Morphological and Genetics Support for a Hitherto Undescribed Spotted Cat Species (Genus Leopardus; Felidae, Carnivora) from the Southern Colombian Andes
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Mitochondrial DNA
2.3. Nuclear DNA Microsatellites
2.4. Statistical Analyses
2.4.1. Mitochondrial Genes
Phylogenetic Analyses and Temporal Split Estimations
Genetic Distances
2.4.2. Nuclear DNA Microsatellites
Genetic Relationships among Neotropical Cat Species
3. Results
3.1. Description of the Skin of the Nariño Cat
3.2. Mitochondrial Analysis
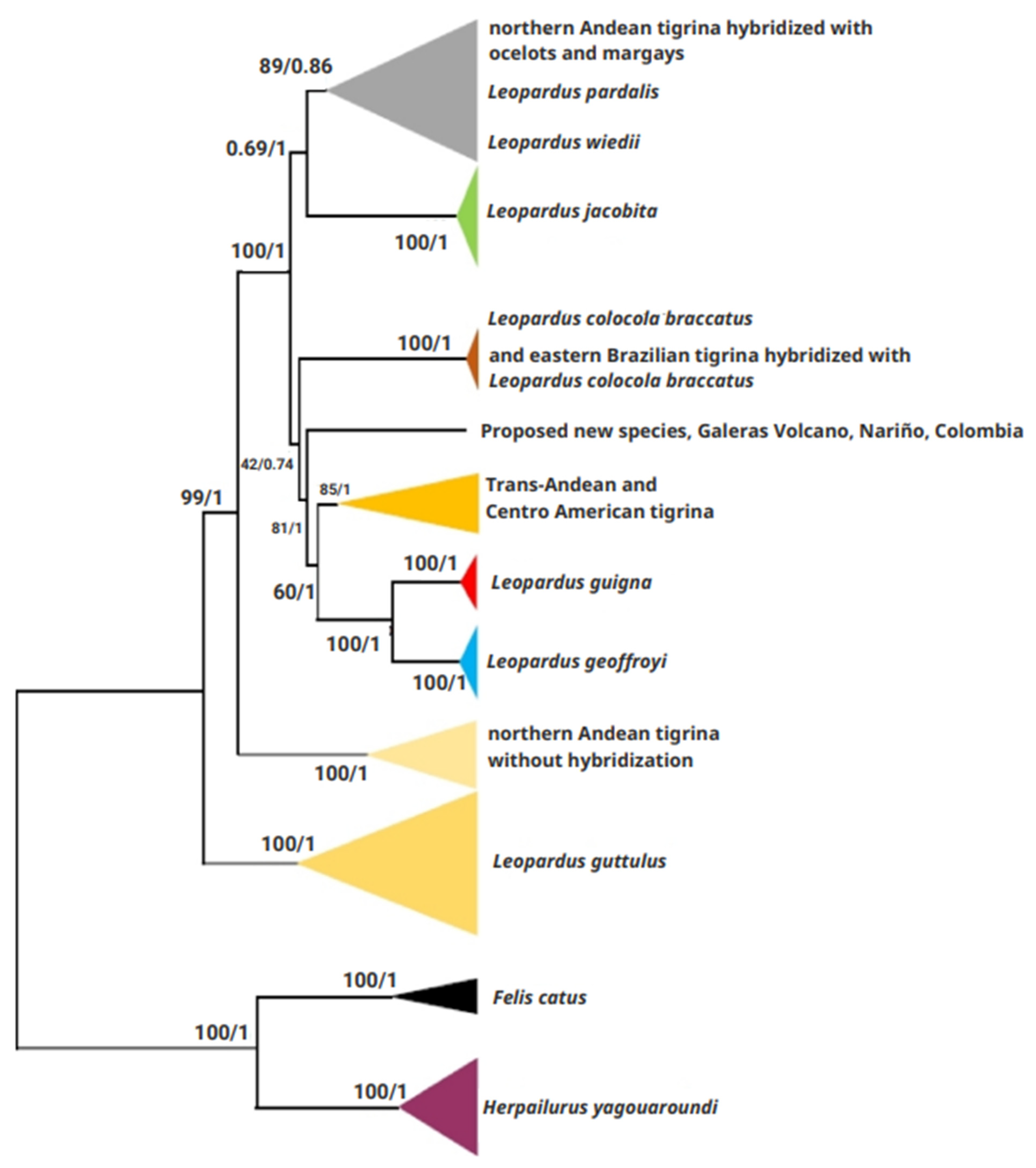
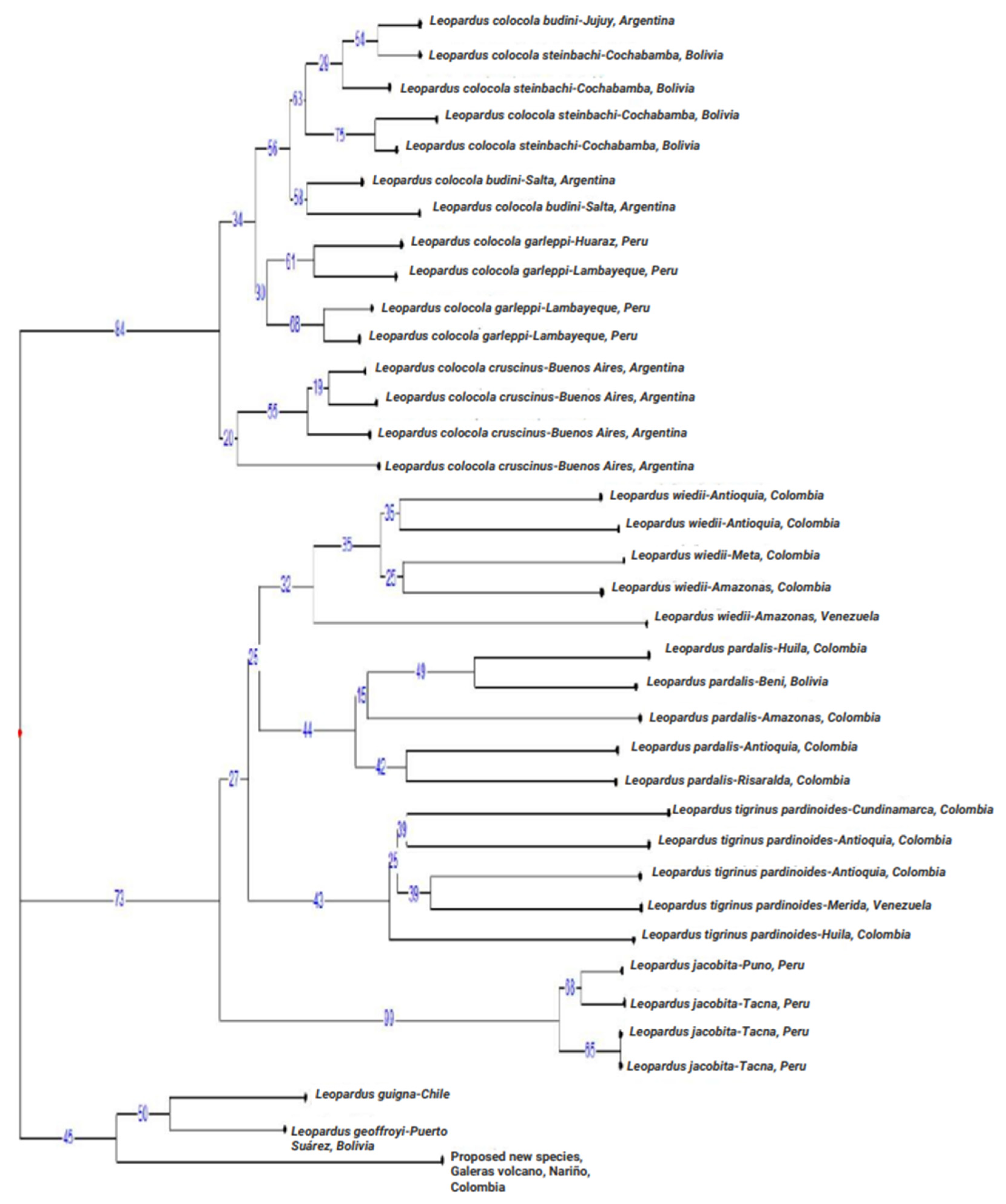
3.2.1. Phylogenetic Analyses of Mitogenomes
3.2.2. Genetic Distance Analysis with Mitogenomes
3.2.3. Phylogenetic Analyses at the mtND5 Gene
3.2.4. Genetic Distance Analysis at the mtND5 and mtCyt-b Genes
3.3. Microsatellites
4. Discussion
4.1. Some Taxonomic Insight of the Tigrina
4.2. The Nariño Cat
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cracraft, J. Species concepts and speciation analysis. In Current ornithology, Volume I; Johnston, R.J., Ed.; Plenum Press: New York, NY, USA, 1983; pp. 177–196. [Google Scholar]
- Groves, C.P. Primate Taxonomy; Smithsonian Institution Press: Washington, DC, USA, 2001. [Google Scholar]
- Boubli, J.P.; Rylands, A.B.; Farias, I.P.; Alfaro, M.E.; Alfaro, J.L. Cebus phylogenetic relationships: A preliminary reassessment of the diversity of the untufted capuchin monkeys. Am. J. Primatol. 2012, 74, 381–393. [Google Scholar] [CrossRef] [PubMed]
- Groves, C.P.; Grubb, P. Ungulate Taxonomy; Johns Hopkins University Press: Baltimore, MD, USA, 2011. [Google Scholar]
- Helgen, K.M.; Kays, R.; Helgen, L.E.; Tsuchiya-Jerep, M.T.N.; Pinto, C.M.; Koepfli, K.-P.; Eizirik, E.; Maldonado, J.E. Taxonomic boundaries and geographic distributions revealed by an integrative systematic overview of the mountain coatis, Nasuella (Carnivora: Procyonidae). Small Carniv. Conserv. 2009, 41, 65–74. [Google Scholar]
- Molina, M.; Molinari, J. Taxonomy of Venezuelan white-tailed deer (Odocoileus, Cervidae, Mammalia), based on cranial and mandibular traits. Can. J. Zool. 1999, 77, 632–645. [Google Scholar] [CrossRef]
- Molinari, J. Variación geográfica en los venados de cola blanca (Cervidae, Odocoileus) de Venezuela, con énfasis en O. margaritae, la especie enana de la Isla de Margarita. Mem. Fund. Salle Cienc. Nat. 2007, 167, 29–72. [Google Scholar]
- Van Roosmalen, M.G.M.; Frenz, L.; Van Hooft, P.; de Iongh, H.H.; Leirs, H. A new species of living peccary (Mammalia: Tayassuidae) from the Brazilian Amazon. Bonn. Zool. Beiträge 2007, 55, 105–112. [Google Scholar]
- Hrbek, T.; Da Silva, V.M.F.; Dutra, N.; Gravena, W.; Martin, A.R.; Farias, I.P. A new species of river dolphin from Brazil or: How little do we know our biodiversity. PLoS ONE 2014, 9, e0083623. [Google Scholar] [CrossRef]
