The Unique Homothallic Mating-Type Loci of the Fungal Tree Pathogens Chrysoporthe syzygiicola and Chrysoporthe zambiensis from Africa
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome Sequencing, Assembly, and Analysis
2.2. Structure of the Mating-Type Loci of C. zambiensis and C. syzygiicola
2.3. Phylogenetic Analysis of Mating-Type Genes
3. Results
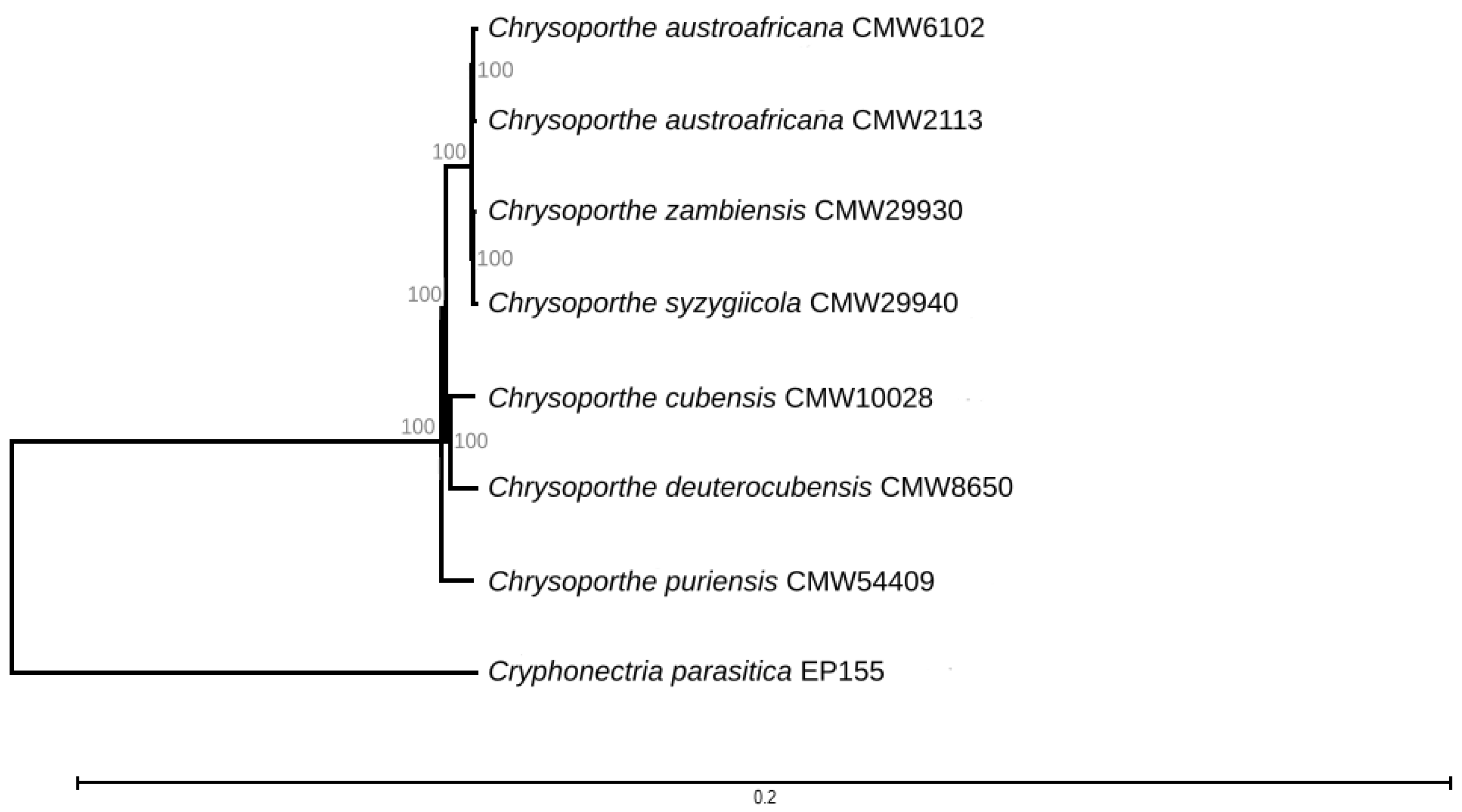
3.1. Genome Sequencing and Taxonomic Confirmation
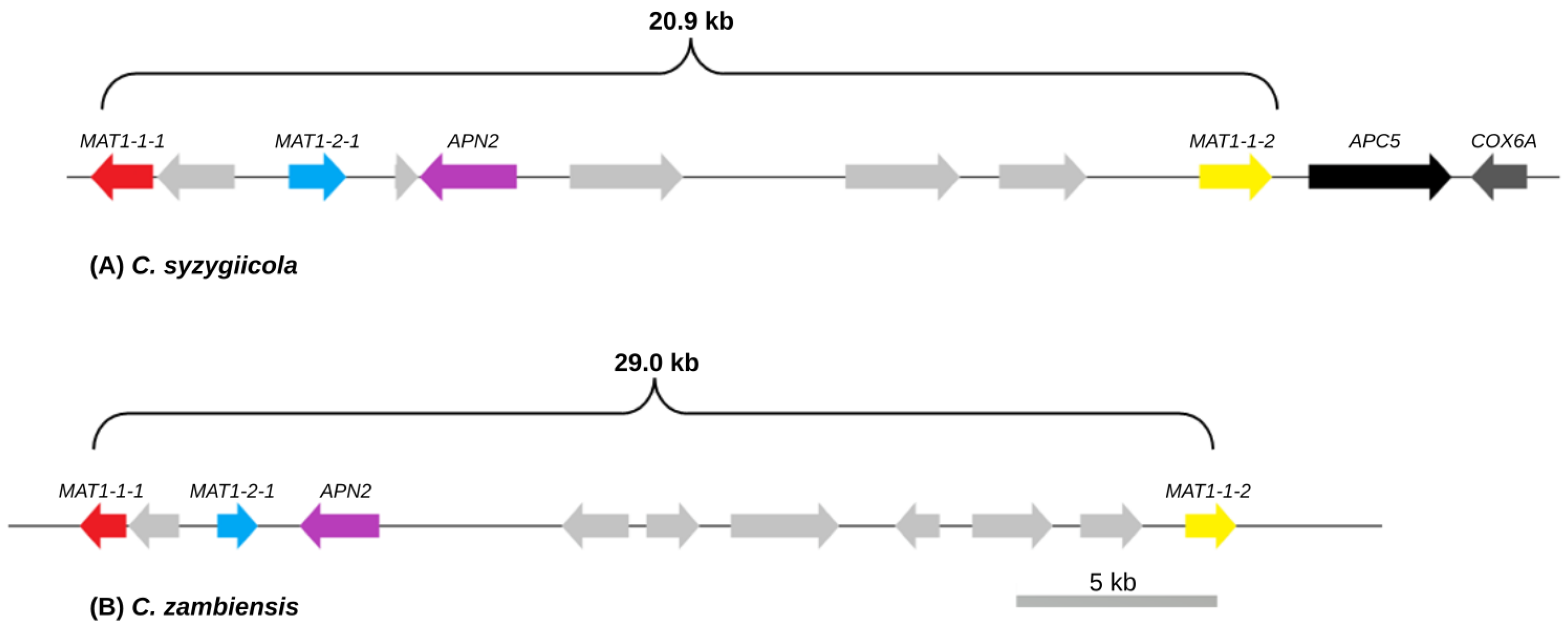
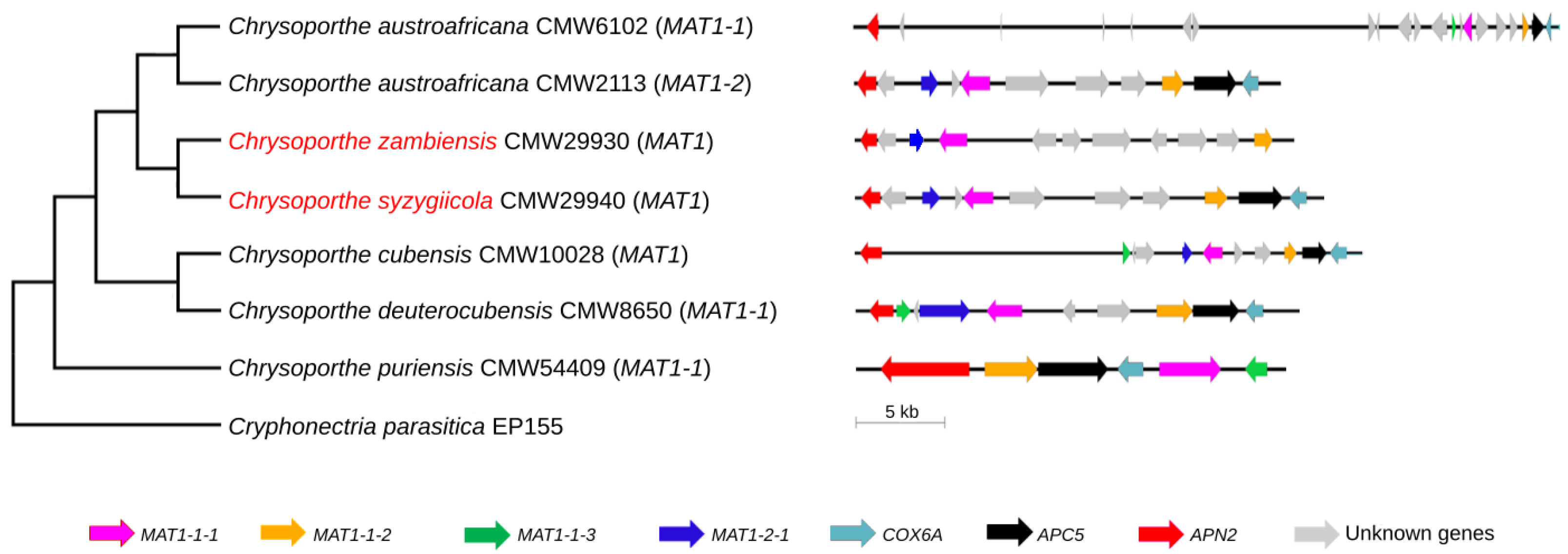
3.2. Mating-Type Genes and Structure of the Mating-Type Loci of C. syzygiicola and C. zambiensis
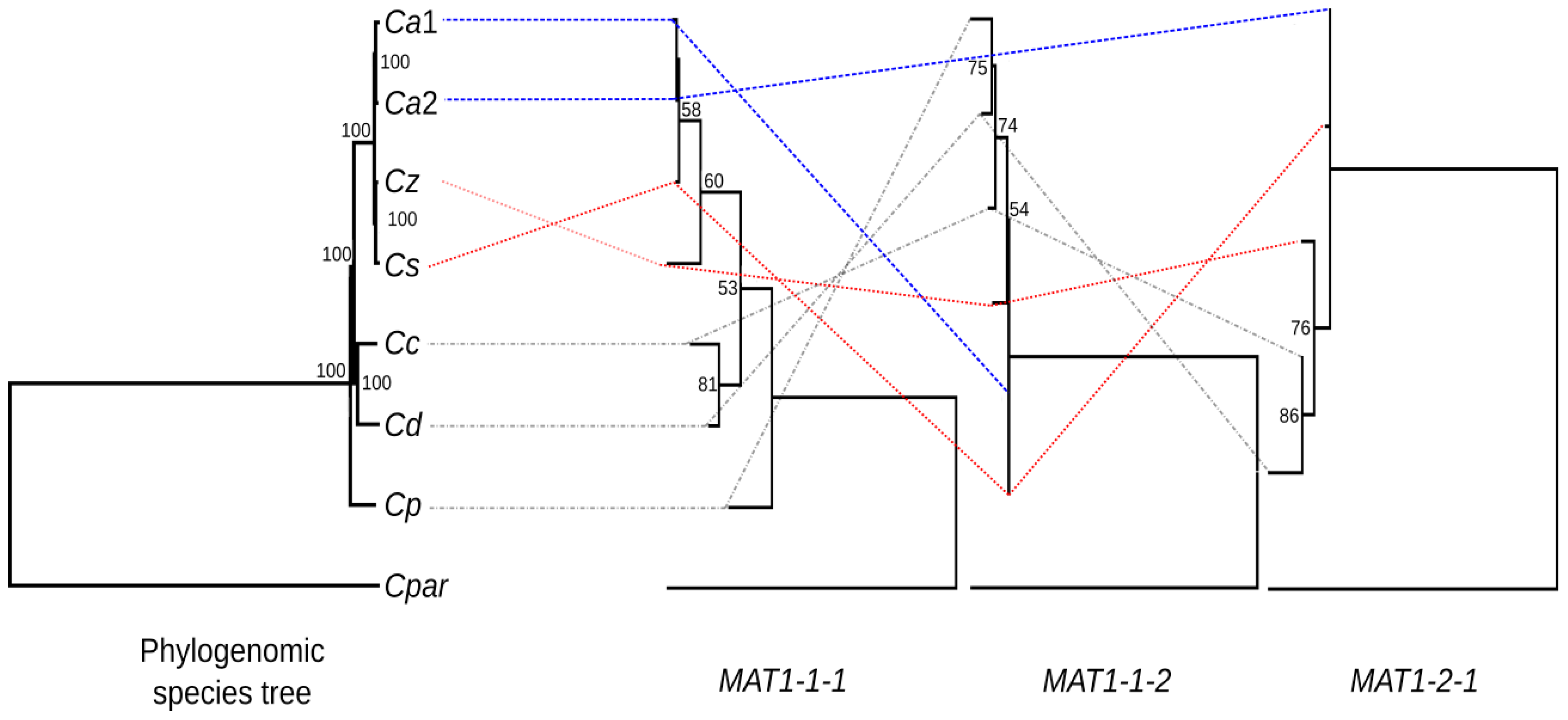
3.3. Phylogenetic Analysis of the Mating-Type Genes
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Debuchy, R.; Turgeon, B.G. Mating-type structure, evolution, and function in Euascomycetes. In The Mycota; Esser, K., Ed.; Springer: Berlin/Heidelberg, Germany, 2006; Volume 1, pp. 293–323. [Google Scholar] [CrossRef]
- Dyer, P.S.; Inderbitzin, P.; Debuchy, R. Mating-type structure, function, regulation and evolution in the Pezizomycotina. In The Mycota; Esser, K., Ed.; Springer: Berlin/Heidelberg, Germany, 2006; Volume 1, pp. 351–358. [Google Scholar] [CrossRef]
- Coppin, E.; Debuchy, R.; Arnaise, S.; Picard, M. Mating types and sexual development in filamentous ascomycetes. Micobiol. Mol. Biol. Rev. 1997, 61, 411–428. [Google Scholar] [CrossRef]
- Wilson, A.M.; Wilken, P.M.; van der Nest, M.A.; Steenkamp, E.T.; Wingfield, M.J.; Wingfield, B.D. Homothallism: An umbrella term for describing diverse sexual behaviours. IMA Fungus 2015, 6, 207–214. [Google Scholar] [CrossRef]
- Attanayake, R.N.; Tennekoon, V.; Johnson, D.A.; Porter, L.D.; del Río-Mendoza, L.; Jiang, D.; Chen, W. Inferring outcrossing in the homothallic fungus Sclerotinia sclerotiorum using linkage disequilibrium decay. Heredity 2014, 113, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Billiard, S.; López-Villavicencio, M.; Hood, M.E.; Giraud, T. Sex, outcrossing and mating types: Unsolved questions in fungi and beyond. J. Evol. Biol. 2012, 25, 1020–1038. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.M.; Wilken, P.M.; Wingfield, M.J.; Wingfield, B.D. Genetic networks that govern sexual reproduction in the Pezizomycotina. Microbiol. Mol. Biol. Rev. 2021, 85, e00020-21. [Google Scholar] [CrossRef]
