Genetic Diversification of Starch Branching Enzymes during Maize Domestication and Improvement
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. DNA Isolation and ZmSBEs Resequencing
2.3. Determination of Maize Kernel Starch Content and Starch Pasting and Gelatinization Properties
2.4. Analysis of Sequence Data
3. Results
3.1. ZmSBE Sequence Polymorphisms
3.2. Analysis of Nucleotide Diversity and Selection of ZmSBE Genes in Teosinte, Landraces, and Inbred Lines
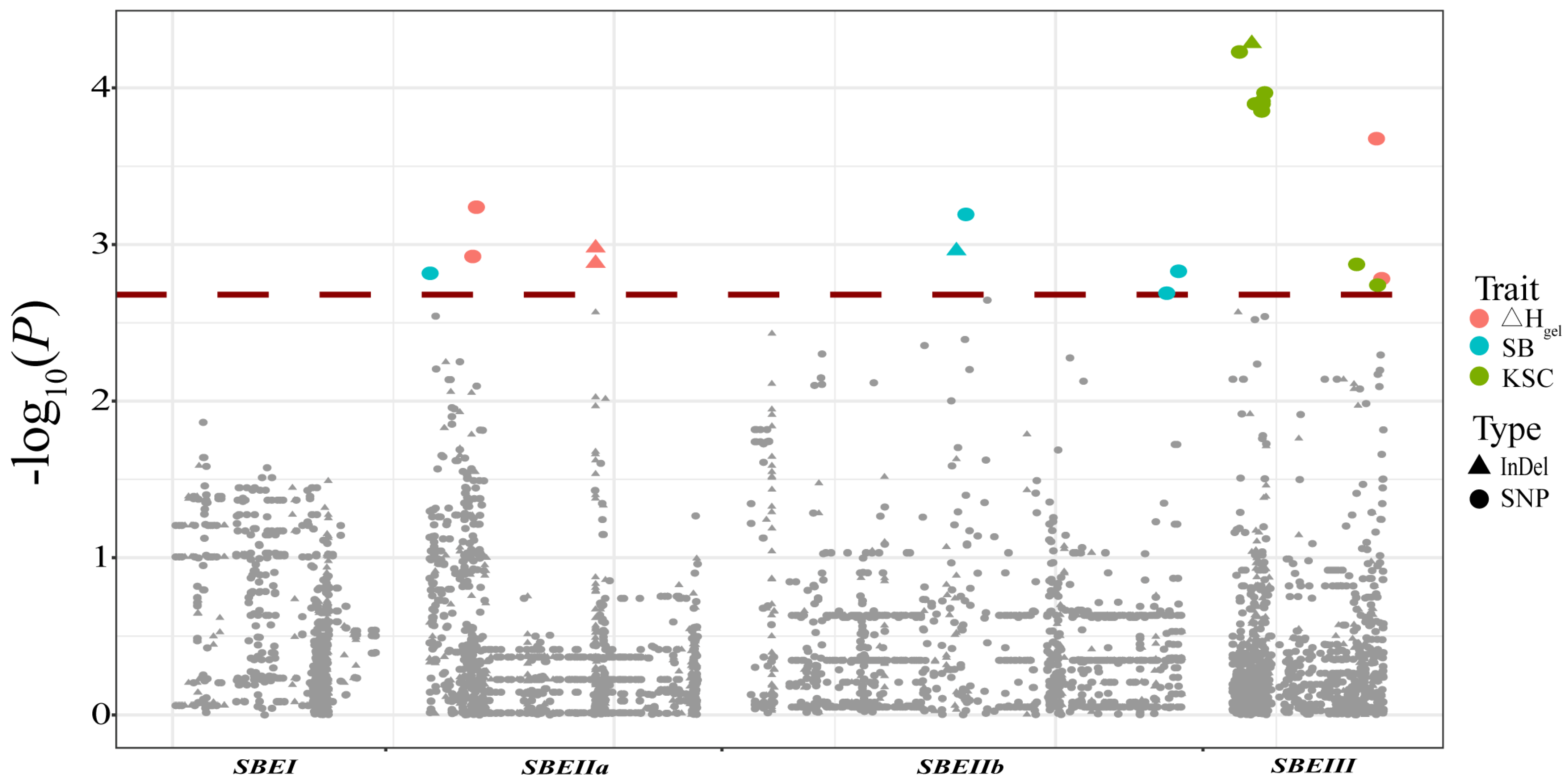
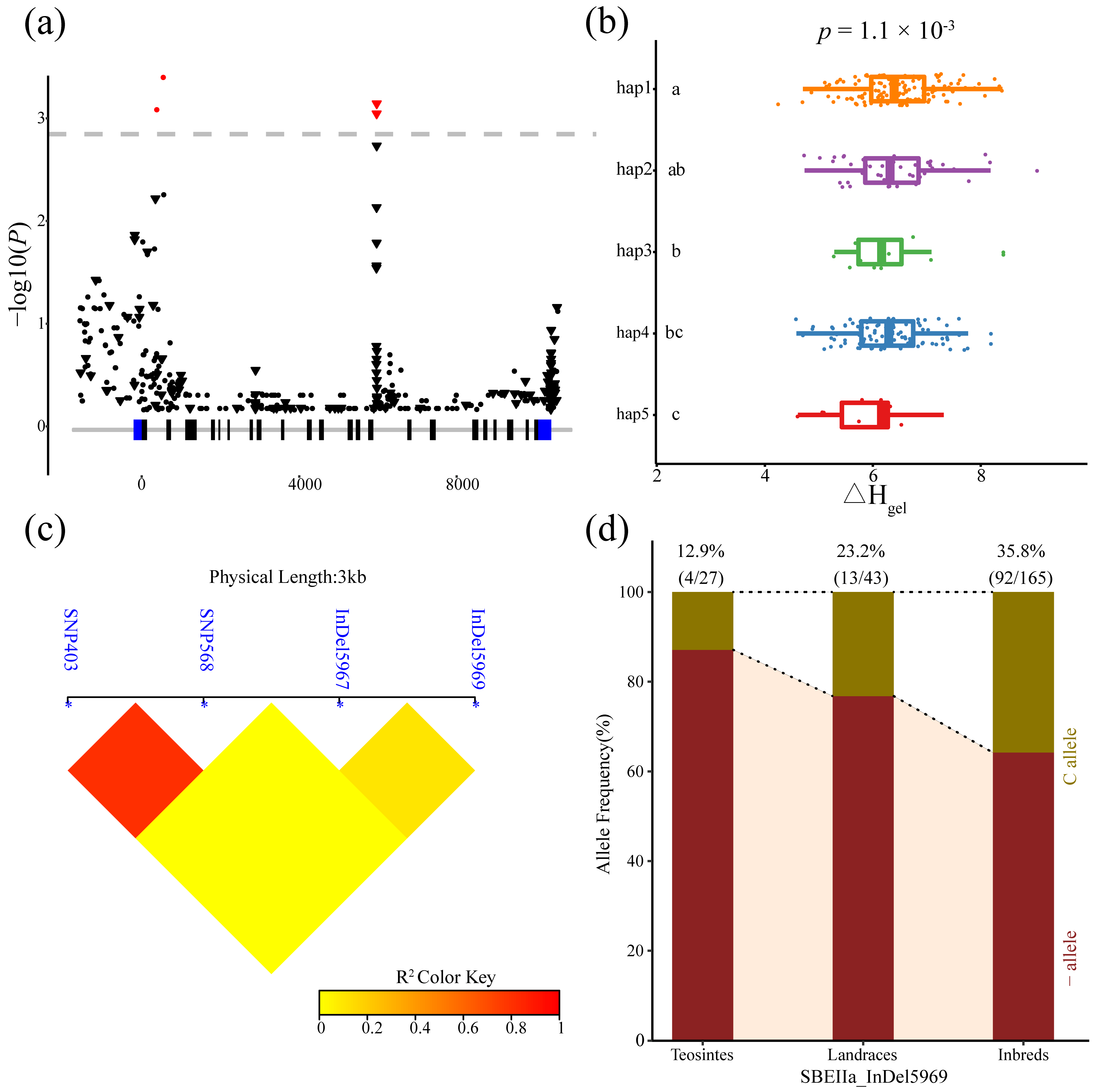
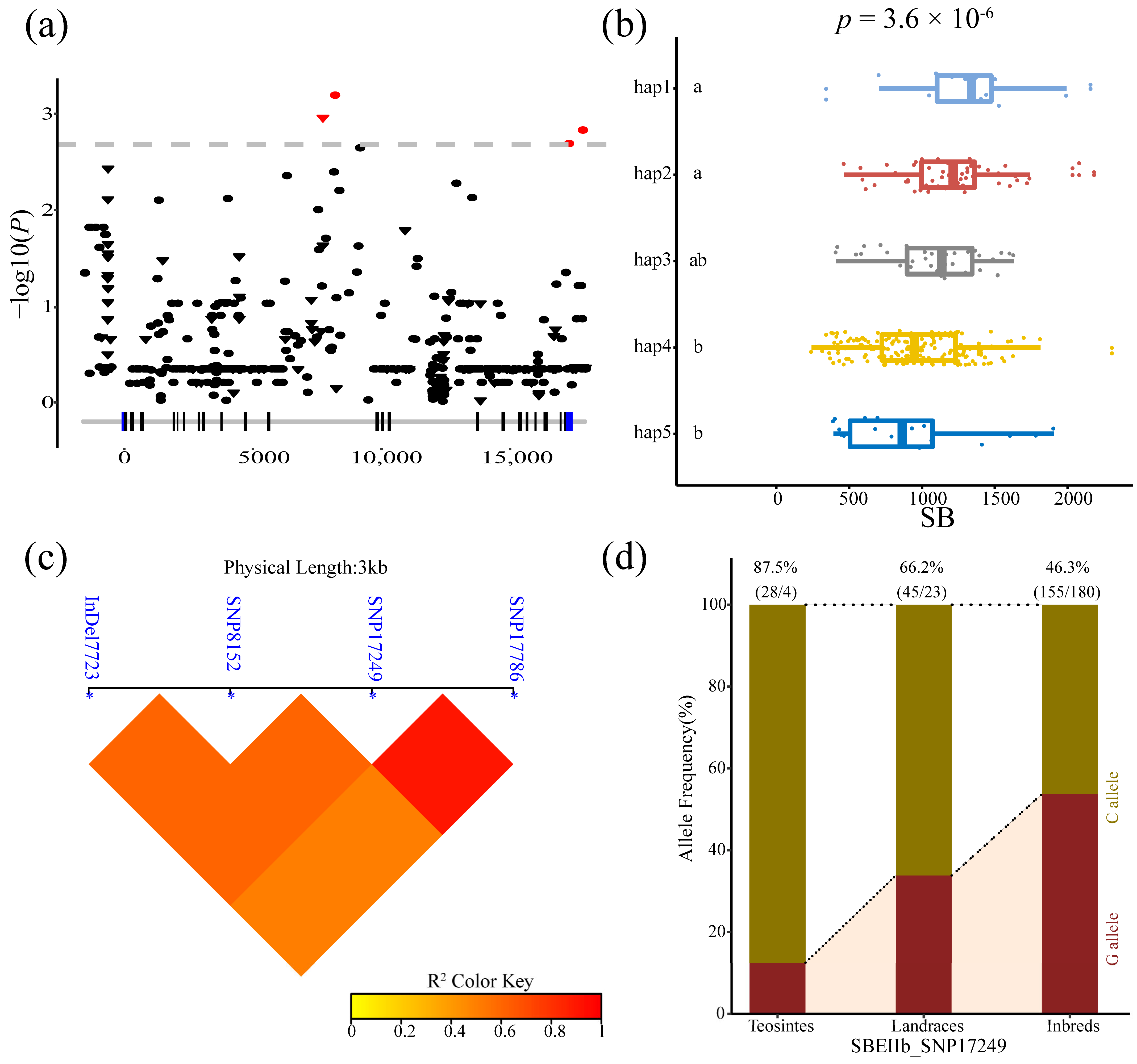
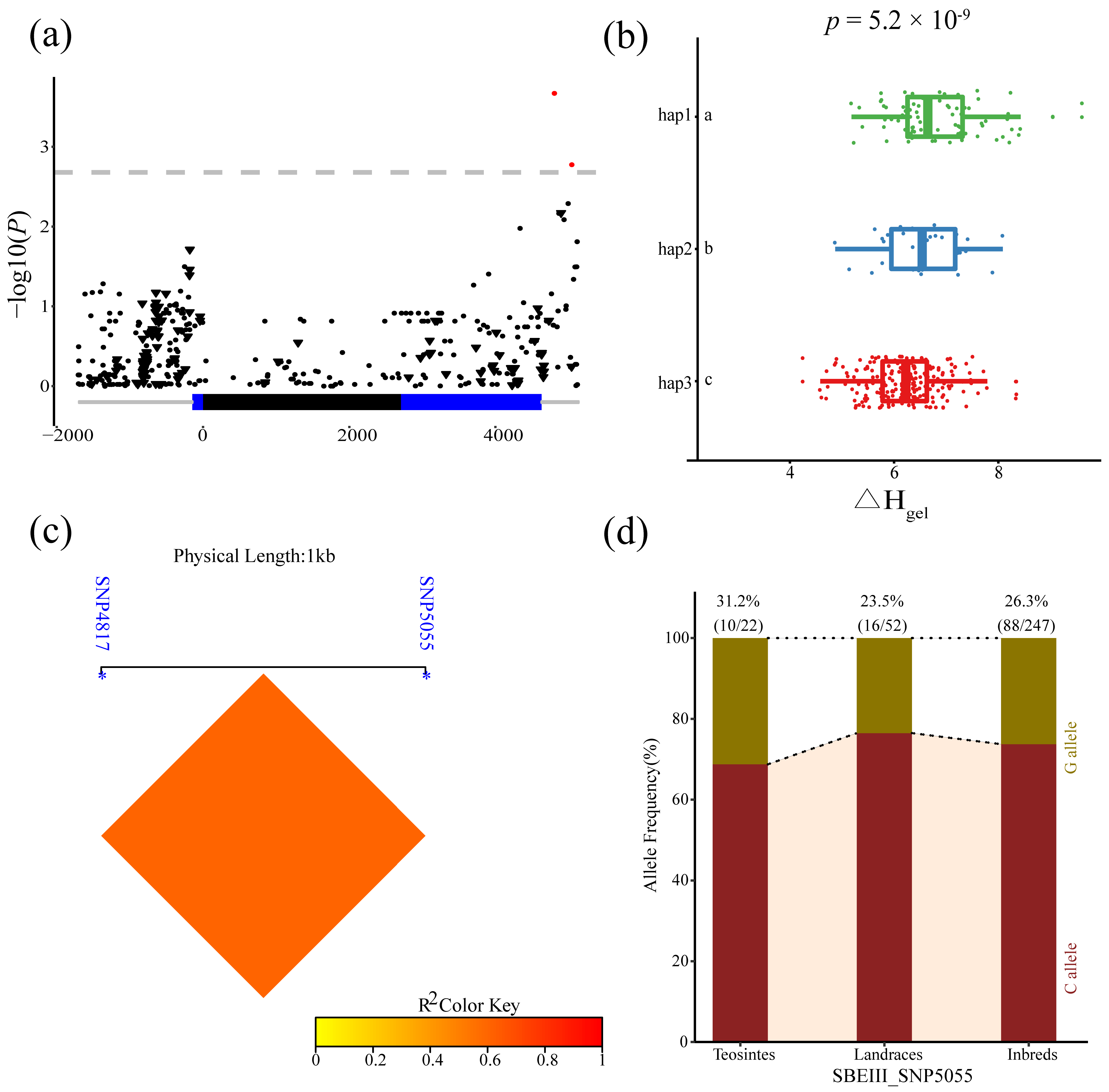
3.3. Analysis of the Association between Phenotypes and ZmSBEs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, J.; Chen, J.; Yi, Q.; Hu, Y.; Liu, H.; Liu, Y.; Huang, Y. Novel role of ZmaNAC36 in co-expression of starch synthetic genes in maize endosperm. Physiol. Mol. Biol. Plants 2014, 84, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Nelson, O.; Pan, D. Starch Synthesis in Maize Endosperms. Ann. Rev. Plant Physiol. Plant Mol. Biol. 1995, 46, 475–496. [Google Scholar] [CrossRef]
- Tetlow, I.J.; Emes, M.J. Starch Biosynthesis in the Developing Endosperms of Grasses and Cereals. Agronomy 2017, 7, 81. [Google Scholar] [CrossRef]
- Martin, C.; Smith, A.M. Starch Biosynthesis. Plant Cell 1995, 7, 971–985. [Google Scholar] [PubMed]
- Huang, L.; Tan, H.; Zhang, C.; Li, Q.; Liu, Q. Starch biosynthesis in cereal endosperms: An updated review over the last decade. Plant Commun. 2021, 2, 22. [Google Scholar] [CrossRef] [PubMed]
