High Expression of MRE11A Is Associated with Shorter Survival and a Higher Risk of Death in CRC Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Tissue Collection
2.2. Gene Expression Analysis and HRD Assessment
2.3. Investigation of Microsatellite Instability
2.4. Inflammation-Related Peripheral Blood Measurements
2.5. Statistical Analyses
3. Results
3.1. Characteristics of CRC Patients
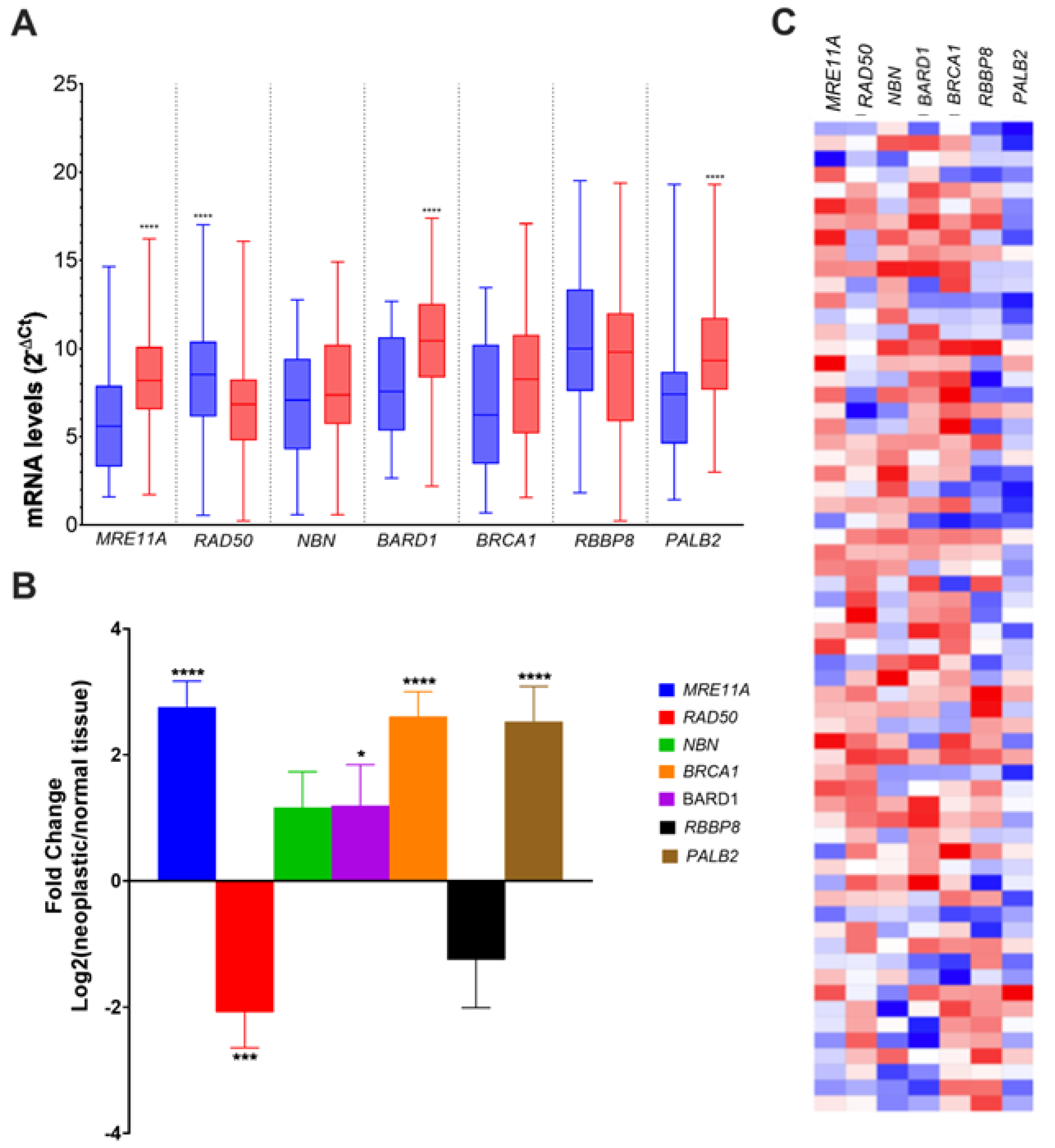
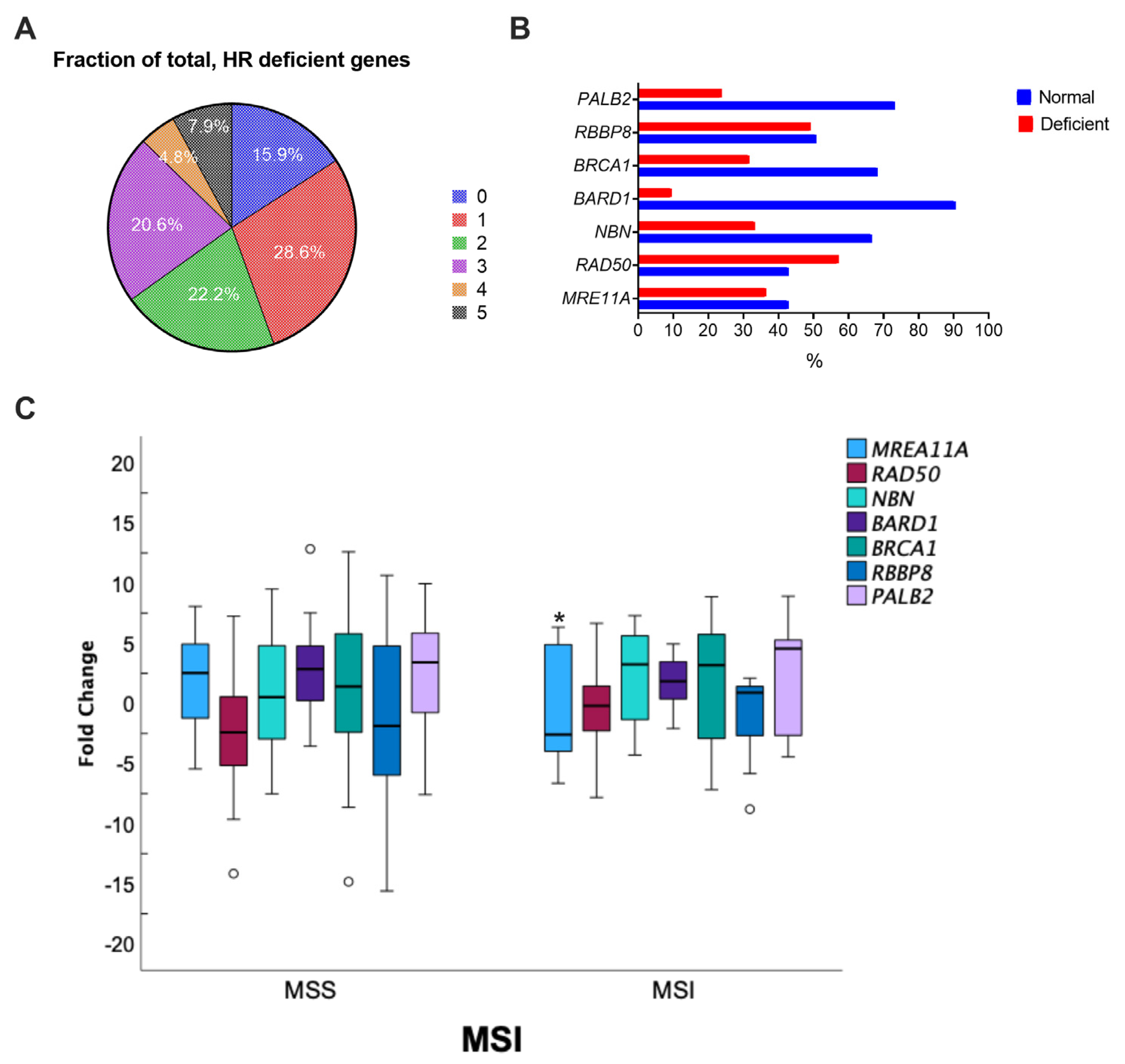
3.2. Alteration of HR Gene Expression in CRC
3.3. Associations between HR Gene Expression and Clinicopathological Features in CRC
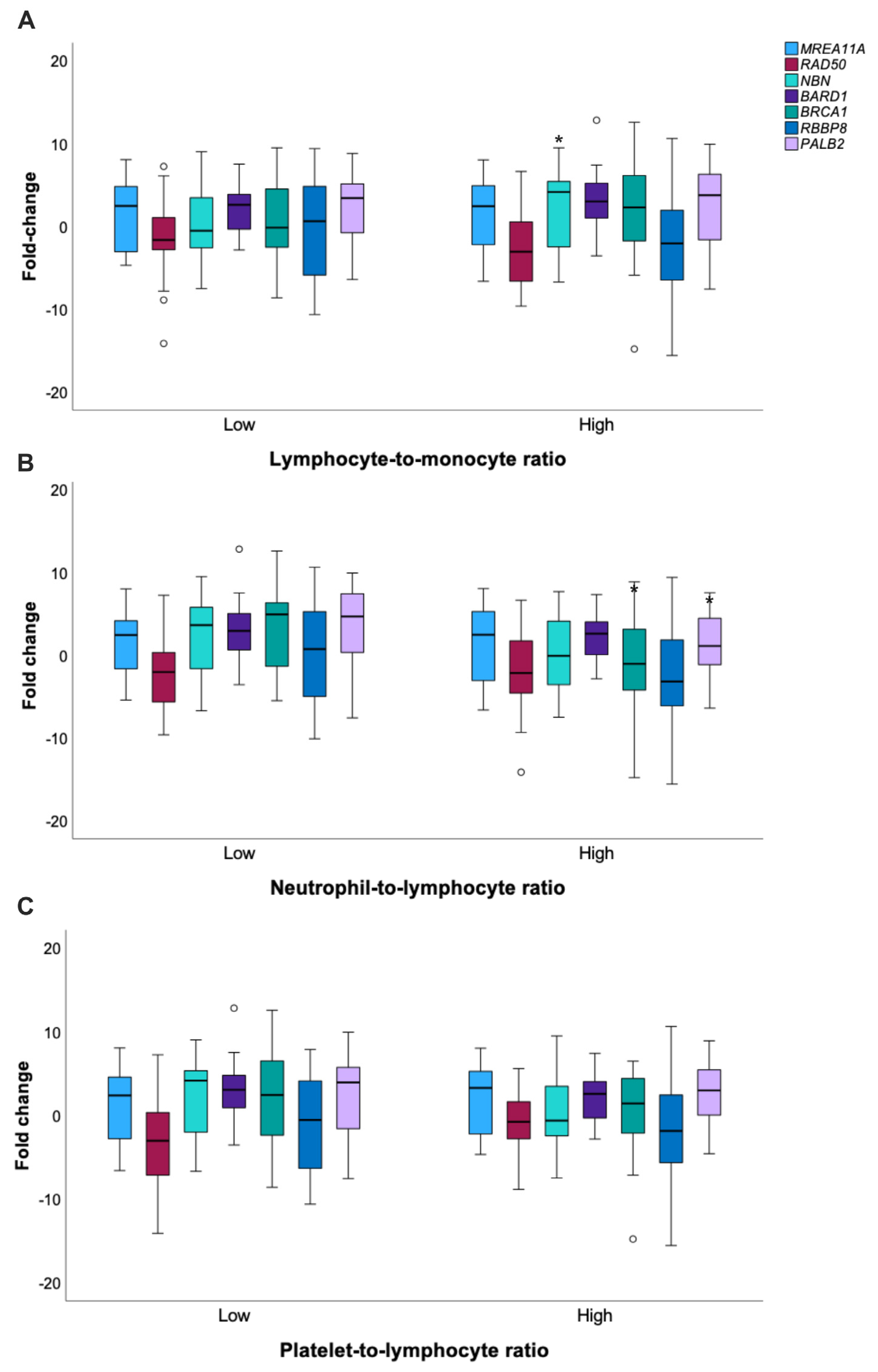
3.4. Analysis of Inflammatory Features in CRC Patients and Crosstalks with HR
3.5. High MRE11A Expression Is Associated with Poor Survival in CRC
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Daley, J.M.; Niu, H.; Miller, A.S.; Sung, P. Biochemical Mechanism of DSB End Resection and Its Regulation. DNA Repair 2015, 32, 66–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, P.; Greenberg, R.A. Noncanonical Views of Homology-Directed DNA Repair. Genes Dev. 2016, 30, 1138–1154. [Google Scholar] [CrossRef] [Green Version]
- Panier, S.; Boulton, S.J. Double-Strand Break Repair: 53BP1 Comes into Focus. Nat. Rev. Mol. Cell Biol. 2014, 15, 7–18. [Google Scholar] [CrossRef]
- Tarsounas, M.; Sung, P. The Antitumorigenic Roles of BRCA1–BARD1 in DNA Repair and Replication. Nat. Rev. Mol. Cell Biol. 2020, 21, 284–299. [Google Scholar] [CrossRef] [PubMed]
- Chartron, E.; Theillet, C.; Guiu, S.; Jacot, W. Targeting Homologous Repair Deficiency in Breast and Ovarian Cancers: Biological Pathways, Preclinical and Clinical Data. Crit. Rev. Oncol./Hematol. 2019, 133, 58–73. [Google Scholar] [CrossRef]
- Ihara, K.; Yamaguchi, S.; Ueno, N.; Tani, Y.; Shida, Y.; Ogata, H.; Domeki, Y.; Okamoto, K.; Nakajima, M.; Sasaki, K.; et al. Expression of DNA Double-Strand Break Repair Proteins Predicts the Response and Prognosis of Colorectal Cancer Patients Undergoing Oxaliplatin-Based Chemotherapy. Oncol. Rep. 2016, 35, 1349–1355. [Google Scholar] [CrossRef] [Green Version]