- Wilson, A.M.; Wilken, P.M.; van der Nest, M.A.; Wingfield, M.J.; Wingfield, B.D. It’s all in the genes: The regulatory pathways of sexual reproduction in filamentous ascomycetes. Genes 2019, 10, 330. [Google Scholar] [CrossRef]
- Casselton, L.A. Mate recognition in fungi. Heredity 2002, 88, 142–147. [Google Scholar] [CrossRef]
- Turgeon, B.G.; Yoder, O.C. Proposed nomenclature for mating type genes of filamentous ascomycetes. Fungal Genet. Biol. 2000, 31, 1–5. [Google Scholar] [CrossRef]
- Wilken, P.M.; Steenkamp, E.T.; Wingfield, M.J.; de Beer, Z.W.; Wingfield, B.D. Which MAT gene? Pezizomycotina (Ascomycota) mating-type gene nomenclature reconsidered. Fungal Biol. Rev. 2017, 31, 199–211. [Google Scholar] [CrossRef]
- Myburg, H.; Gryzenhout, M.; Heath, R.; Roux, J.; Wingfield, B.D.; Wingfield, M.J. Cryphonectria canker on Tibouchina in South Africa. Mycol. Res. 2002, 106, 1299–1306. [Google Scholar] [CrossRef]
- Nakabonge, G.; Roux, J.; Gryzenhout, M.; Wingfield, M.J. Distribution of Chrysoporthe canker pathogens on Eucalyptus and Syzygium spp. in Eastern and Southern Africa. Plant Dis. 2006, 90, 734–740. [Google Scholar] [CrossRef] [PubMed]
- Heath, R.N.; Gryzenhout, M.; Roux, J.; Wingfield, M.J. Discovery of the canker pathogen Chrysoporthe austroafricana on native Syzygium spp. in South Africa. Plant Dis. 2006, 90, 433–438. [Google Scholar] [CrossRef]
- Chen, S.F.; Gryzenhout, M.; Roux, J.; Xie, Y.J.; Wingfield, M.J.; Zhou, X.D. Identification and pathogenicity of Chrysoporthe cubensis on Eucalyptus and Syzygium spp. in South China. Plant Dis. 2010, 94, 1143–1150. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, M.E.S.; van der Merwe, N.A.; Wingfield, M.J.; Wingfield, B.D.; Soares, T.P.F.; Kanzi, A.M.; Ferreira, M.A. Chrysoporthe puriensis sp. nov. from Tibouchina spp. in Brazil: An emerging threat to Eucalyptus. Australas. Plant Pathol. 2021, 50, 29–40. [Google Scholar] [CrossRef]