- Tetlow, I.J. Starch biosynthesis in developing seeds. Seed Sci. Res. 2011, 21, 5–32. [Google Scholar] [CrossRef]
- Mizuno, K.; Kawasaki, T.; Shimada, H.; Satoh, H.; Kobayashi, E.; Okumura, S.; Arai, Y.; Baba, T. Alteration of the structural properties of starch components by the lack of an isoform of starch branching enzyme in rice seeds. J. Biol. Chem. 1993, 268, 19084–19091. [Google Scholar] [CrossRef]
- Gao, M.; Fisher, D.K.; Kim, K.N.; Shannon, J.C.; Guiltinan, M.J. Independent Genetic Control of Maize Starch-Branching Enzymes IIa and IIb (Isolation and Characterization of a Sbe2a cDNA). Plant Physiol. 1997, 114, 69–78. [Google Scholar] [CrossRef]
- Regina, A.; Kosar-Hashemi, B.; Li, Z.; Pedler, A.; Mukai, Y. Starch branching enzyme IIb in wheat is expressed at low levels in the endosperm compared to other cereals and encoded at a non-syntenic locus. Planta 2005, 222, 899–909. [Google Scholar] [CrossRef]
- Han, Y.; Sun, F.J.; Rosales-Mendoza, S.; Korban, S.S. Three orthologs in rice, Arabidopsis, and Populus encoding starch branching enzymes (SBEs) are different from other SBE gene families in plants. Gene 2007, 401, 123–130. [Google Scholar] [CrossRef]
- Kang, G.; Li, S.; Zhang, M.; Peng, H.; Wang, C.; Zhu, Y.; Guo, T. Molecular cloning and expression analysis of the starch-branching enzyme III gene from common wheat (Triticum aestivum). Biochem. Genet. 2013, 51, 377–386. [Google Scholar] [CrossRef]
- Flint-Garcia, S.A.; Bodnar, A.L.; Scott, M.P. Wide variability in kernel composition, seed characteristics, and zein profiles among diverse maize inbreds, landraces, and teosinte. Theor. Appl. Genet. 2009, 119, 1129–1142. [Google Scholar] [CrossRef]
