cancer Inst. 2022, 114, 436–445. [Google Scholar] [CrossRef]
- Lynn, R.C.; Weber, E.W.; Sotillo, E.; Gennert, D.; Xu, P.; Good, Z.; Anbunathan, H.; Lattin, J.; Jones, R.; Tieu, V.; et al. c-Jun overexpression in CAR T cells induces exhaustion resistance. Nature 2019, 576, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Park, J.J.; Peng, L.; Yang, Q.; Chow, R.D.; Dong, M.B.; Lam, S.Z.; Guo, J.; Tang, E.; Zhang, Y.; et al. A genome-scale gain-of-function CRISPR screen in CD8 T cells identifies proline metabolism as a means to enhance CAR-T therapy. Cell Metab. 2022, 34, 595–614. [Google Scholar] [CrossRef]
- Blache, U.; Weiss, R.; Boldt, A.; Kapinsky, M.; Blaudszun, A.R.; Quaiser, A.; Pohl, A.; Miloud, T.; Burgaud, M.; Vucinic, V.; et al. Advanced Flow Cytometry Assays for Immune Monitoring of CAR-T Cell Applications. Front. Immunol. 2021, 12, 658314. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, L.; Haas, E.R.; Vyas, V.; Urak, R.; Forman, S.J.; Wang, X. Single-cell analysis by mass cytometry reveals CD19 CAR T cell spatiotemporal plasticity in patients. OncoImmunology 2022, 11, 2040772. [Google Scholar] [CrossRef] [PubMed]
- Volpe, A.; Nagle, V.L.; Lewis, J.S.; Ponomarev, V. Predicting CAR-T cell Immunotherapy Success through ImmunoPET. Clin. Cancer Res. 2021, 27, 911–912. [Google Scholar] [CrossRef]
- Simonetta, F.; Alam, I.S.; Lohmeyer, J.K.; Sahaf, B.; Good, Z.; Chen, W.; Xiao, Z.; Hirai, T.; Scheller, L.; Engels, P.; et al. Molecular Imaging of Chimeric Antigen Receptor T Cells by ICOS-ImmunoPET. Clin. Cancer Res. 2021, 27, 1058–1068. [Google Scholar] [CrossRef]
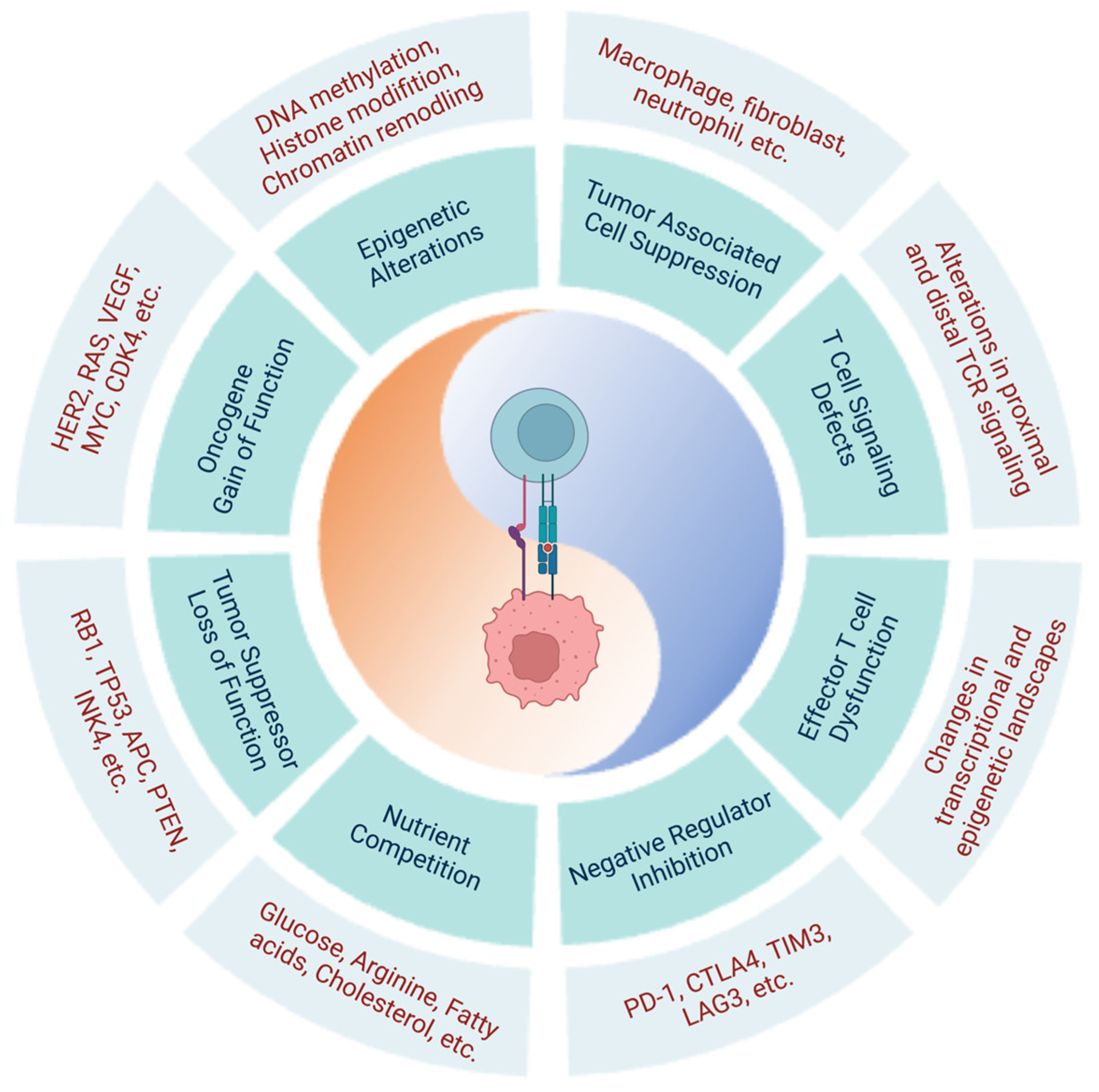

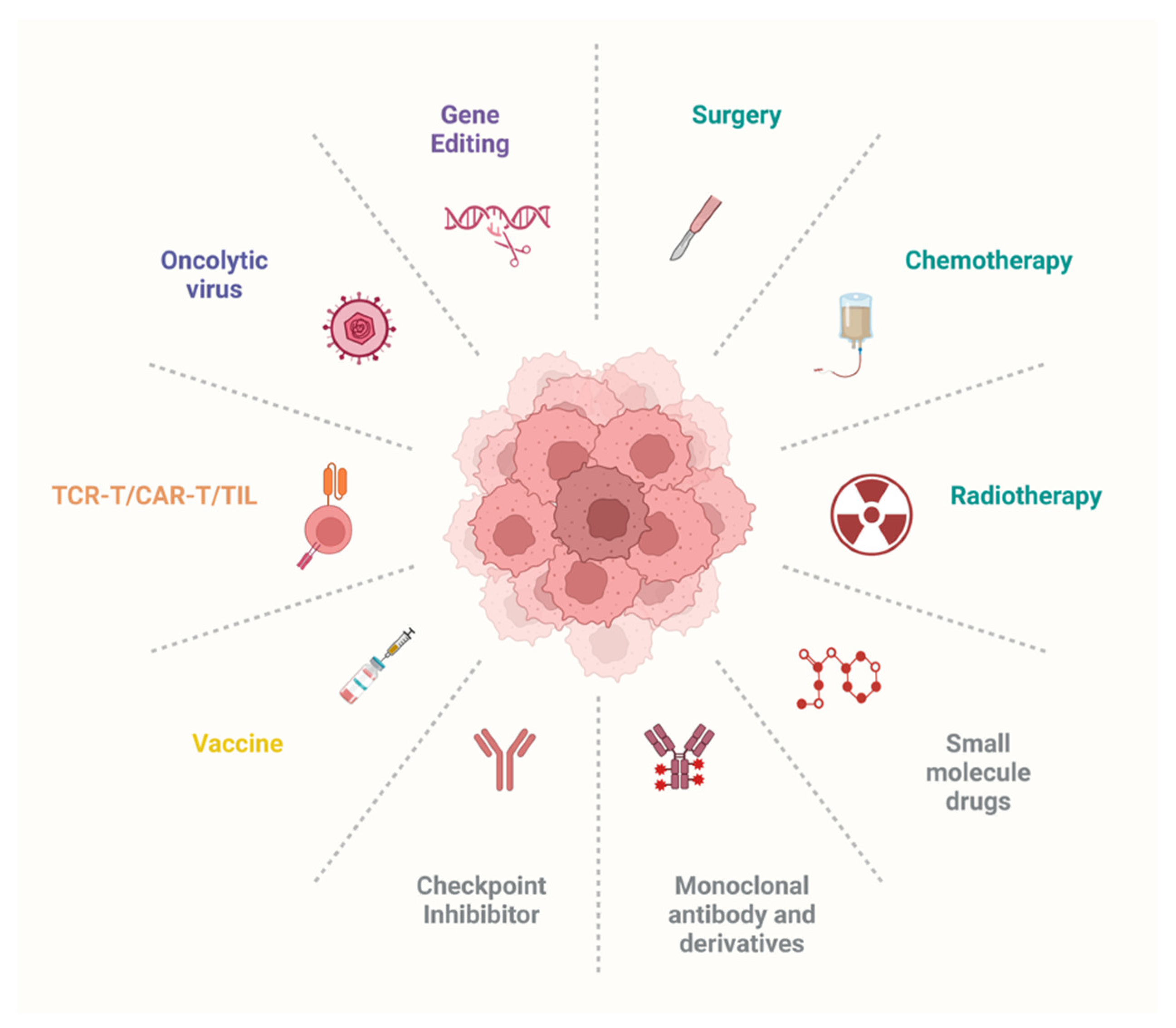
| NCT ID | Official Study Title | Target Antigen | Trial Phase | Targeted Conditions | Related Publications |
|---|---|---|---|---|---|
| Clinical trials of CAR T therapies | |||||
| NCT04030195 | A Phase 1/2a, Open-label, Dose-escalation, Dose-expansion, Parallel Assignment Study to Evaluate the Safety and Clinical Activity of PBCAR20A in Subjects with Relapsed/Refractory (r/r) Non-Hodgkin Lymphoma (NHL), or r/r Chronic Lymphocytic Leukemia (CLL), or Small Lymphocytic Lymphoma (SLL) | Cluster of differentiation 20 (CD20) | Phase I/IIa | Non-Hodgkin lymphoma, chronic lymphocytic leukemia, small lymphocytic lymphoma | |