- Van Heerden, S.W.; Wingfield, M.J. Genetic diversity of Cryphonectria cubensis isolates in South Africa. Mycol. Res. 2001, 105, 94–99. [Google Scholar] [CrossRef]
- Chungu, D.; Gryzenhout, M.; Muimba-Kankolongo, A.; Wingfield, M.J.; Roux, J. Taxonomy and pathogenicity of two novel Chrysoporthe species from Eucalyptus grandis and Syzygium guineense in Zambia. Mycol. Prog. 2010, 9, 379–393. [Google Scholar] [CrossRef]
- Gryzenhout, M.; Myburg, H.; van der Merwe, N.A.; Wingfield, B.D.; Wingfield, M.J. Chrysoporthe, a new genus to accommodate Cryphonectria cubensis. Stud. Mycol. 2004, 50, 119–142. [Google Scholar]
- Kanzi, A.M.; Steenkamp, E.T.; van der Merwe, N.A.; Wingfield, B.D. The mating system of the Eucalyptus canker pathogen Chrysoporthe austroafricana and closely related species. Fungal Genet. Biol. 2019, 123, 41–52. [Google Scholar] [CrossRef]
- Pöggeler, S.; Kück, U. Comparative analysis of the mating-type loci from Neurospora crassa and Sordaria macrospora: Identifcation of novel transcribed ORFs. Mol. Gen. Genet. 2000, 263, 292–301. [Google Scholar] [CrossRef]
- McGuire, I.C.; Marra, R.E.; Turgeon, B.G.; Milgroom, M.G. Analysis of mating-type genes in the chestnut blight fungus, Cryphonectria parasitica. Fungal Genet. Biol. 2001, 34, 131–144. [Google Scholar] [CrossRef]
- Duong, T.A.; de Beer, Z.W.; Wingfield, B.D.; Wingfield, M.J. Characterization of the mating-type genes in Leptographium procerum and Leptographium profanum. Fungal Biol. 2013, 117, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, M.E.S.; Kanzi, A.M.; van der Merwe, N.A.; Wingfield, M.J.; Wingfield, B.D.; Silva, G.A.; Ferreira, M.A. Genetic variability in populations of Chrysoporthe cubensis and Chr. puriensis in Brazil. Australas. Plant Pathol. 2022, 51, 175–191. [Google Scholar] [CrossRef]
- Steenkamp, E.T.; Wingfield, B.D.; Coutinho, T.A.; Wingfield, M.J.; Marasas, W.F.O. Differentiation of Fusarium subglutinans f. sp. pini by histone gene sequence data. Appl. Environ. Microbiol. 1999, 65, 3401–3406. [Google Scholar]