- Lin, P.C.; Yeh, Y.M.; Chan, R.H.; Lin, B.W.; Chen, P.C.; Pan, C.C.; Shen, M.R. Sequential and Co-Occurring DNA Damage Response Genetic Mutations Impact Survival in Stage III Colorectal Cancer Patients Receiving Adjuvant Oxaliplatin-Based Chemotherapy. BMC Cancer 2021, 21, 217. [Google Scholar] [CrossRef]
- Lord, C.J.; Ashworth, A. PARP Inhibitors: Synthetic Lethality in the Clinic. Science 2017, 355, 1152–1158. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA A Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Wang, R.; Wang, M.J.; Ping, J. Clinicopathological Features and Survival Outcomes of Colorectal Cancer in Young Versus Elderly. Medicine 2015, 94, e1402. [Google Scholar] [CrossRef]
- Li, H.; Fu, G.; Wei, W.; Huang, Y.; Wang, Z.; Liang, T.; Tian, S.; Chen, H.; Zhang, W. Re-Evaluation of the Survival Paradox Between Stage IIB/IIC and Stage IIIA Colon Cancer. Front. Oncol. 2020, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Mo, S.; Zhu, L.; Xu, T.; Cai, G. The Survival and Clinicopathological Differences between Patients with Stage IIIA and Stage II Rectal Cancer: An Analysis of 12,036 Patients in the SEER Database. Oncotarget 2016, 7, 79787–79796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Cutsem, E.; Cervantes, A.; Adam, R.; Sobrero, A.; Van Krieken, J.H.; Aderka, D.; Aguilar, E.A.; Bardelli, A.; Benson, A.; Bodoky, G.; et al. ESMO Consensus Guidelines for the Management of Patients with Metastatic Colorectal Cancer. Ann. Oncol. 2016, 27, 1386–1422. [Google Scholar] [CrossRef] [PubMed]
- Guinney, J.; Dienstmann, R.; Wang, X.; De Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The Consensus Molecular Subtypes of Colorectal Cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef] [PubMed]
- Sveen, A.; Cremolini, C.; Dienstmann, R. Predictive Modeling in Colorectal Cancer: Time to Move beyond Consensus Molecular Subtypes. Ann. Oncol. 2019, 30, 1682–1685. [Google Scholar] [CrossRef]