- Li, P.; Wei, J.; Wang, H.; Fang, Y.; Yin, S.; Xu, Y.; Liu, J.; Yang, Z.; Xu, C. Natural Variation and Domestication Selection of ZmPGP1 Affects Plant Architecture and Yield-Related Traits in Maize. Genes 2019, 10, 664. [Google Scholar] [CrossRef] [PubMed]
- Fulton, T.M.; Chunwongse, J.; Tanksley, S.D. Microprep Protocol for Extraction of DNA from Tomato and other Herbaceous Plants. Plant Mol. Biol. Report. 1995, 13, 207–209. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Asan; Xu, Y.; Jiang, H.; Tyler-Smith, C.; Xue, Y.; Jiang, T.; Wang, J.; Wu, M.; Liu, X.; Tian, G.; et al. Comprehensive comparison of three commercial human whole-exome capture platforms. Genome Biol. 2011, 12, R95. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J. DNA sequence polymorphism analysis using DnaSP. Methods Mol. Biol. 2009, 537, 337–350. [Google Scholar] [PubMed]
- Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Fu, Y.X.; Li, W.H. Statistical Tests of Neutrality of Mutations. Genetics 1993, 133, 693–709. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Buckler, E.S. TASSEL: Software for Association Mapping of Complex Traits in Diverse Samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, E.; Jiang, Y.; Xu, S.; Pan, L.; Chen, Q.; Xu, C. Sequence polymorphisms in Zmisa2 gene are significantly associated with starch pasting and gelatinization properties in maize (Zea mays L.). Mol. Breed. 2014, 34, 1833–1842. [Google Scholar] [CrossRef]
