Differential Detection of Alternaria alternata Haplotypes Isolated from Carya illinoinensis Using PCR-RFLP Analysis of Alt a1 Gene Region
Abstract
1. Introduction
2. Material and Methods
2.1. Sampling and Isolation of Alternaria
2.2. DNA Extraction and PCR Amplification
2.3. Alt a1 PCR-RFLP Analyses
2.4. Principal Coordinate Analyses
2.5. DNA Sequence and Phylogenetic Analysis
3. Results
3.1. A. alternata Sampled from Pecan Tissues
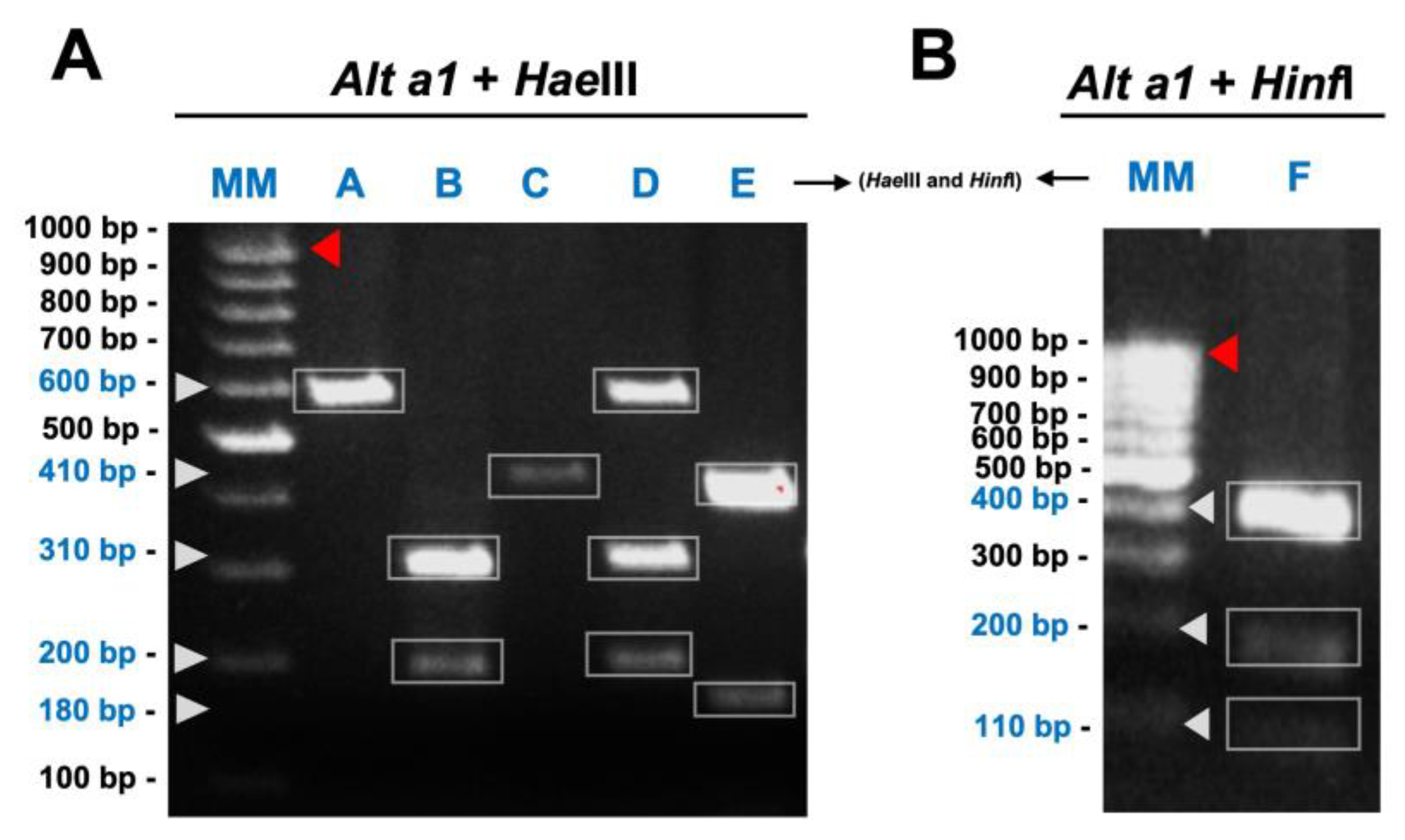
3.2. Alt a1 PCR-RFLP Analyses
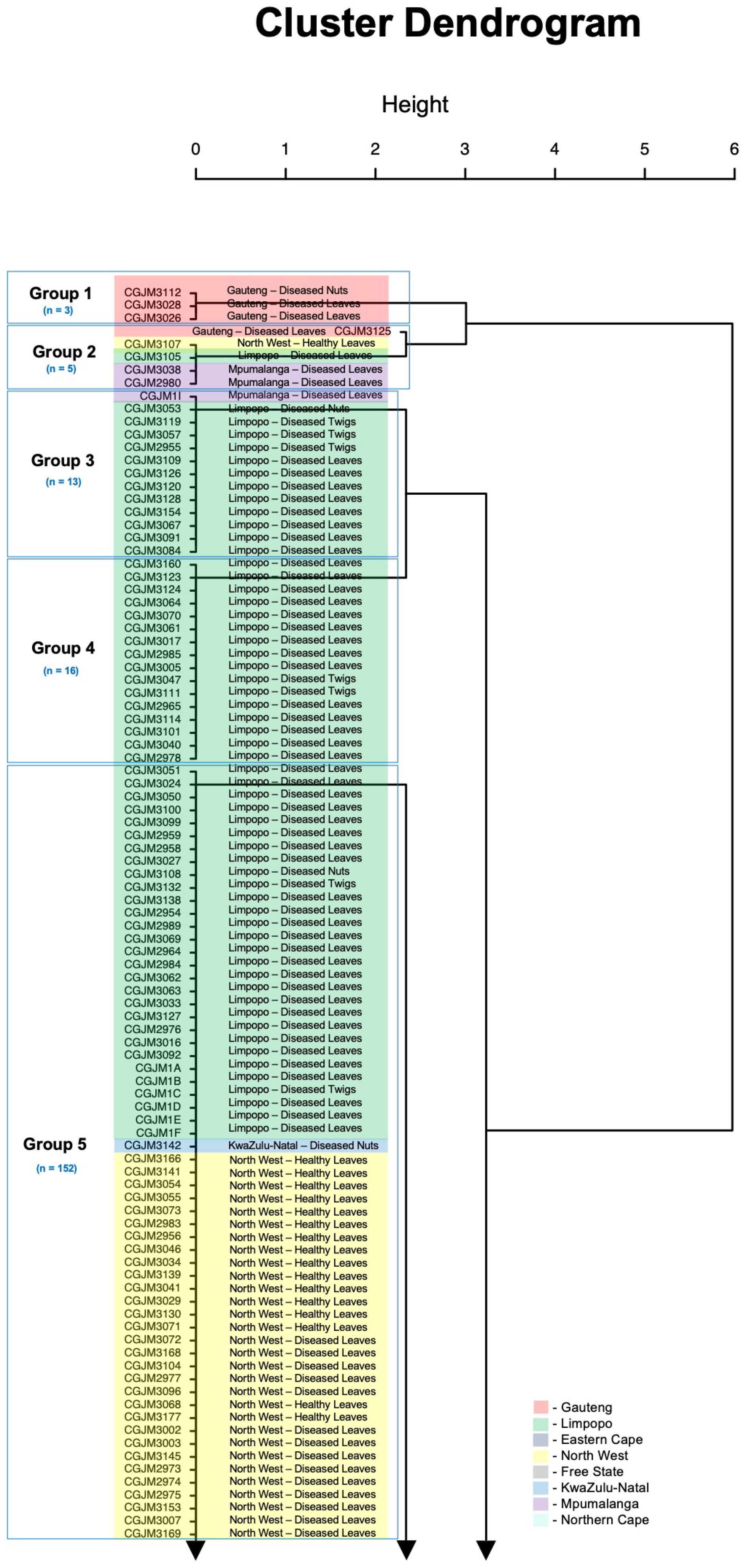
3.3. Principal Coordinate Analyses
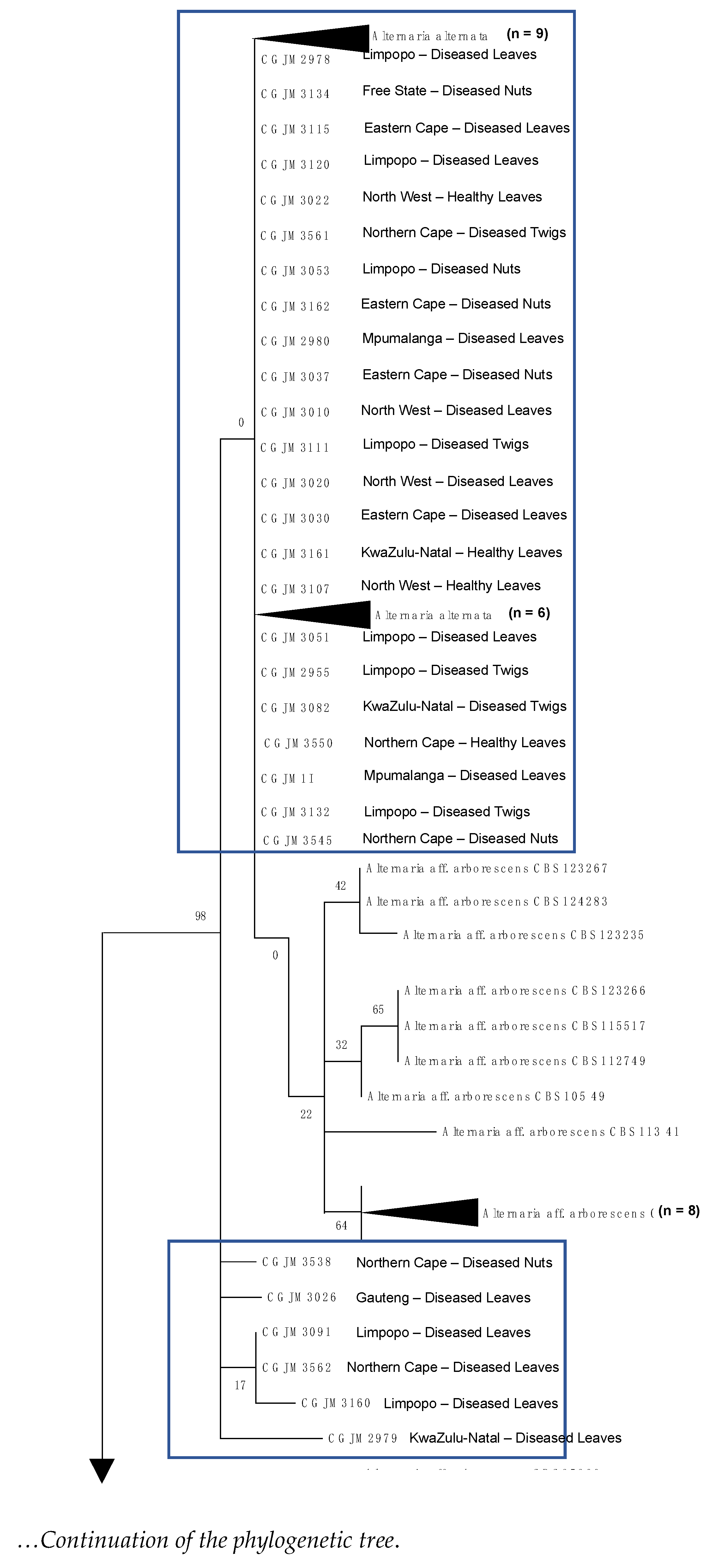
3.4. DNA Sequence and Phylogenetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Woudenberg, J.; Groenewald, J.; Binder, M.; Crous, P. Alternaria Redefined. Stud. Mycol. 2013, 75, 171–212. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Seidl, M.F.; Groenewald, J.Z.; De Vries, M.; Stielow, J.B.; Crous, P.W. Studies in Mycology. Stud. Mycol. 2014, 77, ii. [Google Scholar] [CrossRef][Green Version]
