Characterization and Genome Study of a Newly Isolated Temperate Phage Belonging to a New Genus Targeting Alicyclobacillus acidoterrestris
Abstract
1. Introduction
2. Materials and Methods
2.1. Source of Bacterial Host Strains
2.2. Bacteriophage Isolation
2.3. Purification of Bacteriophages
2.4. Determination of the Bacterial Hosts Range
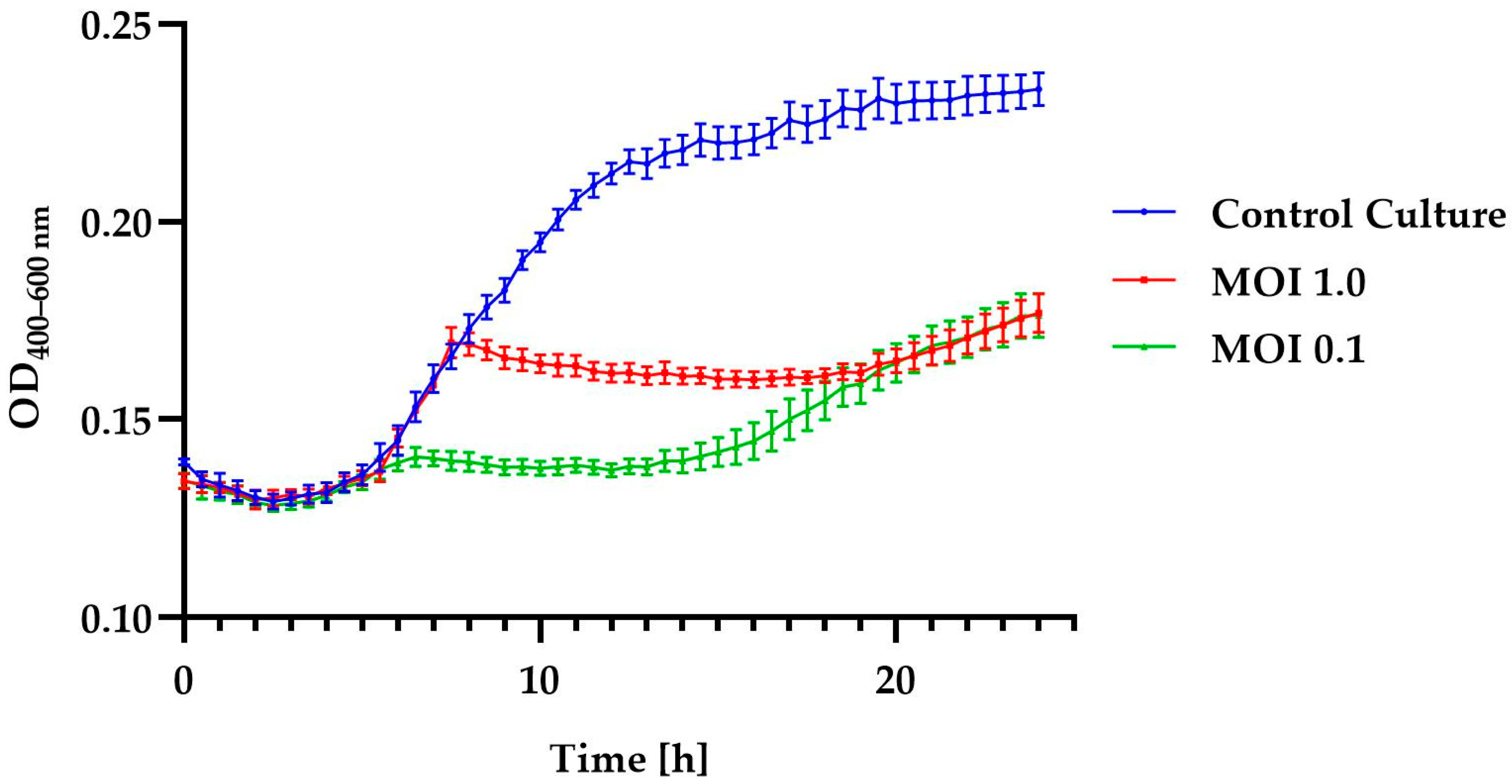
2.5. Changes in the Growth Kinetics of A. acidoterrestris Strain KKP 3133 after Phage Infection
2.6. Effect of Selected Factors on the Preservation of the Phage Activity
2.7. Determination of Morphological Characteristics of Phage
2.8. Extraction of Bacteriophage Genomic DNA
2.9. Genome Sequencing and Bioinformatics Analysis