- Cozzuol, M.A.; Clozato, C.L.; Holanda, E.C.; Rodrigues, F.H.G.; Nienow, S.D.S.; De Thoisy, B.; Redondo, R.A.F.; Santos, F.R. A new species of tapir from the Amazon. J. Mammal. 2013, 94, 1331–1345. [Google Scholar] [CrossRef]
- Ruiz-García, M.; Castellanos, A.; Bernal, L.A.; Pinedo-Castro, M.; Kaston, F.; Shostell, J.M. Mitogenomics of the mountain tapir (Tapirus pinchaque, Tapiridae, Perissodactyla, Mammalia) in Colombia and Ecuador: Phylogeography and insights into the origin and systematics of the South American tapirs. Mamm. Biol. 2016, 81, 163–175. [Google Scholar] [CrossRef]
- Ruiz-García, M.; Castellanos, A.; Kaston, F.; Pinedo-Castro, M.; Shostell, J.M. New insights into the molecular evolution of Tapirus pinchaque (Tapiridae, Perissodactyla) and the rising and fall of Tapirus kabomani as a full species. Genes 2023. submitted. [Google Scholar]
- Anderson, R.P.; Handley, C.O. A new species of three-toed sloth (Mammalia: Xenarthra) from Panama, with a review of the genus Bradypus. Proc. Biol. Soc. Wash. 2001, 114, 1–33. [Google Scholar]
- Helgen, K.M.; Pinto, C.M.; Kays, R.; Helgen, L.; Tsuchiya, M.; Quinn, A.; Wilson, D.; Maldonado, J. Taxonomic revision of the olingos (Bassaricyon), with description of a new species, the Olinguito. ZooKeys 2013, 324, 1–83. [Google Scholar] [CrossRef]
- Schneider, H.; Bernardi, J.A.R.; Da Cunha, D.B.; Tagliaro, C.H.; Vallinoto, M.; Ferrari, S.F.; Sampaio, I. A molecular analysis of the evolutionary relationships in the Callitrichinae, with emphasis on the position of the dwarf marmoset. Zool. Scr. 2011, 41, 1–10. [Google Scholar] [CrossRef]
- Costa-Araújo, R.; de Melo, F.R.; Canale, G.R.; Hernández-Rangel, S.M.; Messias, M.R.; Rossi, R.V.; Silva, F.E.; da Silva, M.N.F.; Nash, S.D.; Boubli, J.P.; et al. The Munduruku marmoset: A new monkey species from southern Amazonia. Peer J. 2019, 7, e7019. [Google Scholar] [CrossRef]
- Costa-Araujo, R.; Silva, J.S., Jr.; Boubli, J.P.; Rossi, R.V.; Canale, G.R.; Melo, F.R.; Bertuol, F.; Silva, F.E.; Silva, D.A.; Nash, S.D.; et al. An integrative analysis uncovers a new, pseudo-cryptic species of Amazonian marmoset (Primates: Callitrichidae: Mico) from the arc of deforestation. Sci. Rep. 2021, 11, 15665. [Google Scholar] [CrossRef]
- Moratelli, R.; Wilson, D.; Novaes, R.L.M.; Helgen, K.M.; Gutiérrez, E. Caribbean Myotis (Chiroptera, Vespertilionidae), with description of a new species from Trinidad and Tobago. J. Mammal. 2017, 98, 994–1008. [Google Scholar] [CrossRef]
- Brito, J.; Koch, C.; Tinoco, N.; Pardiñas, U.F.J. A new species of Mindomys (Rodentia, Cricetidae) with remarks on external traits as indicators of arboreality in sigmodontine rodents. Evol. Sist. 2022, 6, 35–55. [Google Scholar] [CrossRef]