- Woudenberg, J.H.C.; Seidl, M.F.; Groenewald, J.Z.; de Vries, M.; Stielow, J.B.; Thomma, B.P.H.J.; Crous, P.W. Alternaria Section Alternaria: Species, Formae Speciales or Pathotypes? Stud. Mycol. 2015, 82, 1–21. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Simmons, E.G.; Roberts, R.G. Alternaria Themes and Variations (73). Mycotaxon 1993, 48, 109–140. [Google Scholar]
- Simmons, E.G. Alternaria Themes and Variations (236–243). Mycotaxon 1999, 70, 325–369. [Google Scholar]
- Ellis, M. Dematiaceous Hyphomycetes; Commonwealth Mycological Institute: Kew, UK, 1971. [Google Scholar]
- Simmons, E.G. Alternaria: An Identification Manual; CBS Biodiversity Series 6; CBS Fungal Biodiversity Centre: Utrecht, The Netherlands, 2007. [Google Scholar]
- Woudenberg, J.H.C.; Groenewald, J.Z.; van der Merwe, N.A.; Crous, P.W. Diversity and Movement of Indoor Alternaria alternata across the Mainland USA. Fungal Genet. Biol. 2015, 81, 62–72. [Google Scholar] [CrossRef][Green Version]
- Ashraf, S.; Zuhaib, M. Fungal Biodiversity: A Potential Tool in Plant Disease Management. In Management of Microbial Resources in the Environment; Malik, A., Alves, M., Grohmann, E., Eds.; Springer: Dordrecht, The Netherlands, 2013; pp. 69–90. ISBN 9789400759312. [Google Scholar]