- Jalili, V.; Afgan, E.; Gu, Q.; Clements, D.; Blankenberg, D.; Goecks, J.; Taylor, J.; Nekrutenko, A. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2020 update. Nucleic Acids Res. 2020, 48, W395–W402. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 22–736. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Stanke, M.; Morgenstern, B. AUGUSTUS: A web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005, 33, W465–W467. [Google Scholar] [CrossRef]
- Stanke, M.; Tzvetkova, A.; Morgenstern, B. AUGUSTUS at EGASP: Using EST, protein and genomic alignments for improved gene prediction in the human genome. Genome Biol. 2006, 7, S11. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. Bmc Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed]
- McGowan, J.; O’Hanlon, R.; Owens, R.A.; Fitzpatrick, D.A. Comparative denomic and proteomic analyses of three widespread Phytophthora species: Phytophthora chlamydospora, Phytophthora gonapodyides and Phytophthora pseudosyringae. Microorganisms 2020, 8, 653. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Quang Minh, B. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Gertz, E.M.; Yu, Y.K.; Agarwala, R.; Schäffer, A.A.; Altschul, S.F. Composition-based statistics and translated nucleotide searches: Improving the TBLASTN module of BLAST. BMC Biol. 2006, 4, 41. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Ke, X.; Li, Z.; Chen, J.; Gao, X.; Huang, L. Unconventional recombination in the mating type locus of heterothallic apple canker pathogen Valsa mali. Genes Genomes Genet. 2017, 7, 1259–1265. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- Finn, R.D.; Attwood, T.K.; Babbitt, P.C.; Bateman, A.; Bork, P.; Bridge, A.J.; Chang, H.Y.; Dosztányi, Z.; El-Gebali, S.; Fraser, M.; et al. InterPro in 2017—Beyond protein family and domain annotations. Nucleic Acids Res. 2017, 45, D190–D199. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Van der Nest, M.A.; Chávez, R.; De Vos, L.; Duong, T.A.; Gil-Durán, C.; Ferreira, M.A.; Lane, F.A.; Levicán, G.; Santana, Q.C.; Steenkamp, E.T.; et al. IMA genome—F14: Draft genome sequences of Penicillium roqueforti, Fusarium sororula, Chrysoporthe puriensis, and Chalaropsis populi. IMA Fungus 2021, 12, 5. [Google Scholar] [CrossRef]