| NCT03958656 | A Phase I Clinical Trial of T-cells Expressing an Anti-SLAMF7 CAR for Treating Multiple Myeloma | Signaling lymphocytic activation molecule F7 (SLAM7) | Phase I | Multiple myeloma | |
| NCT03338972 | A Phase I Study of Adoptive Immunotherapy for Advanced B Cell Maturation Antigen (BCMA)+ Multiple Myeloma with Autologous CD4+ and CD8+ T Cells Engineered to Express a BCMA-Specific Chimeric Antigen Receptor | BCMA | Phase I | Multiple myeloma | |
| NCT03289455 | A Single-Arm, Open-Label, Multi-Centre, Phase I/II Study Evaluating the Safety and Clinical Activity Of AUTO3, a CAR T Cell Treatment Targeting CD19 And CD22 in Pediatric And Young Adult Patients With Relapsed or Refractory B Cell Acute Lymphoblastic Leukemia | CD19, CD22 | Phase I/II | B cell acute lymphoblastic leukemias | |
| NCT03049449 | Anti-CD30 CAR T Cells with Fully-human Binding Domains for Treating CD30-expressing Lymphomas Including Anaplastic Large Cell Lymphomas | CD30 | Phase 1 | Lymphomas | |
| NCT03019055 | Phase 1/1b Study of Redirected Autologous T Cells Engineered to Contain an Anti CD19 and Anti CD20 scFv Coupled to CD3ζ and 4-1BB Signaling Domains in Patients with Relapsed and/or Refractory CD19 or CD20 Positive B Cell Malignancies | CD19, CD20 | Phase I/Ib | Lymphoma, non-Hodgkin lymphoma, B cell chronic lymphocytic leukemia, small lymphocytic lymphoma | |
| NCT02761915 | A Cancer Research UK Phase I Trial of Anti-GD2 Chimeric Antigen Receptor (CAR) Transduced T-cells (1RG-CART) in Patients with Relapsed or Refractory Neuroblastoma | disialoganglioside (GD2) | Phase I | Neuroblastoma | [14] |
| NCT02659943 | T Cells Expressing a Fully-Human Anti-CD19 Chimeric Antigen Receptor for Treating B cell Malignancies | CD19 | Phase I | B cell lymphoma, non-Hodgkin’s lymphoma | [15] |
| NCT02215967 | A Phase I Clinical Trial of T-Cells Targeting B Cell Maturation Antigen for Previously Treated Multiple Myeloma | BCMA | Phase I | Multiple myeloma | [16,17,18] |
| NCT02030847 | Phase II Study of Redirected Autologous T Cells Engineered to Contain Anti-CD19 Attached to TCR and 4-1BB Signaling Domains in Patients with Chemotherapy Resistant or Refractory Acute Lymphoblastic Leukemia | CD19 | Phase II | B cell acute lymphoblastic leukemias | [19] |