- Miranda, F.R.; Casali, D.M.; A Perini, F.; Machado, F.; Santos, F.R. Taxonomic review of the genus Cyclopes Gray, 1821 (Xenarthra: Pilosa), with the revalidation and description of new species. Zool. J. Linn. Soc. 2018, 183, 687–721. [Google Scholar] [CrossRef]
- Wurster-Hill, D.H.; Centerwall, W.R. The interrelationships of chromosome banding patterns in Procyonids, Viverrids and Felids. Cytogenet. Cell Genet. 1982, 34, 178–192. [Google Scholar] [CrossRef]
- Collier, G.E.; O’Brien, S.J. A molecular phylogeny of the Felidae: Immunological distance. Evolution 1985, 39, 437–487. [Google Scholar] [CrossRef]
- Salles, L.O. Felid phylogenetics: Extant taxa and skull morphology (Felidae, Aeluroidae). Am. Mus. Novit. 1992, 3047, 1–67. [Google Scholar]
- Pecon Slattery, J.; Johnson, W.E.; Goldman, D.; O’Brien, S.J. Phylogenetic reconstruction of South American felids defined by protein electrophoresis. J. Mol. Evol. 1994, 39, 296–305. [Google Scholar] [CrossRef] [PubMed]
- Pecon Slattery, J.; O’Brien, S.J. Patterns of Y and X chromosome DNA sequence divergence during the Felidae radiation. Genetics 1994, 148, 1245–1255. [Google Scholar] [CrossRef] [PubMed]
- Johnson, W.E.; O’Brien, S.J. Phylogenetic reconstruction of the Felidae using 16S rRNA and NADH-5 mitochondrial genes. J. Mol. Evol. 1997, 44, S98–S116. [Google Scholar] [CrossRef] [PubMed]
- Johnson, W.E.; Culver, M.; Iriarte, J.; Eizirik, E.; Seymour, K.; O’Brien, S. Tracking the evolution of the elusive Andean mountain cat (Oreailurus jacobita) from mitochondrial DNA. J. Hered. 1998, 89, 227–232. [Google Scholar] [CrossRef]
- Johnson, W.E.; Dratch, P.A.; Martenson, J.S.; O’Brien, S. Resolution of recent radiations within three evolutionary lineages of Felidae using mitochondrial restriction fragment length polymorphism variation. J. Mamm. Evol. 1996, 3, 97–120. [Google Scholar] [CrossRef]
- Johnson, W.E.; Eizirik, E.; Pecon-Slattery, J.; Murphy, W.J.; Antunes, A.; Teeling, E.; O’Brien, S.J. The late Miocene radiation of modern Felidae: A genetic assessment. Science 2006, 311, 73–77. [Google Scholar] [CrossRef]
- Trigo, T.C.; Freitas, T.R.O.; Kunzler, G.; Cardoso, L.; Silva, J.C.R.; Johnson, W.E.; O’Brien, S.J.; Bonatto, S.L.; Eizirik, E. Inter-species hybridization among Neotropical cats of the genus Leopardus, and evidence for an introgressive hybrid zone between L. geoffroyi and L. tigrinus in southern Brazil. Mol. Ecol. 2008, 17, 4317–4333. [Google Scholar] [CrossRef]
- Trigo, T.C.; Schneider, A.; Oliveira, T.G.; Lehugeur, L.M.; Silveira, L.; Freitas, T.R.; Eizirik, E. Molecular data reveal complex hybridization and a cryptic species of Neotropical wild cat. Curr. Biol. 2013, 23, 2528–2533. [Google Scholar] [CrossRef]
- Trigo, T.C.; Tirelli, F.P.; De Freitas, T.R.O.; Eizirik, E. Comparative assessment of genetic and morphological variation at an extensive hybrid zone between two wild cats in Southern Brazil. PLoS ONE 2014, 9, e108469. [Google Scholar] [CrossRef]