- Wingfield, B.D.; Ades, P.K.; Al-Naemi, F.A.; Beirn, L.A.; Bihon, W.; Crouch, J.A.; de Beer, Z.W.; De Vos, L.; Duong, T.A.; Fields, C.J.; et al. IMA Genome—F4: Draft genome sequences of Chrysoporthe austroafricana, Diplodia scrobiculata, Fusarium nygamai, Leptographium lundbergii, Limonomyces culmigenus, Stagonosporopsis tanaceti, and Thielaviopsis punctulata. IMA Fungus 2015, 6, 233–248. [Google Scholar] [CrossRef]
- Wingfield, B.D.; Barnes, I.; de Beer, Z.W.; De Vos, L.; Duong, T.A.; Kanzi, A.M.; Naidoo, K.; Nguyen, H.D.T.; Santana, Q.C.; Sayari, M.; et al. IMA Genome—F5: Draft genome sequences of Ceratocystis eucalypticola, Chrysoporthe cubensis, C. deuterocubensis, Davidsoniella virescens, Fusarium temperatum, Graphilbum fragrans, Penicillium nordicum, and Thielaviopsis musarum. IMA Fungus 2015, 6, 493–506. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Sullivan, T.D.; Walton, E.; Floyd Averette, A.; Sakthikumar, S.; Cuomo, C.A.; Klein, B.S.; Heitman, J. Identification of the mating-type (MAT) locus that controls sexual reproduction of Blastomyces dermatitidis. Eukaryot. Cell 2013, 12, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Bihon, W.; Wingfield, M.J.; Slippers, B.; Duong, T.A.; Wingfield, B.D. MAT gene idiomorphs suggest a heterothallic sexual cycle in a predominantly asexual and important pine pathogen. Fungal Genet. Biol. 2014, 62, 55–61. [Google Scholar] [CrossRef]
- Nagel, J.H.; Wingfield, M.J.; Slippers, B. Evolution of the mating types and mating strategies in prominent genera in the Botryosphaeriaceae. Fungal Genet. Biol. 2018, 114, 24–33. [Google Scholar] [CrossRef]
- Kück, U.; Böhm, J. Mating type genes and cryptic sexuality as tools for genetically manipulating industrial molds. Appl. Microbiol. Biotechnol. 2013, 97, 9609–9620. [Google Scholar] [CrossRef]