| NCT01583686 | Phase I/II Study of Metastatic Cancer Using Lymphodepleting Conditioning Followed by Infusion of Anti-mesothelin Gene Engineered Lymphocytes | Mesothelin | Phase I/II | Cervical, pancreatic, ovarian, lung cancers; mesothelioma | [20,21] |
| NCT00924287 | Phase I/II Study of Metastatic Cancer That Expresses Her-2 Using Lymphodepleting Conditioning Followed by Infusion of Anti-Her-2 Gene Engineered Lymphocytes | HER-2 | Phase I/II | Metastatic cancers | [22,23] |
| NCT01460901 | Phase I Study of Donor Derived, Gene Modified, Multi-virus-specific, Cytotoxic T-Lymphocytes Redirected to GD2 for Relapsed/Refractory Neuroblastoma Post-allo Stem Cell Transplantation with Submyeloblative Conditioning | disialoganglioside (GD2) | Phase I | Neuroblastoma | |
| NCT01454596 | A Phase I/II Study of the Safety and Feasibility of Administering T Cells Expressing Anti-EGFRvIII Chimeric Antigen Receptor to Patients with Malignant Gliomas Expressing EGFRvIII | Epidermal growth factor receptor variant III (EGFRvIII) | Phase I/II | Malignant glioma, glioblastoma, brain cancer, gliosarcoma | |
| NCT01029366 | Pilot Study of Redirected Autologous T-cells Engineered to Contain Anti-CD19 Attached to TCR and 4-1BB Signaling Domains in Patient with Chemotherapy Resistant or Refractory CD19+ Leukemia and Lymphoma | CD19 | Phase I | Leukemias, lymphomas | [19,24,25] |
| Clinical trials of TCR T therapies | |||||
| NCT02992743 | A Pilot Study of NY-ESO-1c259T Cells in Subjects with Advanced Myxoid/Round Cell Liposarcoma | New York esophageal antigen-1 (NY-ESO-1) | Phase II | Liposarcoma | |
| NCT02650986 | A Phase I/IIa Study of TGFß Blockade in TCR-Engineered T Cell Cancer Immunotherapy in Patients with Advanced Malignancies | NY-ESO-1 | Phase I/IIa | Various carcinomas, neoplasms, sarcomas | |
| NCT02588612 | A Pilot Open-Label Clinical Trial Evaluating the Safety and Efficacy of Autologous T Cells Expressing Enhanced TCRs Specific for NY-ESO-1 in Subjects with Stage IIIb or Stage IV Non-Small Cell Lung Cancer (NSCLC) | NY-ESO-1 | Phase I | Non-small cell lung cancer | |
| NCT02280811 | A Phase I/II Study of T Cell Receptor Gene Therapy Targeting HPV-16 E6 for HPV-Associated Cancers | HPV-16 E6 | Phase I/II | (HPV)-16+ cancers (cervical, vulvar, vaginal, penile, anal, and oropharyngeal cancers) | [26,27,28] |