- Li, G.; Davis, B.W.; Eizirik, E.; Murphy, W.J. Phylogenomic evidence for ancient hybridization in the genomes of living cats (Felidae). Genome Res. 2016, 26, 1057–1068. [Google Scholar] [CrossRef]
- García-Perea, R. The pampas cat group (Genus Lynchailurus Severtzov, 1958 (Carnivora, Felidae), a systematic and biogeographic review. Am. Mus. Novit. 1994, 3096, 1–35. [Google Scholar]
- von Schreber, J.C.D. Die Säugethiere in Abbildungen nach der Natur mit Beschreibungen; Verlegts Wolfgang Walther: Erlangen, Geramgy, 1775; 2(14), pl. 99, 100; 2(15): Pl. 101, 101B, 105, 106; 2(16): Pl. 108. [Google Scholar]
- von Schreber, J.C.D. Die Säugethiere in Abbildungen nach der Natur mit Beschreibungen; Wolfgang: Erlangen, Geramgy, 1777; v. 2, p. 13, pl. 110B; v. 3, pt. 22, 384–387, 392; v. 3, pt. 23, p. 393–394; 396–397; 407; v. 3, pt. 24, p. 412–414; v. 3, pt. 25, pl. 104B, 109B. [Google Scholar]
- Buffon, G. L’Histoire Naturelle, Générale et Particulière, avec la Description du Cabinet du Roi. Tome XIII (Quadrupèdes X); L’Imprimerie Royale: Paris, France, 1765; pp. 1–368. [Google Scholar]
- Gray, J.E. Notes on certain species of cats in the collection of the British Museum. Proc. Zool. Soc. Lond. 1867, 14, 394–405. [Google Scholar]
- Gray, J.E. The Bogota cat (Felis pardinoides, Gray). Ann. Mag. Nat. Hist. Ser. 1874, 13, 475. [Google Scholar] [CrossRef]
- Hensel, R.F. Beitrage zur Kenntniss der Saugethiere Sud-Brasiliens; Abhandlungen der Königlich Preussischen Akademie der Wissenschaft: Berlin, Germany, 1872; pp. 1–130. [Google Scholar]
- Allen, J.A. New South American mammals. Bull. Amer. Mus. Nat. Hist. 1915, 34, 625–634. [Google Scholar]
- Cabrera, A. Catálogo de los mamíferos de América del Sur. I (Metatheria, Unguiculata, Carnívora). Rev. Del Mus. Argent. De Cienc. Nat. Zool. 1957, 4, 1–307. [Google Scholar]
- Cabrera, A. Los félidos vivientes de la República de Argentina. Rev. Del Mus. Argent. De Cienc. Nat. Zool. 1961, 6, 161–247. [Google Scholar]
- Wilson, D.E.; Reeder, D.A.M. Mammal Species of the World. A Taxonomic and Geographic Reference, 3rd ed.; Johns Hopkins University Press: Baltimore, MD, USA, 2005. [Google Scholar]
- Nascimento, F.O.; Feijó, A. Taxonomic revision of the tigrina Leopardus tigrinus (Schreber, 1775) species group (Carnivora, Felidae). Papéis Avulsos De Zool. 2017, 57, 231–264. [Google Scholar] [CrossRef]
- Ruiz-García, M.; Pinedo-Castro, M.; Shostell, J.M. Small spotted bodies with multiple specific mitochondrial DNAs: Existence of diverse and differentiated tigrina lineages or species (Leopardus spp: Felidae, Mammalia) throughout Latin America. Mitochondrial DNA Part A 2018, 29, 993–1014. [Google Scholar] [CrossRef]