- Dienstmann, R.; Vermeulen, L.; Guinney, J.; Kopetz, S.; Tejpar, S.; Tabernero, J. Consensus Molecular Subtypes and the Evolution of Precision Medicine in Colorectal Cancer. Nat. Rev. Cancer 2017, 17, 79–92. [Google Scholar] [CrossRef]
- Heeke, A.L.; Pishvaian, M.J.; Lynce, F.; Xiu, J.; Brody, J.R.; Chen, W.-J.; Baker, T.M.; Marshall, J.L.; Isaacs, C. Prevalence of Homologous Recombination–Related Gene Mutations Across Multiple Cancer Types. JCO Precis. Oncol. 2018, 2, 1–13. [Google Scholar] [CrossRef]
- Reilly, N.M.; Novara, L.; Di Nicolantonio, F.; Bardelli, A. Exploiting DNA Repair Defects in Colorectal Cancer. Mol. Oncol. 2019, 13, 681–700. [Google Scholar] [CrossRef] [Green Version]
- AlDubayan, S.H.; Giannakis, M.; Moore, N.D.; Han, G.C.; Reardon, B.; Hamada, T.; Mu, X.J.; Nishihara, R.; Qian, Z.; Liu, L.; et al. Inherited DNA-Repair Defects in Colorectal Cancer. Am. J. Hum. Genet. 2018, 102, 401–414. [Google Scholar] [CrossRef] [Green Version]
- Knijnenburg, T.A.; Wang, L.; Zimmermann, M.T.; Chambwe, N.; Gao, G.F.; Cherniack, A.D.; Fan, H.; Shen, H.; Way, G.P.; Greene, C.S.; et al. Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep. 2018, 23, 239–254.e6. [Google Scholar] [CrossRef] [Green Version]
- Weiser, M.R. AJCC 8th Edition: Colorectal Cancer. Ann. Surg. Oncol. 2018, 25, 1454–1455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Min, A.; Kim, K.; Jeong, K.; Choi, S.; Kim, S.; Suh, K.J.; Lee, K.H.; Kim, S.; Im, S.A. Homologous Repair Deficiency Score for Identifying Breast Cancers with Defective DNA Damage Response. Sci. Rep. 2020, 10, 12506. [Google Scholar] [CrossRef] [PubMed]
- Luchini, C.; Bibeau, F.; Ligtenberg, M.J.L.; Singh, N.; Nottegar, A.; Bosse, T.; Miller, R.; Riaz, N.; Douillard, J.Y.; Andre, F.; et al. ESMO Recommendations on Microsatellite Instability Testing for Immunotherapy in Cancer, and Its Relationship with PD-1/PD-L1 Expression and Tumour Mutational Burden: A Systematic Review-Based Approach. Ann. Oncol. 