- Encinas-Basurto, D.; Valenzuela-Quintanar, M.I.; Sánchez-Estrada, A.; Tiznado-Hernández, M.E.; Rodríguez-Félix, A.; Troncoso-Rojas, R. Alterations in Volatile Metabolites Profile of Fresh Tomatoes in Response to Alternaria alternata (Fr.) Keissl. 1912 Infection. Chil. J. Agric. Res. 2017, 77, 194–201. [Google Scholar] [CrossRef][Green Version]
- Zhao, K.; Shao, B.; Yang, D.; Li, F. Natural Occurrence of Four Alternaria Mycotoxins in Tomato- and Citrus-Based Foods in China. J. Agric. Food Chem. 2015, 63, 343–348. [Google Scholar] [CrossRef]
- Liu, L.; Ma, M.; Liu, Z.; Zhang, L.; Zhou, J. Community Structure of Fungal Pathogens Causing Spikelet Rot Disease of Naked Oat from Different Ecological Regions of China. Sci. Rep. 2021, 11, 1243. [Google Scholar] [CrossRef]
- Wang, F.; Saito, S.; Michailides, T.J.; Xiao, C.L. Postharvest Use of Natamycin to Control Alternaria Rot on Blueberry Fruit Caused by Alternaria alternata and A. arborescens. Postharvest Biol. Technol. 2021, 172, 111383. [Google Scholar] [CrossRef]
- Ahmed, M.Z.; Saeed, S.; Hassan, A.; Ghuffar, S.; Abdullah, A.; Haque, K.; Anwaar, H.A.; Iqbal, R.; Asif, M.A.; Shafique, M.S. Alternaria Alternata Causing Alternaria Leaf Spot of Cucumis melo (Muskmelon) in Pakistan. Plant Dis. 2021, 105, 1853. [Google Scholar] [CrossRef] [PubMed]