- Kück, U.; Pöggeler, S.; Nowrousian, M.; Nolting, N.; Engh, I. Sordaria macrospora, a model system for fungal development. In Mycota XV; Anke, T., Weber, D., Eds.; Physiology and Genetics; Springer: Berlin/Heidelberg, Germany, 2009; Chapter 2; pp. 17–39. [Google Scholar]
- Kanematsu, S.; Adachi, Y.; Ito, T. Mating-type loci of heterothallic Diaporthe spp.: Homologous genes are present in opposite mating-types. Curr. Genet. 2007, 52, 11–22. [Google Scholar] [CrossRef]
- Martin, S.H.; Wingfield, B.D.; Wingfield, M.J.; Steenkamp, E.T. Structure and evolution of the Fusarium mating type locus: New insights from the Gibberella fujikuroi complex. Fungal Genet. Biol. 2011, 48, 731–740. [Google Scholar] [CrossRef]
- Yokoyama, E.; Yamagishi, K.; Hara, A. Structures of the mating-type loci of Cordyceps takaomontana. Appl. Environ. Microbiol. 2003, 69, 5019–5022. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, E.; Yamagishi, K.; Hara, A. Heterothallism in Cordyceps takaomontana. Fems Microbiol. Lett. 2005, 250, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Wilken, P.M.; Steenkamp, E.T.; Wingfield, M.J.; de Beer, Z.W.; Wingfield, B.D. DNA loss at the Ceratocystis fimbriata mating locus results in self-sterility. PLoS ONE 2014, 9, e92180. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Q.; Wingfield, B.D.; Wingfield, M.J.; Barnes, I.; Fourie, A.; Crous, P.W.; Chen, S.F. Mating genes in Calonectria and evidence for a heterothallic ancestral state. Persoonia 2020, 45, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Yong, M.; Yu, J.; Pan, X.; Yu, M.; Cao, H.; Qi, Z.; Du, Y.; Zhang, R.; Song, T.; Yin, X.; et al. MAT1-1-3, a mating type gene in the Villosiclava virens, is required for fruiting bodies and sclerotia formation, asexual development and pathogenicity. Front. Microbiol. 2020, 11, 1137. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, F.E.; Duhamel, M.; Carpentier, F.; Hood, M.E.; Foulongne-Oriol, M.; Silar, P.; Malagnac, F.; Grognet, P.; Giraud, T. Recombination suppression and evolutionary strata around mating-type loci in fungi: Documenting patterns and understanding evolutionary and mechanistic causes. New Phytol. 2020, 229, 2470–2491. [Google Scholar] [CrossRef]