2019, 30, 1232–1243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sylman, J.L.; Mitrugno, A.; Atallah, M.; Tormoen, G.W.; Shatzel, J.J.; Yunga, S.T.; Wagner, T.H.; Leppert, J.T.; Mallick, P.; McCarty, O.J.T. The Predictive Value of Inflammation-Related Peripheral Blood Measurements in Cancer Staging and Prognosis. Front. Oncol. 2018, 8, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Breast Cancer Association Consortium. Breast Cancer Risk Genes—Association Analysis in More than 113,000 Women. N. Engl. J. Med. 2021, 384, 428–439. [Google Scholar] [CrossRef]
- Hu, C.; Hart, S.N.; Gnanaolivu, R.; Huang, H.; Lee, K.Y.; Na, J.; Gao, C.; Lilyquist, J.; Yadav, S.; Boddicker, N.J.; et al. A Population-Based Study of Genes Previously Implicated in Breast Cancer. N. Engl. J. Med. 2021, 384, 440–451. [Google Scholar] [CrossRef]
- Arai, H.; Elliott, A.; Xiu, J.; Wang, J.; Battaglin, F.; Kawanishi, N.; Soni, S.; Zhang, W.; Millstein, J.; Sohal, D.; et al. The Landscape of Alterations in DNA Damage Response Pathways in Colorectal Cancer. Clin. Cancer Res. 2021, 27, 3234–3242. [Google Scholar] [CrossRef]
- Tomasini, P.P.; Guecheva, T.N.; Leguisamo, N.M.; Péricart, S.; Brunac, A.C.; Hoffmann, J.S.; Saffi, J. Analyzing the Opportunities to Target DNA Double-Strand Breaks Repair and Replicative Stress Responses to Improve Therapeutic Index of Colorectal Cancer. Cancers 2021, 13, 3130. [Google Scholar] [CrossRef]
- Pan, W.; Lu, K.; Wang, W.; Yao, J.; Hou, Y. PALB2 as a Potential Prognostic Biomarker for Colorectal Cancer. Comput. Biol. Chem. 2020, 87, 107289. [Google Scholar] [CrossRef]
- Ho, V.; Chung, L.; Singh, A.; Lea, V.; Abubakar, A.; Lim, S.H.; Ng, W.; Lee, M.; de Souza, P.; Shin, J.S.; et al. Overexpression of the MRE11-RAD50-NBS1 (MRN) Complex in Rectal Cancer Correlates with Poor Response to Neoadjuvant Radiotherapy and Prognosis. BMC Cancer 2018, 18, 869. [Google Scholar] [CrossRef]
- Fan, C.W.; Kopsida, M.; Liu, Y.B.; Zhang, H.; Gao, J.F.; Arbman, G.; Cao, S.Y.W.; Li, Y.; Zhou, Z.G.; Sun, X.F. Prognostic Heterogeneity of MRE11 Based on the Location of Primary Colorectal Cancer Is Caused by Activation of Different Immune Signals. Front. Oncol. 2020, 9, 1–11. [Google Scholar] [CrossRef]