- Moraes-Barros, N.; Morgante, J.S. A simple protocol for the extraction and sequence analysis of DNA from study skin of museum collections. Genet. Mol. Biol. 2007, 30, 1181–1185. [Google Scholar] [CrossRef]
- Thalmann, O.; Hebler, J.; Poinar, H.N.; Pääbo, S.; Vigilant, L. Unreliable mtDNA data due to nuclear insertions: A cautionary tale from analysis of humans and other apes. Mol. Ecol. 2004, 13, 321–335. [Google Scholar] [CrossRef]
- Raaum, R.L.; Sterner, K.N.; Noviello, C.M.; Stewart, C.-B.; Disotell, T.R. Catarrhine primate divergence dates estimated from complete mitochondrial genomes: Concordance with fossil and nuclear DNA evidence. J. Hum. Evol. 2005, 48, 237–257. [Google Scholar] [CrossRef] [PubMed]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef]
- Larkin, A. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Ruiz-García, M.; García-Perea, R.; Corrales, C.; Murillo, A.; Álvarez, D.; Pinedo-Castro, M.; Shostell, J.M. Determination of microsatellite DNA mutation rates, mutation models and mutation bias in four main Felidae lineages (European wild cat, Felis silvestris; Ocelot, Leopardus pardalis; Puma, Puma concolor; Jaguar, Panthera onca). In Molecular Population Genetics, Phylogenetics, Evolutionary Biology and Conservation of the Neotropical Carnivores; Ruiz-García, M., Shostell, J.M., Eds.; Nova Science Publishers: New York, NY, USA, 2013; pp. 333–412. [Google Scholar]
- Menotti-Raymond, M.A.; O’Brien, S.J. Evolutionary conservation of ten Microsatellite loci in four species of Felidae. J. Hered. 1995, 86, 319–322. [Google Scholar] [CrossRef]
- Menotti-Raymond, M.; David, V.A.; Lyons, L.; Schäffer, A.A.; Tomlin, J.F.; Hutton, M.K.; O’Brien, S.J. A genetic linkage map of microsatellites in the domestic cat (Felis catus). Genomics 1999, 57, 9–23. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Tanabe, A.S. Kakusan4 and Aminosan: Two programs for comparing nonpartitioned, proportional and separate models for combined molecular phylogenetic analyses of multilocus sequence data. Mol. Ecol. Resour. 2011, 11, 914–921. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Autom. Control. 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Posada, D.; Buckley, T.R. Model selection and model averaging in phylogenetics: Advantages of akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst. Biol. 2004, 53, 793–808. [Google Scholar] [CrossRef]
- Lanave, C.; Preparata, G.; Saccone, C.; Serio, G. A new method for calculating evolutionary substitution rates. J. Mol. Evol. 1984, 20, 86–93. [Google Scholar] [CrossRef]
- Tavaré, S. Some Probabilistic and Statistical Problems in the Analysis of DNA Sequences. Lect. Math. Life Sci. 1986, 17, 57–86. [Google Scholar]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J. TreeAnnotator v1.8.0. 2013. Available online: https://beast.bio.ed.ac.uk/ (accessed on 7 November 2021).