- Praveen, B.; Nagaraja, A.; Prasanna Kumar, M.K.; Pramesh, D.; Palanna, K.B.; Buella, P.P. First Report of Alternaria Alternata Causing Leaf Blight on Little Millet (Panicum sumatrense) in India. Plant Dis. 2021, 105, 1202. [Google Scholar] [CrossRef] [PubMed]
- Kgatle, M.G.; Truter, M.; Ramusi, T.M.; Flett, B.; Aveling, T.A.S. Alternaria alternata, the Causal Agent of Leaf Blight of Sunflower in South Africa. Eur. J. Plant Pathol. 2018, 151, 677–688. [Google Scholar] [CrossRef][Green Version]
- Van Der Waals, J.E.; Pitsi, B.E.; Marais, C.; Wairuri, C.K. First Report of Alternaria Alternata Causing Leaf Blight of Potatoes in South Africa. Plant Dis. 2011, 95, 363. [Google Scholar] [CrossRef]
- Achilonu, C.C.; Marais, G.J.; Ghosh, S.; Gryzenhout, M. Multigene Phylogeny and Pathogenicity Trials Revealed Alternaria alternata as the Causal Agent of Black Spot Disease and Seedling Wilt of Pecan (Carya illinoinensis) in South Africa. Pathogens 2023, 12, 672. [Google Scholar] [CrossRef]
- Sikuka, W.; Bonsu, K. Positive Outlook for South African Tree Nut Production and Trade; Global Agricultural Information Network: Pretoria, South Africa, 2020.