- Patterson-Fortin, J.; Jadhav, H.; Pantelidou, C.; Phan, T.; Grochala, C.; Mehta, A.K.; Guerriero, J.L.; Wulf, G.M.; Wolpin, B.M.; Stanger, B.Z.; et al. Polymerase θ Inhibition Activates the CGAS-STING Pathway and Cooperates with Immune Checkpoint Blockade in Models of BRCA-Deficient Cancer. Nat. Commun. 2023, 14, 1390. [Google Scholar] [CrossRef] [PubMed]
- Belan, O.; Sebald, M.; Adamowicz, M.; Anand, R.; Vancevska, A.; Neves, J.; Grinkevich, V.; Hewitt, G.; Segura-Bayona, S.; Bellelli, R.; et al. POLQ Seals Post-Replicative SsDNA Gaps to Maintain Genome Stability in BRCA-Deficient Cancer Cells. Mol. Cell 2022, 82, 4664–4680.e9. [Google Scholar] [CrossRef] [PubMed]
- Azar, I.; Al Masalmeh, N.; Esfandiarifard, S.; Virk, G.; Kiwan, W.; Frank Shields, A.; Mehdi, S.; Philip, P.A. The Impact of Primary Tumor Sidedness on Survival in Early-onset Colorectal Cancer by Stage: A National Veterans Affairs Retrospective Analysis. Cancer Med. 2021, 10, 2987–2995. [Google Scholar] [CrossRef]
- Malakorn, S.; Ouchi, A.; Hu, C.-Y.; Sandhu, L.; Dasari, A.; You, Y.-Q.N.; Kopetz, E.S.; Ellis, L.M.; Chang, G.J. Tumor Sidedness, Recurrence, and Survival After Curative Resection of Localized Colon Cancer. Clin. Color. Cancer 2021, 20, e53–e60. [Google Scholar] [CrossRef]
- Petrelli, F.; Tomasello, G.; Borgonovo, K.; Ghidini, M.; Turati, L.; Dallera, P.; Passalacqua, R.; Sgroi, G.; Barni, S. Prognostic Survival Associated With Left-Sided vs. Right-Sided Colon Cancer: A Systematic Review and Meta-Analysis. JAMA Oncol. 2017, 3, 211–219. [Google Scholar] [CrossRef] [PubMed]

| Variable | n (%) | p Value |
|---|---|---|
| Total cases | 63 (100) | |
| Age (years; mean ± SD) | 64 ± 11 | |
| Age (years) | 0.705 | |
| ≤65 | 33 (52) | |
| >65 | 30 (47) | |
| Gender | 0.032 | |
| Female | 23 (36.5) | |
| Male | 40 (63.5) | |
| Primary Tumor location | 0.033 | |
| Colon | 40 (64) | |
| Rectum | 23 (36) | |