- Rambaut, A.; Suchard, M.A.; Xie, W.; Drummond, A.J. Tracer v1.6. 2013. Available online: https://tree.bio.ed.ac.uk/software/tracer (accessed on 7 November 2012).
- Drummond, A.J.; Ho, S.Y.W.; Phillips, M.J.; Rambaut, A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006, 4, e88. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree v1.4. 2012. Available online: https://tree.bio.ed.ac.uk/software/figtree/ (accessed on 7 November 2021).
- Soibelzon, L.H.; Prevosti. Fossils of South American land carnivores (Carnivora, Mammalia). In Molecular Population Genetics, Evolutionary Biology and Biological Conservation of Neotropical Carnivores; Ruiz-García, M., Shostell, J.M., Eds.; Nova Science Publisher: New York, NY, USA, 2013; pp. 509–527. [Google Scholar]
- Pennington, R.T.; Dick, C.W. Diversification of the Amazonian flora and its relation to key geological and environmental events: A molecular perspective. In Amazonia, Landscape and Species Evolution: A Look into the Past; Hoorn, C., Wesselingh, F., Eds.; Wiley-Blackwell: Oxford, UK, 2010; pp. 373–385. [Google Scholar]
- Bandelt, H.-J.; Forster, P.; Rohl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Morral, N.; Bertrantpetit, J.; Estivill, X.; Nunes, V.; Casals, T.; Giménez, J.; Reis, A.; Varon-Mateeva, R.; Macek, M., Jr.; Kalaydjieva, L.; et al. The origin of the major cystic fibrosis mutation (delta F508) in European populations. Nat. Genet. 1994, 7, 169–175. [Google Scholar] [CrossRef]
- Culver, M.; Johnson, W.E.; Pecon-Slattery, J.; O’Brien, S. Genomic ancestry of the American puma (Puma concolor). J. Hered. 2000, 91, 186–197. [Google Scholar] [CrossRef] [PubMed]
- López, J.V.; Culver, M.; Stephens, J.C.; O’Brien, S.J. Rates of nuclear and cytoplasmic mitochondrial DNA sequence divergence in mammals. Mol. Biol. Evol. 1997, 14, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Posada, D.; Crandall, K.A. Intraspecific gene genealogies: Trees grafting into networks. Trends Ecol. Evol. 2001, 16, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Avise, J.C. Molecular Markers, Natural History, and Evolution; Chapman and Hall: New York, NY, USA, 1994. [Google Scholar]
- Bradley, R.D.; Baker, R.J. A test of the genetic species concept: Cytochrome-b sequences and mammals. J. Mammal. 2001, 82, 960–973. [Google Scholar] [CrossRef]
- Kartavtsev, Y. Divergence at Cyt-b and Co-1 mtDNA genes on different taxonomic levels and genetics of speciation in animals. Mitochondrial DNA 2011, 22, 55–65. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Ratnasingham, S.; de Waard, J.R. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. B 2003, 270 (Suppl. S1), S96–S99. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Stoeckle, M.Y.; Zemlak, T.; Francis, C.M. Identification of birds through DNA barcodes. PLoS Biol. 2004, 2, e312. [Google Scholar] [CrossRef]