| NCT02111850 | A Phase I/II Study of the Treatment of Metastatic Cancer That Expresses MAGE-A3 Using Lymphodepleting Conditioning Followed by Infusion of HLA-DP0401/0402 Restricted Anti-MAGE-A3 TCR-Gene Engineered Lymphocytes and Aldesleukin | Melanoma antigen family A, 3 (MAGE-A3) | Phase I/II | Cervical, renal, urothelial, breast cancers; melanoma | |
| NCT04015336 | A Phase II Study of E7 TCR T Cell Induction Immunotherapy for Stage II and Stage III HPV-Associated Oropharyngeal Cancer | E7 (HPV oncoprotein) | Phase II | Papillomavirus infections, oropharyngeal neoplasms | |
| NCT01567891 | A Phase I/IIa, Open Label Clinical Trial Evaluating the Safety and Efficacy of Autologous T Cells Expressing Enhanced TCRs Specific for NY-ESO-1 in Patients with Recurrent or Treatment Refractory Ovarian Cancer. | NY-ESO-1 | Phase I/IIa | Ovarian cancer | |
| NCT00393029 | Phase II Study of Metastatic Cancer That Overexpresses p53 Using Lymphodepleting Conditioning Followed by Infusion of Anti-p53 T Cell Receptor (TCR)-Gene Engineered Lymphocytes | Protein 53 (p53) | Phase II | Metastatic cancers overexpressing p53 | |
| NCT01343043 | A Pilot Study of Genetically Engineered NY-ESO-1 Specific NY-ESO-1ᶜ2⁵⁹T in HLA-A2+ Patients with Synovial Sarcoma (NY-ESO-1) | NY-ESO-1 | Phase I | Neoplasms | [29,30,31] |
| NCT00509288 | Phase II Study of Metastatic Melanoma Using Lymphodepleting Conditioning Followed by Infusion of Anti-MART-1 F5 TCR-Gene Engineered Lymphocytes | Melanoma antigen recognized by T-cells (MART-1) | Phase II | Melanoma, skin cancer | [32,33,34,35] |
| Molecule | Alteration | Consequence | Associated Malignancy | References | ||
|---|---|---|---|---|---|---|
| Negative regulators | Cell Surface | PD-1 | Overexpressed in T cells upon chronic antigen exposure; denotes exhausted phenotype | Limited T cell survival and function; associated with T cell dysfunction | Oncogenesis | [11,12,48,51,56,57,58,63,81,88,89,90,91,92,93,94,95,96,97,99,128,130,146] |
| TIM-3 | ||||||
| LAG-3 | ||||||
| CTLA-4 | ||||||
| BTLA | ||||||
| TIGIT | ||||||
| CD244 | ||||||
| CD160 | ||||||
| Intracellular | SHP-1 | Decreased expression of SHP-1 | Hyperactive TYK2 and JAK1 kinases | Lymphomagenesis | [106,107] | |
| SHP-2 | Lower levels of phosphorylated SHP-2 | Metastasis; increased release of inflammatory cytokines; accumulation of MDSCs | Melanoma (murine model) | [108] | ||