- Zedan, D. South Africa—The Pecan Industry’s next Paradigm Shift. SA Pecan 2018, 80, 34–35. [Google Scholar]
- Lemmer, B.Y.W. SA Pecan Production Still in a Boom Phase; Ryan, J., Mnyandu, E., Chetty, S., Moodley, P., Eds.; Farmer’s Weekly, Press Council: Johannesburg, South Africa, 2020; pp. 28–29. [Google Scholar]
- Vogel, E.; Donat, M.G.; Alexander, L.V.; Meinshausen, M.; Ray, D.K.; Karoly, D.; Meinshausen, N.; Frieler, K. The Effects of Climate Extremes on Global Agricultural Yields. Environ. Res. Lett. 2019, 14, 54010. [Google Scholar] [CrossRef]
- Zhao, J.; Li, S.; Jiang, T.; Liu, Z.; Zhang, W.; Jian, G.; Qi, F. Chilling Stress—The Key Predisposing Factor for Causing Alternaria alternata Infection and Leading to Cotton (Gossypium hirsutum L.) Leaf Senescence. PLoS ONE 2012, 7, e36126. [Google Scholar] [CrossRef][Green Version]
- Upadhyay, A.; Kadam, U.S.; Chacko, P.M.; Aher, L.; Karibasappa, G. Microsatellite Analysis to Differentiate Clones of Thompson Seedless Grapevine. Indian J. Hortic. 2010, 67, 260–263. [Google Scholar]
- Wei, X.; Xue, L.; Li, C. The First Report of Leaf Spot Caused by Alternaria Alternata on Italian Ryegrass (Lolium multiflorum) in China. Plant Dis. 2020, 39, 1. [Google Scholar] [CrossRef]
- Meng, J.W.; Zhu, W.; He, M.H.; Wu, E.J.; Duan, G.H.; Xie, Y.K.; Jin, Y.J.; Yang, L.N.; Shang, L.P.; Zhan, J. Population Genetic Analysis Reveals Cryptic Sex in the Phytopathogenic Fungus Alternaria alternata. Sci. Rep. 2015, 5, 18250. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lawrence, D.P.; Gannibal, P.B.; Peever, T.L.; Pryor, B.M. The Sections of Alternaria: Formalizing Species-Group Concepts. Mycologia 2013, 105, 530–546. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pryor, B.; Michailides, T. Morphological, Pathogenic, and Molecular Characterization of Alternaria Isolates Associated with Alternaria Late Blight of Pistachio. Phytopathology 2002, 92, 406–416. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Meyer, R.; Höfelein, C.; Lüthy, J.; Candrian, U. Polymerase Chain Reaction—Restriction Fragment Length Polymorphism Analysis: A Simple Method for Species Identification in Food. J. AOAC Int. 1995, 78, 1542–1551. [Google Scholar] [CrossRef] [PubMed]
- Chavhan, R.L.; Hinge, V.R.; Kadam, U.S.; Kalbande, B.B.; Chakrabarty, P.K. Real-Time PCR Assay for Rapid, Efficient and Accurate Detection of Paramyrothecium Roridum a Leaf Spot Pathogen of Gossypium Species. J. Plant Biochem. Biotechnol. 2018, 27, 199–207. [Google Scholar] [CrossRef]
- Chavhan, R.L.; Sable, S.; Narwade, A.V.; Hinge, V.R.; Kalbande, B.B.; Mukherjee, A.K.; Chakrabarty, P.K.; Kadam, U.S. Multiplex Molecular Marker-Assisted Analysis of Significant Pathogens of Cotton (Gossypium Sp.). Biocatal. Agric. Biotechnol. 2023, 47, 102557. [Google Scholar] [CrossRef]
- Atoui, A.; Khoury, A. El PCR-RFLP for Aspergillus Species. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2017; Volume 1542, pp. 313–320. [Google Scholar]
- Zelmat, L.; Mansi, J.M.; Aouzal, S.; Gaboun, F.; Khayi, S.; Ibriz, M.; El Guilli, M.; Mentag, R. Genetic Diversity and Population Structure of Moroccan Isolates Belong to Alternaria spp. Causing Black Rot and Brown Spot in Citrus. Int. J. Genomics 2021, 2021, 9976969. [Google Scholar]