| Laterality (Colon) | 0.027 | |
| Right-sided | 13 (33) | |
| Left-sided | 27 (67) | |
| Preoperative CEA, ng/mL | <0.001 | |
| ≤5 | 45 (71) | |
| >5 | 18 (29) | |
| pT | <0.001 | |
| T1-T2 | 16 (25) | |
| T3-T4 | 47 (75) | |
| Nodal metastasis | 0.257 | |
| No | 36 (57) | |
| Yes | 27 (43) | |
| Tumor Grade | 0.432 | |
| Low | 23 (54) | |
| Moderate/high | 40 (46) | |
| Lymphatic invasion | 0.529 | |
| No | 34 (54) | |
| Yes | 29 (46) | |
| Perineural invasion | <0.001 | |
| No | 47 (75) | |
| Yes | 16 (25) | |
| TNM stage | 0.378 | |
| I | 12 (19) | |
| II | 23 (36) | |
| III | 20 (32) | |
| IV | 8 (13) | |
| Chemotherapy | 0.131 | |
| Neoadjuvant | 17 (27) | |
| Adjuvant | 23 (36) | |
| Both | 24 (36) | |
| MMR status | <0.001 | |
| pMMR/MSS | 54 (86) | |
| dMMR/MSI | 9 (14) | |
| Disease recurrence | 0.060 | |
| No | 45 (71) | |
| Yes | 18 (29) | |
| Survival | 0.259 | |
| Alive | 34 (54) | |
| Dead | 29 (46) |
| MRE11A | RAD50 | NBN | BARD1 | BRCA1 | RBBP8 | PALB2 | ||
|---|---|---|---|---|---|---|---|---|
| Gender | x2 | 0.535 | 0.205 | 0.034 | 0.521 | 0.535 | 0.476 | 0.877 |
| p | 0.464 | 0.650 | 0.853 | 0.471 | 0.464 | 0.490 | 0.349 | |
| Age at diagnosis | x2 | 1.801 | 0.005 | 0.000 | 3.391 | 0.067 | 0.014 | 5.219 |
| p | 0.180 | 0.942 | 1.000 | 0.046a * | 0.796 | 0.904 | 0.022 * | |
| Tumor site | x2 | 7.747 | 1.284 | 0.548 | 0.029 | 0.154 | 0.128 | 0.877 |
| p | 0.008 * | 0.257 | 0.459 | 0.865 | 0.695 | 0.721 | 0.349 | |
| Sidedness | x2 | 3.790 | 2.196 | 0.005 | 0.620 | 0.005 | 0.014 | 0.114 |
| p | 0.042 * | 0.138 | 0.941 | 0.431 | 0.941a | 0.906 | 0.73 | |
| CEA | x2 | 1.802 | 1.429 | 0.800 | 0.022 | 0.022 | 1.429 | 0.089 |
| p | 0.148 | 0.185 | 0.276 | 0.560 | 0.560 | 0.185 | 0.489 | |
| Tumor size | x2 | 3.704 | 0.406 | 1.429 | 1.429 | 1.429 | 0.229 | 1.429 |
| p | 0.040 * | 0.360 | 0.180 | 0.180 | 0.180 | 0.421 | 0.180 | |
| Histological tumor grade | x2 | 2.301 | 0.365 | 2.191 | 0.521 | 0.912 | 1.471 | 0.877 |
| p | 0.129 | 0.546 | 0.139 | 0.471 | 0.340 | 0.225 | 0.349 | |