- Zhang, E.; Yang, Z.; Wang, Y.; Hu, Y.; Song, X.; Xu, C. Nucleotide polymorphisms and haplotype diversity of RTCS gene in China elite maize inbred lines. PLoS ONE 2013, 8, e56495. [Google Scholar] [CrossRef]
- Li, X.; Zhu, C.; Yeh, C.T.; Wu, W.; Takacs, E.M.; Petsch, K.A.; Tian, F.; Bai, G.; Buckler, E.S.; Muehlbauer, G.J.; et al. Genic and nongenic contributions to natural variation of quantitative traits in maize. Genome Res. 2012, 22, 2436–2444. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Warburton, M.; Crouch, J. Association Mapping for Enhancing Maize (Zea mays L.) Genetic Improvement. Crop Sci. 2011, 51, 433–449. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, E.; Li, J.; Jiang, Y.; Wang, Y.; Hu, Y.; Xu, C. Analyses of sequence polymorphism and haplotype diversity of LEAFY genes revealed post-domestication selection in the Chinese elite maize inbred lines. Mol. Biol. Rep. 2014, 41, 1117–1125. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Zhao, H.; Ren, L.; Song, W.; Zeng, B.; Guo, J.; Wang, B.; Liu, Z.; Chen, J.; Li, W.; et al. Genome-wide genetic changes during modern breeding of maize. Nat. Genet. 2012, 44, 812–815. [Google Scholar] [CrossRef]
- Liang, Y.; Liu, Q.; Wang, X.; Huang, C.; Xu, G.; Hey, S.; Lin, H.-Y.; Li, C.; Xu, D.; Wu, L.; et al. ZmMADS69 functions as a flowering activator through the ZmRap2.7-ZCN8 regulatory module and contributes to maize flowering time adaptation. New Phytol. 2019, 221, 2335–2347. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Liu, X.; Ge, S.; Jensen, J.D.; Hu, F.; Li, X.; Dong, Y.; Gutenkunst, R.N.; Fang, L.; Huang, L.; et al. Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 2012, 30, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Jiang, Y.; Wang, Z.; Gou, Z.; Lyu, J.; Li, W.; Yu, Y.; Shu, L.; Zhao, Y.; Ma, Y.; et al. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat. Biotechnol. 2015, 33, 408–414. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Peng, Z.; Yang, X.; Wang, W.; Fu, J.; Wang, J.; Han, Y.; Chai, Y.; Guo, T.; Yang, N. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat. Genet. 2013, 45, 43–50. [Google Scholar] [CrossRef]
- Kandianis, C.B.; Yan, J.; Harjes, C.E.; Bai, L.; Kim, E.H.; Yang, X.; Skinner, D.J.; Fu, Z.; Mitchell, S.; Li, Q. Rare genetic variation at Zea mays crtRB1 increases beta-carotene in maize grain. Nat. Genet. 2010, 42, 322. [Google Scholar]
- Yang, Z.; Ma, S.; Hu, Y.; Zhang, E.; Xie, Z.; Xu, S.; Liu, L.; Deng, L.; Xu, C.; Huang, J. Association Analysis of the Maize Gene ZmYS1 with Kernel Mineral Concentrations. Plant Mol. Biol. Report. 2015, 33, 1327–1335. [Google Scholar] [CrossRef]
- Doebley, J. The Genetics of Maize Evolution. Ann. Rev. Genet. 2004, 38, 37–59. [Google Scholar] [CrossRef]
- Yamasaki, M.; Wright, S.I.; Mcmullen, M. Genomic screening for artificial selection during domestication and improvement in maize. Ann. Bot. 2007, 100, 967–973. [Google Scholar] [CrossRef]
- Chen, W.; Chen, L.; Zhang, X.; Yang, N.; Guo, J.; Wang, M.; Ji, S.; Zhao, X.; Yin, P.; Cai, L.; et al. Convergent selection of a WD40 protein that enhances grain yield in maize and rice. Science 2022, 375, eabg7985. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, M.; Tenaillon, M.I.; Vroh Bi, I.; Schroeder, S.G.; Sanchez-Villeda, H.; Doebley, J.F.; Gaut, B.S.; McMullen, M.D. A Large-Scale Screen for Artificial Selection in Maize Identifies Candidate Agronomic Loci for Domestication and Crop Improvement. Plant Cell 2005, 17, 2859–2872. [Google Scholar] [CrossRef] [PubMed]