- Fraser, J.A.; Stajich, J.E.; Tarcha, E.J.; Cole, G.T.; Inglis, D.O.; Sil, A.; Heitman, J. Evolution of the mating type locus: Insights gained from the dimorphic primary fungal pathogens Histoplasma capsulatum, Coccidioides immitis, and Coccidioides posadasii. Eukaryot. Cell 2007, 6, 622–629. [Google Scholar] [CrossRef]
- Roux, J.; Nkuekam, G.K.; Marincowitz, S.; van der Merwe, N.A.; Uchida, J.; Wingfield, M.J.; Chen, S.F. Cryphonectriaceae associated with rust-infected Syzygium jambos in Hawaii. MycoKeys 2020, 76, 49–79. [Google Scholar] [CrossRef]
- Kanzi, A.M.; Trollip, C.; Wingfield, M.J.; Barnes, I.; van der Nest, M.A.; Wingfield, B.D. Phylogenomic incongruence in Ceratocystis: A clue to speciation? BMC Genom. 2020, 21, 362. [Google Scholar] [CrossRef]
- Kanzi, A.M.; Wingfield, B.D.; Steenkamp, E.T.; Naidoo, S.; van der Merwe, N.A. Intron derived size polymorphism in the mitochondrial genomes of closely related Chrysoporthe species. PLoS ONE 2016, 11, e0156104. [Google Scholar] [CrossRef]
- Heitman, J.; Carter, D.A.; Dyer, P.S.; Soll, D.R. Sexual reproduction of human fungal pathogens. Cold Spring Harb. Perspect. Med. 2014, 4, 1–19. [Google Scholar] [CrossRef]
| Assembly Metric | C. zambiensis | C. syzygiicola |
|---|---|---|
| Genome size (bp) | 48,317,394 | 42,500,337 |
| Number of contigs | 211 | 233 |
| GC content (%) | 56.57 | 55.43 |
| N50 (bp) | 691,378 | 617,420 |
| L50 | 19 | 21 |
| BUSCO Statistic | C. zambiensis | C. syzygiicola |
|---|---|---|
| Overall completeness (%) | 96.2 | 95 |
| Complete BUSCO genes (C) | 3672 | 3631 |
| Single copy orthologs (S) | 3664 | 3625 |
| Duplicated orthologs (D) | 8 | 6 |
| Fragmented orthologs (F) | 34 | 39 |
| Missing orthologs (M) | 111 | 147 |
| NCBI | C. syzygiicola | C. zambiensis | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Species | Isolate | Assembly | MAT1-1-1 | MAT1-1-2 | MAT1-2-1 | AVG | MAT1-1-1 | MAT1-1-2 | MAT1-2-1 | AVG |