- Kim, K.; Lee, H.M.; Kim, Y.H. Morphogenetic Alterations of Alternaria alternata Exposed to Dicarboximide Fungicide, Iprodione. Plant Pathol. J. 2017, 33, 95–100. [Google Scholar] [CrossRef][Green Version]
- Sabbagh, S.K. Genetic Variability of Paecilomyces variotii Isolates, the Causal Agent of Dieback Disease in Pistachio, Using ITS-RFLP Analysis. Mycol. Iran. 2016, 3, 111–120. [Google Scholar] [CrossRef]
- Zhao, J.; Ma, G.P.; Liu, Y.Y.; Wu, X.H. Alternaria Species Infecting Potato in Southern China. Can. J. Plant Pathol. 2018, 40, 312–317. [Google Scholar] [CrossRef]
- Hong, S.G.; Cramer, R.A.; Lawrence, C.B.; Pryor, B.M. Alt. a 1 Allergen Homologs from Alternaria and Related Taxa: Analysis of Phylogenetic Content and Secondary Structure. Fungal Genet. Biol. 2005, 42, 119–129. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Volume 2, 3559p. [Google Scholar]
- Peakall, R.; Smouse, P.E. GenALEx 6.5: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research-an Update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Diguta, C.F.; Vincent, B.; Guilloux-Benatier, M.; Alexandre, H.; Rousseaux, S. PCR ITS-RFLP: A Useful Method for Identifying Filamentous Fungi Isolates on Grapes. Food Microbiol. 2011, 28, 1145–1154. [Google Scholar] [CrossRef]
- Ozkilinc, H.; Sevinc, U. Molecular Phylogenetic Species in Alternaria Pathogens Infecting Pistachio and Wild Relatives. 3 Biotech. 2018, 8, 250. [Google Scholar] [CrossRef]
- Hirakue, A.; Sugiyama, S. Relationship between Foliar Endophytes and Apple Cultivar Disease Resistance in an Organic Orchard. Biol. Control. 2018, 127, 139–144. [Google Scholar] [CrossRef]
- Eschenbrenner, C.J.; Feurtey, A.; Stukenbrock, E.H. Population Genomics of Fungal Plant Pathogens and the Analyses of Rapidly Evolving Genome Compartments. Methods Mol. Biol. 2020, 2090, 337–355. [Google Scholar] [CrossRef][Green Version]
- Meng, J.W.; He, D.C.; Zhu, W.; Yang, L.N.; Wu, E.J.; Xie, J.H.; Shang, L.P.; Zhan, J. Human-Mediated Gene Flow Contributes to Metapopulation Genetic Structure of the Pathogenic Fungus Alternaria alternata from Potato. Front. Plant. Sci. 2018, 9, 198–210. [Google Scholar] [CrossRef][Green Version]
- Bouwmester, A. SA Pecan: Celebrating 25 Years Together. SA Pecan 2017, 78, 2–3. [Google Scholar]
- Johnson, J.D.; Black, M.C. Pecan Disease Identification and Control. In Texas Pecan Handbook; Stein, L.A., McEachern, G.R., Nesbitt, M.L., Eds.; Texas AgriLife Extension Service: College Station, TX, USA, 2012; pp. 128–179. [Google Scholar]
- Von Broembsen, S. Pecan Diseases: Prevention and Control. Okla. Coop. Ext. Serv. 2016, 7642–7647. [Google Scholar]
- Latz, M.A.C.; Kerrn, M.H.; Sørensen, H.; Collinge, D.B.; Jensen, B.; Brown, J.K.M.; Madsen, A.M.; Jørgensen, H.J.L. Succession of the Fungal Endophytic Microbiome of Wheat Is Dependent on Tissue-Specific Interactions between Host Genotype and Environment. Sci. Total Environ. 2021, 759, 143804. [Google Scholar] [CrossRef] [PubMed]