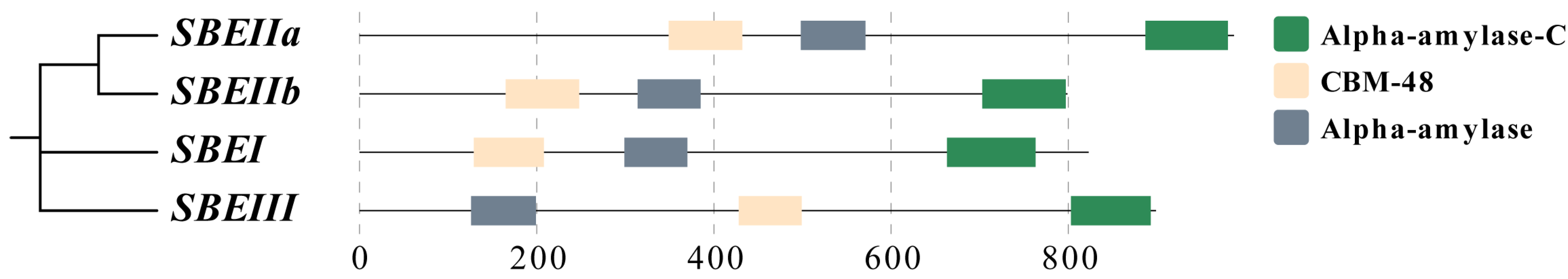

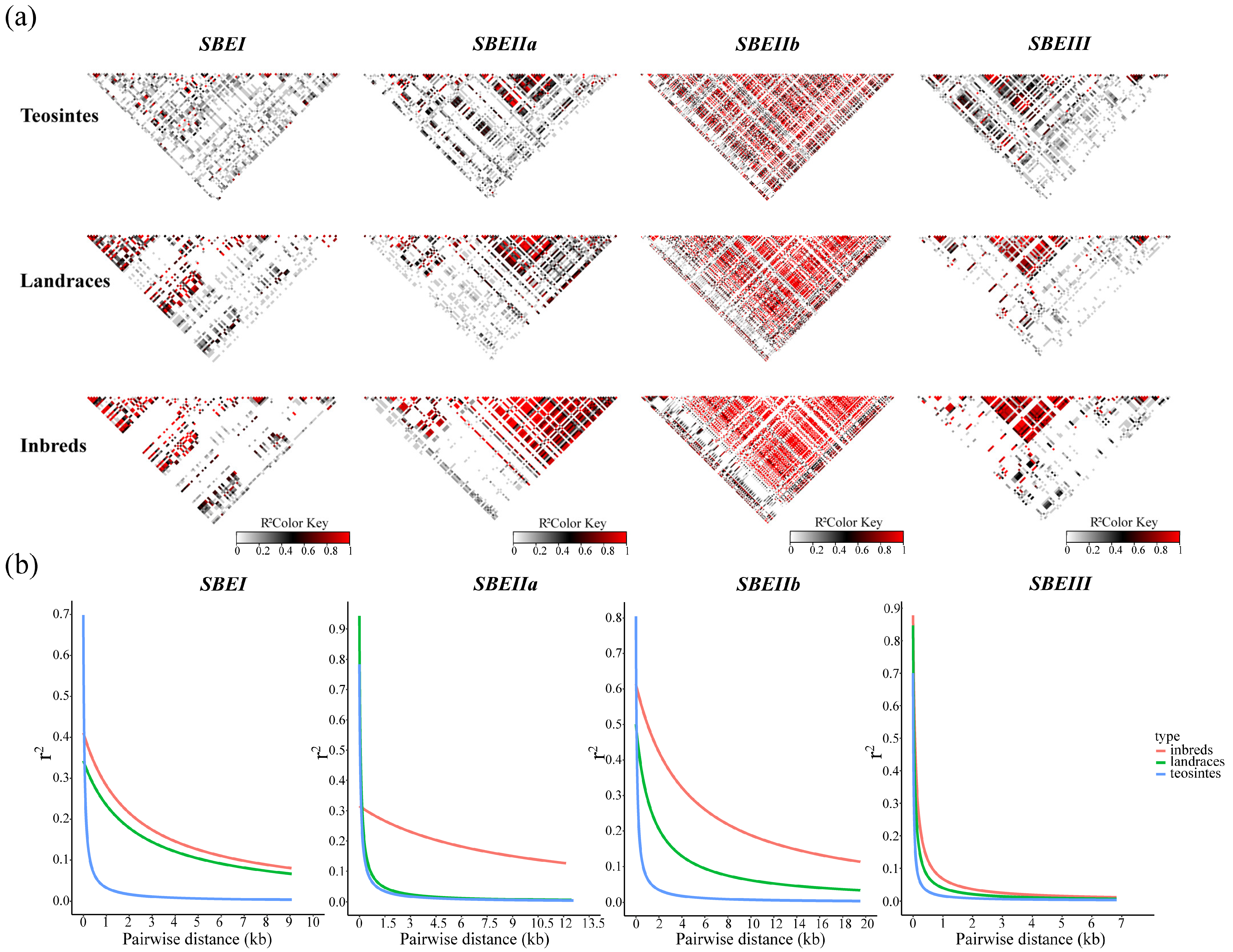
| Parameters | SBEI. | SBEIIa | SBEIIb | SBEIII |
|---|---|---|---|---|
| Chr | 5 | 2 | 5 | 8 |
| Gene length | 5694 | 10,473 | 17,049 | 2940 |
| Total length of amplicons (bp) | 9105 | 12,656 | 19,561 | 6831 |
| Number of all of the sequence variants | 1102 | 1519 | 1616 | 1044 |
| Frequency of all of the sequence variants | 8.3 | 8.3 | 12.0 | 6.5 |
| Number of nucleotide substitutions (bp) | 916 | 1302 | 1353 | 903 |
| Frequency of polymorphic sites per bp | 9.9 | 9.7 | 14.5 | 7.6 |
| Number of indels | 186 | 217 | 263 | 141 |
| Frequency of indels per bp | 50.0 | 58.8 | 76.9 | 47.6 |
| Average indels length | 5.4 | 4.9 | 3.9 | 4.7 |
| Gene | Pop. | C | π | θ | Tajima’s D | D* | F* |
|---|---|---|---|---|---|---|---|
| SBEI | Inbreds | 0.952 | 0.00397 | 0.00583 | −0.981 | −0.739 | −1.023 |
| Landraces | 0.943 | 0.01098 | 0.01075 | 0.074 | −0.324 | −0.194 | |
| Teosintes | 0.870 | 0.02304 | 0.03078 | −0.974 | −1.271 | −1.387 | |
| ALL | 0.843 | 0.00711 | 0.02005 | −1.976 * | −6.655 ** | −4.700 ** | |
| SBEIIa | Inbreds | 0.948 | 0.00737 | 0.00734 | 0.010 | −1.569 | −0.850 |
| Landraces | 0.943 | 0.01031 | 0.0108 | −0.161 | −0.988 | −0.780 | |
| Teosintes | 0.879 | 0.01874 | 0.02777 | −1.260 | −1.614 | −1.772 | |
| ALL | 0.862 | 0.00853 | 0.01892 | −1.685 | −7.167 ** | −4.747 ** | |