| Transcription & epigenetic factors | DUSP2 (PAC1) | Highly expressed in TILs | Recruitment of Mi-2β nucleosome-remodeling and histone-deacetylase complex; facilitated T cell exhaustion and loss of proliferative and effector functions | Various cancers | [134,135] | |
| Blimp-1 | Overexpressed in exhausted CD8+ T cells | Increased expression of inhibitory receptors; repression of memory CD8+ T cell differentiation | Various cancers | [54,84,128,131,132,146,147] | ||
| EOMES | Directly controls expression of TIM-3; antagonization of T-bet | |||||
| BATF | Expression of inhibitory receptors; repression of TCF1, a transcription factor required for memory T cell differentiation | |||||
| T-bet | Decreased expression in dysfunctional T cells | T-bet directly represses expression of PD1 | Various cancers | [54,128,129] | ||
| TOX | Critical for CD8+ T cell exhaustion | Required for gene expression of inhibitory receptors (e.g., Pdcd1, Entpd1, Havcr2, Cd244, and Tigit) | Various cancers | [54,55,57,98,118,119,120,121] | ||
| NR4A1 | Expressed at high levels in tolerant T cells | Various cancers | [110,112,113,114,115,116] | |||
| Critical for development of dysfunctional exhausted T cells | Attenuated T cell effector functions and upregulation of inhibitory receptors | [54,55,57,98,118,119,120,121] | ||||
| TET2 | Mutations | Heightened hypermethylation of T cell signaling/differentiation genes | Lymphomas | [75,77,78,143,144,145] | ||
| DNMT3A | ||||||
| IDH2 |
| Gene | Tumor Type | Reference | ||
|---|---|---|---|---|
| Genetics | Oncogenes | BCL2 | B cell leukemia, lymphoma | [148] |
| BCR/ABL | Leukemia | [149,150] | ||
| BRAF | Melanoma, papillary thyroid carcinoma | [151] | ||
| CDK4 | Various tumors | [152] | ||
| C-KIT | Acute myeloid leukemia, myelodysplastic syndrome (MDS), MDS-derived AML | [153] | ||
| EGFR | Lung tumor | [154,155] | ||
| ERBA | Breast cancer | [156] | ||
| FGF5 | Hepatocellular carcinoma, colorectal cancer, prostate cancer | [157,158] | ||
| FMS | Feline McDonough sarcoma | [159] | ||
| FOS | HNSCC, breast cancer, various tumors | [160] | ||
| GLI | Various tumors | [161] | ||
| HER2 | Breast tumor | [162] | ||
| HST | Esophageal cancer, stomach cancer | [163] | ||
| JUN | Various tumors | [164] | ||
| MDM2 | Various tumors | [165] | ||