- Ward, J.H. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Gascuel, O. Concerning the NJ algorithm and its unweighted version, UNJ. Am. Math. Soc. 1997, 37, 149–170. [Google Scholar]
- Gower, J.C. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 1966, 53, 325–338. [Google Scholar] [CrossRef]
- Goldstein, D.B.; Ruiz-Linares, A.; Cavalli-Sforza, L.L.; Feldman, M.W. An evaluation of genetic distances for use with microsatellite loci. Genetics 1995, 139, 463–471. [Google Scholar] [CrossRef]
- Gower, J.C.; Ross, G.J.S. Minimum spanning trees and single cluster analysis. Appl. Stat. 1969, 18, 54–64. [Google Scholar] [CrossRef]
- Rohlf, F.J. Adaptive hierarchical clustering schemes. Syst. Zool. 1970, 19, 145–153. [Google Scholar] [CrossRef]
- Tobe, S.S.; Kitchener, A.C.; Linacre, A.M.T. Reconstructing mammalian phylogenies: A detailed comparison of the Cytochrome b and Cytochrome Oxidasa Subunit I mitochondrial genes. PLoS ONE 2010, 5, e14156. [Google Scholar] [CrossRef]
- Cracraft, J. Speciation and its ontology: The empirical consequences of alternative species concepts for understanding patterns and processes of differentiation. In Speciation and Its Consequences; Otte, D., Endler, J., Eds.; Sinauer Assoc: Sunderland, MA, USA, 1989; pp. 28–59. [Google Scholar]
- Kitchener, A.C.; Breitenmoser-Würsten, C.; Eizirik, E.; Gentry, A.; Werdelin, L.; Wilting, A.; Yamaguchi, N.; Abramov, A.V.; Christiansen, P.; Driscoll, C.; et al. A revised taxonomy of the Felidae. The final report of the Cat Classification Task Force of the IUCN/SSC Cat Specialist Group. Cat News Spec. Issue 2017, 11, 1–80. [Google Scholar]
- Allen, J.A. Notes on the synonymy and nomenclature of the smaller spotted cats of Tropical America. Bull. Am. Mus. Nat. Hist. 1919, 41, 341–419. [Google Scholar]
- Clapperton, C. Quaternary Geology and Geomorphology of South America; Elsevier: Amsterdam, The Netherlands, 1993. [Google Scholar]
- Van Der Hammen, T. La Paleoecología de Suramérica Tropical. Cuarenta Años de Investigación de la Historia del Medio Ambiente y de la Vegetación. Historia, Ecología y Vegetación; Corporación Colombiana para la Amazonía-Araracuara: Bogotá, Colombia, 1992. [Google Scholar]
- Hernández-Camacho, J.; Hurtado, A.; Ortiz, R.; Walschburger, T. Unidades biogeográficas de Colombia. In La Diversidad Biológica de Iberoamérica; Halffter, I.G., Ed.; Acta Zoológica Mexicana, Instituto de Ecología, A.C.: México, Mexico, 1992; pp. 105–151. [Google Scholar]
- Cossíos, E.D.; Lucherini, M.; Ruiz-García, M.; Angers, B. Influence of ancient glacial periods on the Andean fauna: The case of the Pampas cat (Leopardus colocolo). BMC Evol. Biol. 2009, 9, 68. [Google Scholar] [CrossRef]