| SBEIIb | Inbreds | 0.948 | 0.00443 | 0.00533 | −0.519 | −2.571 * | −1.738 |
| Landraces | 0.931 | 0.01085 | 0.01179 | −0.283 | −1.297 | −1.058 | |
| Teosintes | 0.882 | 0.02117 | 0.0269 | −0.826 | −1.360 | −1.397 | |
| ALL | 0.847 | 0.00725 | 0.01749 | −1.797 * | −5.744 ** | −4.095 ** | |
| SBEIII | Inbreds | 0.915 | 0.01325 | 0.01143 | 0.494 | 0.271 | 0.459 |
| Landraces | 0.889 | 0.01906 | 0.02012 | −0.186 | −0.828 | −0.679 | |
| Teosintes | 0.826 | 0.02758 | 0.04001 | −1.203 | −1.222 | −1.443 | |
| ALL | 0.792 | 0.01467 | 0.02709 | −1.404 | −5.439 ** | −3.702 ** |
| Gene | Trait | Section | Variant | Genotype | p-Value | Contribution |
|---|---|---|---|---|---|---|
| SBEIIa | ΔHgel | Intron 14 | Indel 5967 | -/C | 1.32 × 10−3 | 3.17% |
| SBEIIa | ΔHgel | Intron 14 | Indel 5969 | -/C | 1.05 × 10−3 | 4.28% |
| SBEIIa | ΔHgel | Intron 1 | SNP 403 | G/T | 1.19 × 10−3 | 3.21% |
| SBEIIa | ΔHgel | Intron 1 | SNP 568 | G/A | 5.78 × 10−4 | 3.62% |
| SBEIIa | SB | Upstream | SNP 1529 | T/C | 1.53 × 10−3 | 2.99% |
| SBEIIb | SB | Intron 11 | Indel 7723 | -/C | 1.10 × 10−3 | 3.39% |
| SBEIIb | SB | 3’ UTR | SNP 17249 | C/G | 2.05 × 10−3 | 2.83% |
| SBEIIb | SB | Downstream | SNP 17786 | T/A | 1.48 × 10−3 | 3.01% |
| SBEIIb | SB | Intron 11 | SNP 8152 | A/G | 6.42 × 10−4 | 3.95% |
| SBEIII | ΔHgel | Downstream | SNP 4817 | T/C | 2.11 × 10−4 | 4.21% |
| SBEIII | ΔHgel | Downstream | SNP 5055 | C/G | 1.65 × 10−3 | 3.02% |
| SBEIII | KSC | Upstream | Indel 830 | G/- | 5.20 × 10−5 | 4.88% |
| SBEIII | KSC | Upstream | SNP 1391 | G/A | 5.89 × 10−5 | 4.80% |