| C. syzygiicola | CMW29940 | PRJNA971112 | — | — | — | — | 91.29 | 98.11 | 97.12 | 95.50 |
| C. zambiensis | CMW29930 | PRJNA971112 | 91.29 | 98.11 | 97.12 | 95.50 | — | — | — | — |
| C. austroafricana | CMW6102 (MAT1-1) | ASM105115 | 97.51 | 100 | — | 98.76 | 92.57 | 98.11 | — | 95.34 |
| C. austroafricana | CMW2113 (MAT1-2) | ASM1607180 | 98.76 | 100 | 99.64 | 99.47 | 84.57 | 98.11 | 97.48 | 93.39 |
| C. cubensis | CMW10028 | ASM128231 | 84.65 | 97.57 | 97.12 | 93.11 | 87.55 | 96.22 | 97.48 | 93.75 |
| C. deuterocubensis | CMW8650 | ASM151382 | 87.55 | 96.76 | 94.24 | 92.85 | 83.55 | 95.41 | 94.60 | 91.19 |
| C. puriensis | CMW54409 (MAT1-1) | ASM1567895 | 84.23 | 95.41 | — | 89.82 | 83.82 | 94.05 | — | 88.94 |
| Cry. parasitica | EP155 | Crypa2 | 46.47 | 52.15 | 62.23 | 53.62 | 45.23 | 51.61 | 61.87 | 52.90 |
| Per gene average similarity | 84.35 | 91.43 | 90.07 | 81.23 | 90.23 | 89.71 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
van der Merwe, N.A.; Phakalatsane, T.; Wilken, P.M. The Unique Homothallic Mating-Type Loci of the Fungal Tree Pathogens Chrysoporthe syzygiicola and Chrysoporthe zambiensis from Africa. Genes 2023, 14, 1158. https://doi.org/10.3390/genes14061158
van der Merwe NA, Phakalatsane T, Wilken PM. The Unique Homothallic Mating-Type Loci of the Fungal Tree Pathogens Chrysoporthe syzygiicola and Chrysoporthe zambiensis from Africa. Genes. 2023; 14(6):1158. https://doi.org/10.3390/genes14061158
Chicago/Turabian Stylevan der Merwe, Nicolaas A., Tshiamo Phakalatsane, and P. Markus Wilken. 2023. "The Unique Homothallic Mating-Type Loci of the Fungal Tree Pathogens Chrysoporthe syzygiicola and Chrysoporthe zambiensis from Africa" Genes 14, no. 6: 1158. https://doi.org/10.3390/genes14061158
APA Stylevan der Merwe, N. A., Phakalatsane, T., & Wilken, P. M. (2023). The Unique Homothallic Mating-Type Loci of the Fungal Tree Pathogens Chrysoporthe syzygiicola and Chrysoporthe zambiensis from Africa. Genes, 14(6), 1158. https://doi.org/10.3390/genes14061158