- Ruiz-García, M.; Rivas-Sánchez, D.; Lichilín-Ortiz, N. Phylogenetics relationships among four putative taxa of foxes of the Pseudoalopex genus (Canidae, Carnivora) and molecular population genetics of Ps. culpaeus and Ps. sechurae. In Molecular Population Genetics, Evolutionary Biology and Biological Conservation of Neotropical Carnivores; Ruiz-García, M., Shostell, J.M., Eds.; Nova Science Publisher: New York, NY, USA, 2013; pp. 97–128. [Google Scholar]



| Cat taxa | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 |
| 1 | - | 0.9 | 1.2 | 0.8 | 0.9 | 0.9 | 0.8 | 0.4 | 0.3 | 0.8 | 0.7 | 0.7 | 1.5 | 1.6 |
| 2 | 10.6 | - | 1.4 | 1.2 | 1.3 | 1.3 | 1.3 | 1.0 | 1.0 | 1.3 | 1.2 | 1.1 | 1.8 | 2.0 |
| 3 | 15.0 | 16.9 | - | 1.1 | 1.3 | 1.3 | 1.2 | 1.2 | 1.2 | 1.2 | 1.2 | 1.2 | 2.1 | 2.1 |
| 4 | 7.3 | 13.2 | 13.3 | - | 0.9 | 0.9 | 0.7 | 0.7 | 0.9 | 0.7 | 0.8 | 0.7 | 1.5 | 1.7 |
| 5 | 6.4 | 13.6 | 16.3 | 6.9 | - | 0.0 | 1.0 | 1.0 | 1.0 | 0.9 | 1.0 | 0.9 | 1.7 | 1.9 |
| 6 | 6.4 | 13.6 | 16.3 | 6.9 | 0.0 | - | 1.0 | 1.0 | 1.0 | 0.9 | 1.0 | 0.9 | 1.7 | 1.9 |
| 7 | 6.8 | 14.7 | 15.8 | 5.3 | 7.2 | 7.2 | - | 0.8 | 0.9 | 0.5 | 0.9 | 0.9 | 1.7 | 1.9 |
| 8 | 3.3 | 11.9 | 15.2 | 7.2 | 7.7 | 7.7 | 7.2 | - | 0.5 | 0.8 | 0.8 | 0.7 | 1.6 | 1.6 |
| 9 | 2.8 | 10.6 | 15.4 | 8.0 | 6.9 | 6.9 | 7.5 | 4.4 | - | 0.9 | 0.8 | 0.7 | 1.5 | 1.7 |
| 10 | 6.8 | 14.3 | 15.0 | 5.0 | 6.5 | 6.5 | 2.0 | 7.2 | 7.3 | - | 0.8 | 0.8 | 1.5 | 1.8 |
| 11 | 5.7 | 12.8 | 15.3 | 7.7 | 6.9 | 6.9 | 6.9 | 6.4 | 6.3 | 6.7 | - | 0.9 | 1.6 | 1.9 |
| 12 | 6.7 | 13.4 | 14.4 | 5.6 | 6.1 | 6.1 | 6.1 | 7.0 | 7.6 | 5.8 | 7.0 | - | 1.6 | 1.7 |
| 13 | 20.8 | 27.1 | 29.2 | 22.3 | 21.7 | 21.7 | 23.1 | 22.4 | 21.7 | 22.4 | 21.6 | 21.1 | - | 1.1 |
| 14 | 25.6 | 32.8 | 32.4 | 24.9 | 25.9 | 25.9 | 26.8 | 25.2 | 26.6 | 26.1 | 26.3 | 25.1 | 10.9 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruiz-García, M.; Pinedo-Castro, M.; Shostell, J.M. Morphological and Genetics Support for a Hitherto Undescribed Spotted Cat Species (Genus Leopardus; Felidae, Carnivora) from the Southern Colombian Andes. Genes 2023, 14, 1266. https://doi.org/10.3390/genes14061266
Ruiz-García M, Pinedo-Castro M, Shostell JM. Morphological and Genetics Support for a Hitherto Undescribed Spotted Cat Species (Genus Leopardus; Felidae, Carnivora) from the Southern Colombian Andes. Genes. 2023; 14(6):1266. https://doi.org/10.3390/genes14061266
Chicago/Turabian StyleRuiz-García, Manuel, Myreya Pinedo-Castro, and Joseph Mark Shostell. 2023. "Morphological and Genetics Support for a Hitherto Undescribed Spotted Cat Species (Genus Leopardus; Felidae, Carnivora) from the Southern Colombian Andes" Genes 14, no. 6: 1266. https://doi.org/10.3390/genes14061266
APA StyleRuiz-García, M., Pinedo-Castro, M., & Shostell, J. M. (2023). Morphological and Genetics Support for a Hitherto Undescribed Spotted Cat Species (Genus Leopardus; Felidae, Carnivora) from the Southern Colombian Andes. Genes, 14(6), 1266. https://doi.org/10.3390/genes14061266