| Tumor invasive depth | x2 | 3.298 | 0.251 | 5.028 | 0.220 | 0.328 | 2.767 | 0.017 |
| p | 0.039 * | 0.616 | 0.024 * | 0.639 | 0.567 | 0.096 | 0.897 | |
| Nodal metastasis | x2 | 0.001 | 0.022 | 0.829 | 2.017 | 2.872 | 0.024 | 9.314 |
| p | 0.972 | 0.882 | 0.363 | 0.156a | 0.040 * | 0.877 | 0.002 * | |
| TNM stage | x2 | 0.004 | 0.000 | 2.057 | 4.062 | 1.322 | 0.013 | 10.080 |
| p | 0.952 | 1.000 | 0.151 | 0.044 * | 0.250 | 0.910 | 0.001 * | |
| Lymphovascular invasion | x2 | 1.435 | 0.085 | 0.032 | 1.137 | 0.186 | 0.019 | 5.907 |
| p | 0.231 | 0.770 | 0.858 | 0.286a | 0.666 | 0.891 | 0.015 * | |
| Perineural invasion | x2 | 0.961 | 3.379 | 0.168 | 0.220 | 0.002 | 0.423 | 1.516 |
| p | 0.610 | 0.044 * | 0.682 | 0.639 | 0. 961 | 0.358 | 0.173 | |
| M SI | x2 | 3.706 | 0.770 | 0.305 | 0.003 | 0.305 | 0.580 | 2.094 |
| p | 0.049 * | 0.380 | 0.581 | 0.959 | 0.581 | 0.446 | 0.148 |
| Variable | p | Variable | HR (95% CI) | p |
|---|---|---|---|---|
| Age | 0.148 | <65 years | 1.00 | 0.061 |
| >65 years | 3.79 (0.94–5.32) | |||
| CEA | 0.087 | <or = 5 ng/mL | 1.00 | 0.299 |
| >5 ng/mL | 3.18 (0.47–11.65) | |||
| Tumor location | 0.710 | |||
| Sidedness | 0.098 | Left-sided CRC | 1.00 | 0.500 |
| Right-sided CRC | 2.15 (0.23–10.08) | |||
| Tumor size | 0.325 | |||
| Tumor grade | 0.202 | |||
| Lymphovascular invasion | 0.029 | No | 1.00 | 0.588 |
| Yes | 1.73 (0.24–12.68) | |||
| Perineural invasion | 0.206 | |||
| TNM stage | 0.035 | I-II | 1.00 | 0.102 |
| III-IV | 3.75 (0.94–8.54) | |||
| NLR | 0.465 | |||
| LMR | 0.528 | |||
| PLR | 0.207 | |||
| MMR status | 0.140 | MSS | 1.00 | 0.380 |
| MSI | 2.65 (0.31–3.41) | |||
| MRE11A | 0.183 | Relative low expression | 1.00 | 0.384 |
| Relative high expression | 2.31 (0.35–5.06) | |||
| RAD50 | 0.830 | |||
| NBN | 0.475 | |||
| BARD1 | 0.633 | |||
| BRCA1 | 0.278 | |||
| RBBP8 | 0.636 | |||
| PALB2 | 0.269 |
| Univariate Analysis | Multivariate Analysis | |||
|---|---|---|---|---|
| Variable | p | Variable | HR (95% CI) | p |
| Age | 0.306 | |||
| CEA | 0.803 | |||