- Fang, K.; Miao, Y.F.; Chen, L.; Zhou, J.; Yang, Z.P.; Dong, X.F.; Zhang, H.B. Tissue-Specific and Geographical Variation in Endophytic Fungi of Ageratina adenophora and Fungal Associations with the Environment. Front. Microbiol. 2019, 10, 2919. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gabriel, M.F.; Postigo, I.; Gutiérrez-Rodríguez, A.; Suñén, E.; Guisantes, J.A.; Fernández, J.; Tomaz, C.T.; Martínez, J. Alt. A15 Is a New Cross-Reactive Minor Allergen of Alternaria alternata. Immunobiology 2016, 221, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.J.; Meng, Q.; Zeng, L.; Tong, H.R. Phylogenetic and Morphological Characteristics of Alternaria alternata Causing Leaf Spot Disease on Camellia sinensis in China. Australas. Plant Pathol. 2018, 47, 335–342. [Google Scholar] [CrossRef]
- Arie, T.; Kaneko, I.; Yoshida, T.; Noguchi, M.; Nomura, Y.; Yamaguchi, I. Mating-Type Genes from Asexual Phytopathogenic Ascomycetes Fusarium oxysporum and Alternaria alternata. Mol. Plant-Microbe Interact. 2000, 13, 1330–1339. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Berbee, M.L.; Payne, B.P.; Zhang, G.; Roberts, R.G.; Turgeon, B.G. Shared ITS DNA Substitutions in Isolates of Opposite Mating Type Reveal a Recombining History for Three Presumed Asexual Species in the Filamentous Ascomycete Genus Alternaria. Mycol. Res. 2003, 107, 169–182. [Google Scholar] [CrossRef][Green Version]
- Stewart, J.E.; Kawabe, M.; Abdo, Z.; Arie, T.; Peever, T.L. Contrasting Codon Usage Patterns and Purifying Selection at the Mating Locus in Putatively Asexual Alternaria Fungal Species. PLoS ONE 2011, 6, e20083. [Google Scholar] [CrossRef][Green Version]




| Location | Symptomatic Nuts-in-Shuck | Symptomatic Leaves | Symptomatic Shoots | Non-Symptomatic Leaves |
|---|---|---|---|---|
| Gauteng | 1 | 4 | 1 | 0 |
| Limpopo | 2 | 48 | 7 | 0 |
| Kwazulu-Natal | 3 | 1 | 2 | 1 |
| Mpumalanga | 0 | 3 | 0 | 0 |
| Eastern Cape | 3 | 14 | 2 | 4 |
| North West | 1 | 49 | 1 | 38 |
| Free State | 3 | 3 | 0 | 3 |
| Northern Cape | 5 | 11 | 4 | 8 |
| Total * | 18 | 133 | 17 | 54 |
| Percentage | 8.1% | 59.9% | 7.7% | 24.3% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Achilonu, C.C.; Gryzenhout, M.; Marais, G.J.; Ghosh, S. Differential Detection of Alternaria alternata Haplotypes Isolated from Carya illinoinensis Using PCR-RFLP Analysis of Alt a1 Gene Region. Genes 2023, 14, 1115. https://doi.org/10.3390/genes14051115
Achilonu CC, Gryzenhout M, Marais GJ, Ghosh S. Differential Detection of Alternaria alternata Haplotypes Isolated from Carya illinoinensis Using PCR-RFLP Analysis of Alt a1 Gene Region. Genes. 2023; 14(5):1115. https://doi.org/10.3390/genes14051115
Chicago/Turabian StyleAchilonu, Conrad Chibunna, Marieka Gryzenhout, Gert Johannes Marais, and Soumya Ghosh. 2023. "Differential Detection of Alternaria alternata Haplotypes Isolated from Carya illinoinensis Using PCR-RFLP Analysis of Alt a1 Gene Region" Genes 14, no. 5: 1115. https://doi.org/10.3390/genes14051115
APA StyleAchilonu, C. C., Gryzenhout, M., Marais, G. J., & Ghosh, S. (2023). Differential Detection of Alternaria alternata Haplotypes Isolated from Carya illinoinensis Using PCR-RFLP Analysis of Alt a1 Gene Region. Genes, 14(5), 1115. https://doi.org/10.3390/genes14051115