| MET | Hereditary renal papillary carcinoma | [166] | ||
| MYC | Various tumors | [167] | ||
| RAS | Various tumors | [168,169,170,171] | ||
| RET | Papillary thyroid carcinoma | [172] | ||
| SIS | Breast tumor | [173] | ||
| SOS | Ras-related cancer | [174] | ||
| SRC | Various tumors | [175] | ||
| TTG | Various tumors | [176] | ||
| VEGF | Angiogenesis | [177] | ||
| VEGFR | Migration of cancer cells | [178,179] | ||
| Tumor suppressors | APC | Colon/rectum carcinoma, adenomatous polyposis | [180,181] | |
| ARF (p14) | Melanoma | [181] | ||
| BRCA1, BRCA2 | Breast, ovarian, pancreatic carcinomas | [180,181,182,183] | ||
| CDH1 | Gastric cancer | [181] | ||
| CHK 1/2 | Li-Fraumeni syndrome | [181] | ||
| DCC | Colorectal carcinoma | [181] | ||
| DPC4 | Pancreatic carcinoma | [180] | ||
| INK4 (p16) | Melanoma, lung carcinoma, brain tumors, leukemia, lymphoma | [180,181] | ||
| MADR2 | Colon/rectum carcinoma | [180] | ||
| MLH1, MSH2, MSH6 | Colorectal cancer | [181] | ||
| NF1 | Neurofibrosarcoma, neurofibromatosis type I | [180,181] | ||
| NF2 | Meningioma | [180] | ||
| PTC | Basal cell carcinoma | [180] | ||
| PTEN | Brain tumors; melanoma; prostate, endometrial, kidney, lung carcinomas | [180] | ||
| RB1 | Retinoblastoma | [184] | ||
| TP53 | Brain tumors; breast, colon/rectum, esophageal, liver, lung carcinomas; sarcomas; leukemias, lymphomas, Li-Fraumeni syndrome | [180,181] | ||
| VHL | Renal cell carcinoma | [180] | ||
| WT1, WT2 | Wilms’ tumor | [180,185] | ||
| Epigenetics | DNA methylation | AID | Chronic myeloid leukemia | [186] |
| DNMT1 | Colorectal, non-small-cell lung, pancreatic, gastric, breast cancer | [187,188] | ||
| DNMT3A | Myelodysplastic syndromes, acute myeloid leukemia | [189,190,191] | ||
| DNMT3B | ICF syndrome, SNPs in breast and lung adenoma | [192,193] | ||
| IDH1/2 | Glioma, acute myeloid leukemia | [194,195,196] | ||
| MADR2 | Colon/rectum carcinoma | [180] | ||
| MBD1/2 | Lung cancer, breast cancer | [197] | ||
| MLH1, MSH2, MSH6 | Colorectal cancer | [181] | ||
| MLL1/2/3 | Bladder TCC, hematopoietic, non-Hodgkin lymphoma, B cell lymphoma, prostate cancer | [198,199] | ||
| Histone modifiers | BMI-1 | Ovarian, mantle cell lymphomas, Merkel cell carcinomas | [200,201] | |
| CREBBP (CBP/KAT3A) | Gastric and colorectal, epithelial, ovarian, lung, esophageal cancer | [202] | ||