| Tumor location | 0.201 | |||
| Sidedness | <0.001 | Left-sided | 1.00 | 0.006 |
| Right-sided | 4.57 (1.55–13.49) | |||
| Tumor grade | 0.736 | |||
| Lymphovascular invasion | 0.971 | |||
| Perineural invasion | 0.970 | |||
| TNM stage | 0.076 | I-II | 1.00 | 0.187 |
| III-IV | 2.55 (0.63–10.30) | |||
| NLR | 0.548 | |||
| LMR | 0.454 | |||
| PLR | 0.877 | |||
| MMR status | 0.157 | MSS | 1.00 | |
| MSI | 0.907 (0.24–3.36) | 0.884 | ||
| MRE11A | 0.121 | Relative low expression | 1.00 | 0.046 |
| Relative high expression | 3.11 (1.64–15.08) | |||
| RAD50 | 0.498 | |||
| NBN | 0.615 | |||
| BARD1 | 0.607 | |||
| BRCA1 | 0.608 | |||
| RBBP8 | 0.558 | |||
| PALB2 | 0.159 | Relative low expression | 1.00 | 0.044 |
| Relative high expression | 2.06 (1.67–17.63) | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azambuja, D.d.B.; e Gloria, H.d.C.; Montenegro, G.e.S.; Kalil, A.N.; Hoffmann, J.-S.; Leguisamo, N.M.; Saffi, J. High Expression of MRE11A Is Associated with Shorter Survival and a Higher Risk of Death in CRC Patients. Genes 2023, 14, 1270. https://doi.org/10.3390/genes14061270
Azambuja DdB, e Gloria HdC, Montenegro GeS, Kalil AN, Hoffmann J-S, Leguisamo NM, Saffi J. High Expression of MRE11A Is Associated with Shorter Survival and a Higher Risk of Death in CRC Patients. Genes. 2023; 14(6):1270. https://doi.org/10.3390/genes14061270
Chicago/Turabian StyleAzambuja, Daniel de Barcellos, Helena de Castro e Gloria, Gabriel e Silva Montenegro, Antonio Nocchi Kalil, Jean-Sébastien Hoffmann, Natalia Motta Leguisamo, and Jenifer Saffi. 2023. "High Expression of MRE11A Is Associated with Shorter Survival and a Higher Risk of Death in CRC Patients" Genes 14, no. 6: 1270. https://doi.org/10.3390/genes14061270
APA StyleAzambuja, D. d. B., e Gloria, H. d. C., Montenegro, G. e. S., Kalil, A. N., Hoffmann, J.-S., Leguisamo, N. M., & Saffi, J. (2023). High Expression of MRE11A Is Associated with Shorter Survival and a Higher Risk of Death in CRC Patients. Genes, 14(6), 1270. https://doi.org/10.3390/genes14061270