| SBEIII | KSC | Upstream | SNP 245 | G/A | 1.08 × 10−4 | 4.39% |
| SBEIII | KSC | Upstream | SNP 356 | C/T | 1.27 × 10−4 | 4.30% |
| SBEIII | KSC | Upstream | SNP 357 | C/A | 1.22 × 10−4 | 4.32% |
| SBEIII | KSC | Upstream | SNP 381 | C/T | 1.40 × 10−4 | 4.26% |
| SBEIII | KSC | Upstream | SNP 443 | C/T | 1.27 × 10−4 | 4.30% |
| SBEIII | KSC | Upstream | SNP 674 | C/T | 1.27 × 10−4 | 4.30% |
| SBEIII | KSC | 3’ UTR | SNP 3918 | A/T | 1.34 × 10−3 | 2.99% |
| SBEIII | KSC | Downstream | SNP 4867 | C/T | 1.82 × 10−3 | 2.82% |
| SBEIII | KSC | Downstream | SNP 4868 | C/T | 1.82 × 10−3 | 2.82% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.; Yang, T.; Rui, W.; Wang, H.; Wang, Y.; Yang, Z.; Xu, C.; Li, P. Genetic Diversification of Starch Branching Enzymes during Maize Domestication and Improvement. Genes 2023, 14, 1068. https://doi.org/10.3390/genes14051068
Li Q, Yang T, Rui W, Wang H, Wang Y, Yang Z, Xu C, Li P. Genetic Diversification of Starch Branching Enzymes during Maize Domestication and Improvement. Genes. 2023; 14(5):1068. https://doi.org/10.3390/genes14051068
Chicago/Turabian StyleLi, Qi, Tiantian Yang, Wenye Rui, Houmiao Wang, Yunyun Wang, Zefeng Yang, Chenwu Xu, and Pengcheng Li. 2023. "Genetic Diversification of Starch Branching Enzymes during Maize Domestication and Improvement" Genes 14, no. 5: 1068. https://doi.org/10.3390/genes14051068
APA StyleLi, Q., Yang, T., Rui, W., Wang, H., Wang, Y., Yang, Z., Xu, C., & Li, P. (2023). Genetic Diversification of Starch Branching Enzymes during Maize Domestication and Improvement. Genes, 14(5), 1068. https://doi.org/10.3390/genes14051068