| EP300 (P300/KAT3B) | Breast, colorectal, pancreatic cancer | [202] | ||
| EZH2 | Breast, prostate, bladder, colon, pancreas, liver, gastric, uterine tumors, melanoma, lymphoma, myeloma, Ewing’s sarcoma | [203,204] | ||
| G9a | HCC, cervical, uterine, ovarian, breast cancer | [205] | ||
| HDAC2 | Colonic, gastric, endometrial cancer | [206] | ||
| JARID1B/C (KDM5C) | Testicular cancer, breast cancer, RCCC | [207] | ||
| LSD1 | Prostate | [207] | ||
| PCAF | Epithelial | [202] | ||
| PRMT1/5 | Breast, gastric cancer | [202] | ||
| SIRT1, HDAC5/7A | Breast, colorectal, prostate cancer | [202] | ||
| UTX (KDM6A) | Bladder, breast, kidney, lung, pancreas, esophagus, colon, uterus, brain, hematological malignancies | [207] | ||
| Chromatin remodelers | ARID1A (BAF250A) | Ovarian clear cell carcinomas, endometrioid carcinomas, endometrial carcinomas | [208,209] | |
| ARID2 (BAF200) | Primary pancreatic adenocarcinomas | [210] | ||
| BRD7 | Bladder TCC | [211] | ||
| BRM (SMARCA2) | Prostate, basal cell carcinoma | [212,213] | ||
| CHD4/5 | Colorectal and gastric cancer, ovarian, prostate, neuroblastoma, hematopoietic | [214,215,216] | ||
| CHD7 | Gastric and colorectal cancer | [217] | ||
| P400/Tip60 | Lymphoma, colon, head and neck, breast cancer | [218] | ||
| PBRM1 (BAF180) | Breast tumors | [219] | ||
| SNF5 (SMARCB1, INI1) | Lung, rhabdoid, medulloblastoma | [220] | ||
| SRCAP | Prostate | [221] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, C.; Liu, X.; Chang, C.-Y.; Wang, H.Y.; Wang, R.-F. The Interplay between T Cells and Cancer: The Basis of Immunotherapy. Genes 2023, 14, 1008. https://doi.org/10.3390/genes14051008
Chen C, Liu X, Chang C-Y, Wang HY, Wang R-F. The Interplay between T Cells and Cancer: The Basis of Immunotherapy. Genes. 2023; 14(5):1008. https://doi.org/10.3390/genes14051008
Chicago/Turabian StyleChen, Christina, Xin Liu, Che-Yu Chang, Helen Y. Wang, and Rong-Fu Wang. 2023. "The Interplay between T Cells and Cancer: The Basis of Immunotherapy" Genes 14, no. 5: 1008. https://doi.org/10.3390/genes14051008
APA StyleChen, C., Liu, X., Chang, C.-Y., Wang, H. Y., & Wang, R.-F. (2023). The Interplay between T Cells and Cancer: The Basis of Immunotherapy. Genes, 14(5), 1008. https://doi.org/10.3390/genes14051008