2.10. Statistical Analysis
3. Results and Discussion
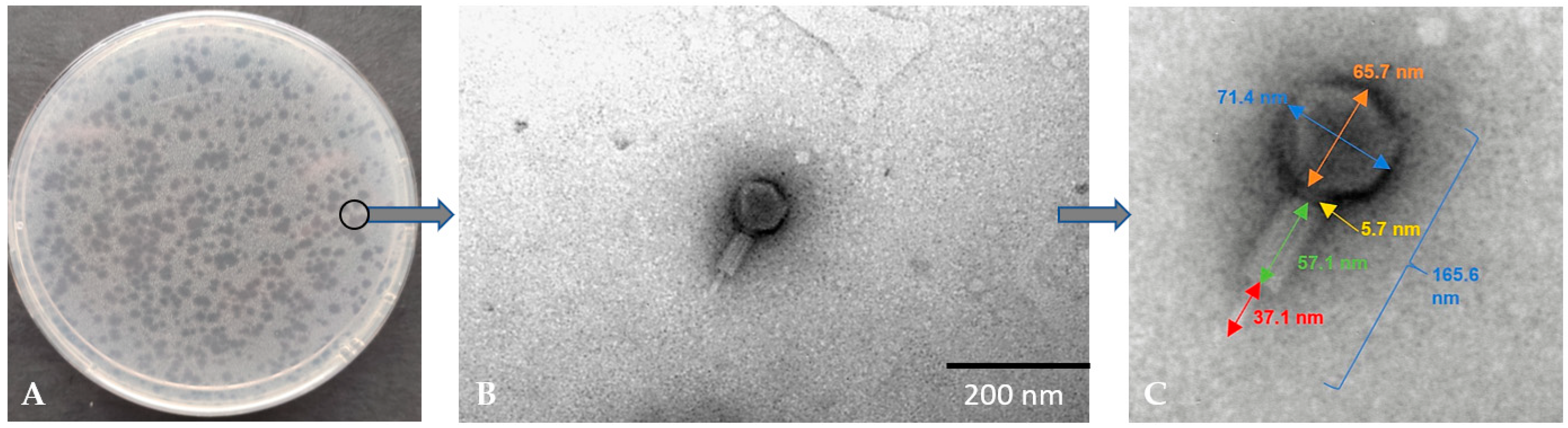
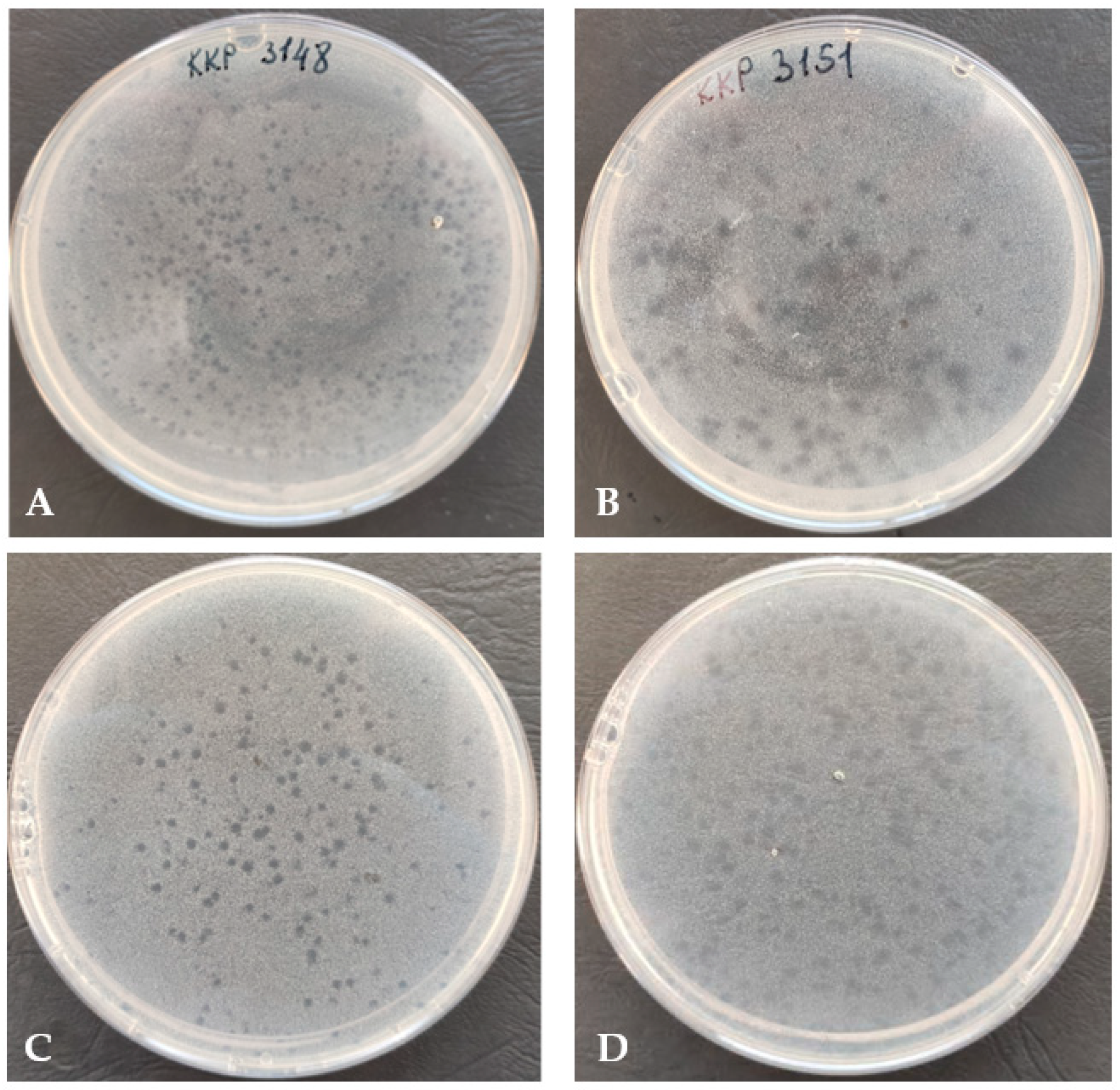
3.1. Determination of Plaque and TEM Morphology of Bacteriophage
3.2. Range of Bacterial Hosts
3.3. Evaluation of the Activity of Phage against Bacterial Hosts
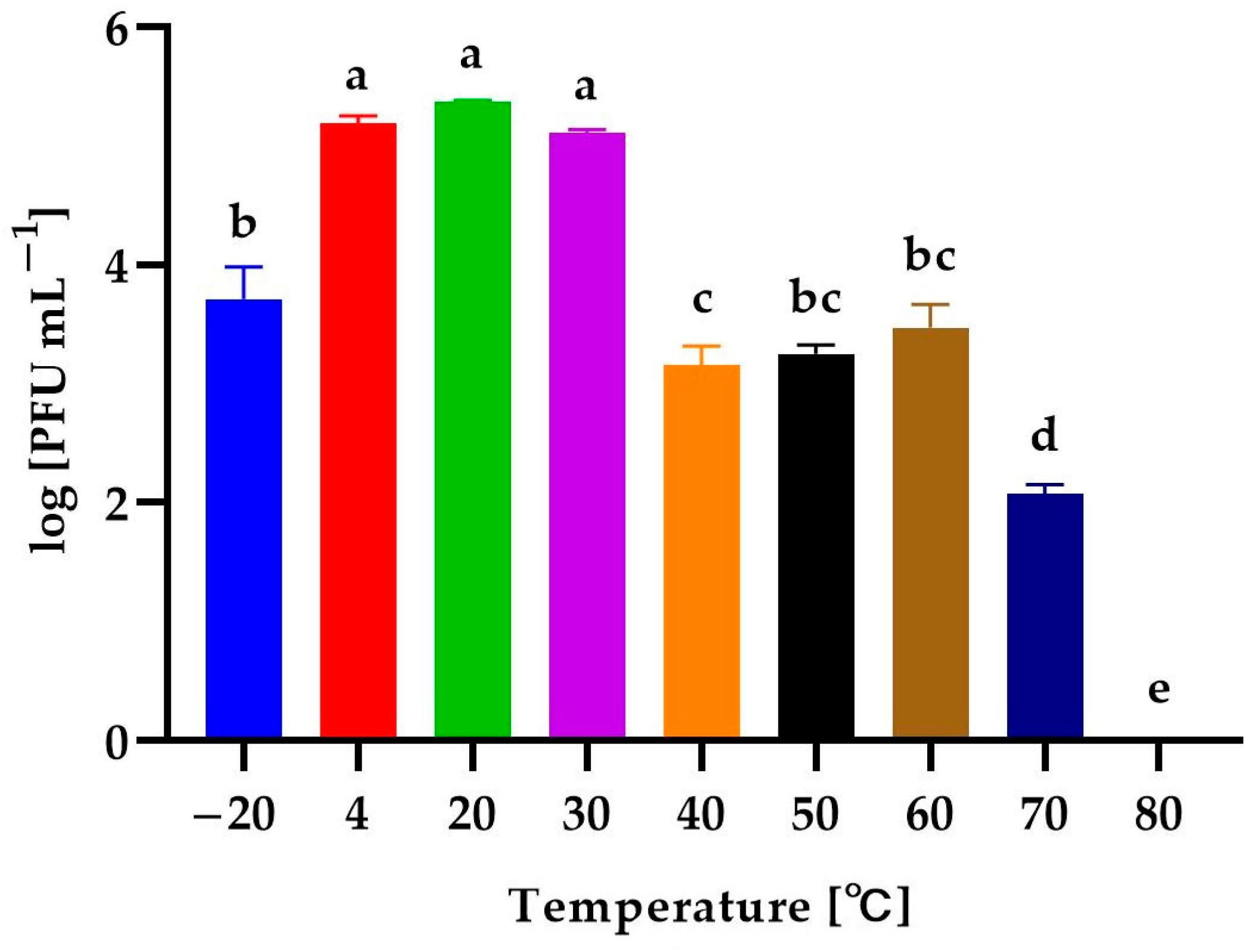
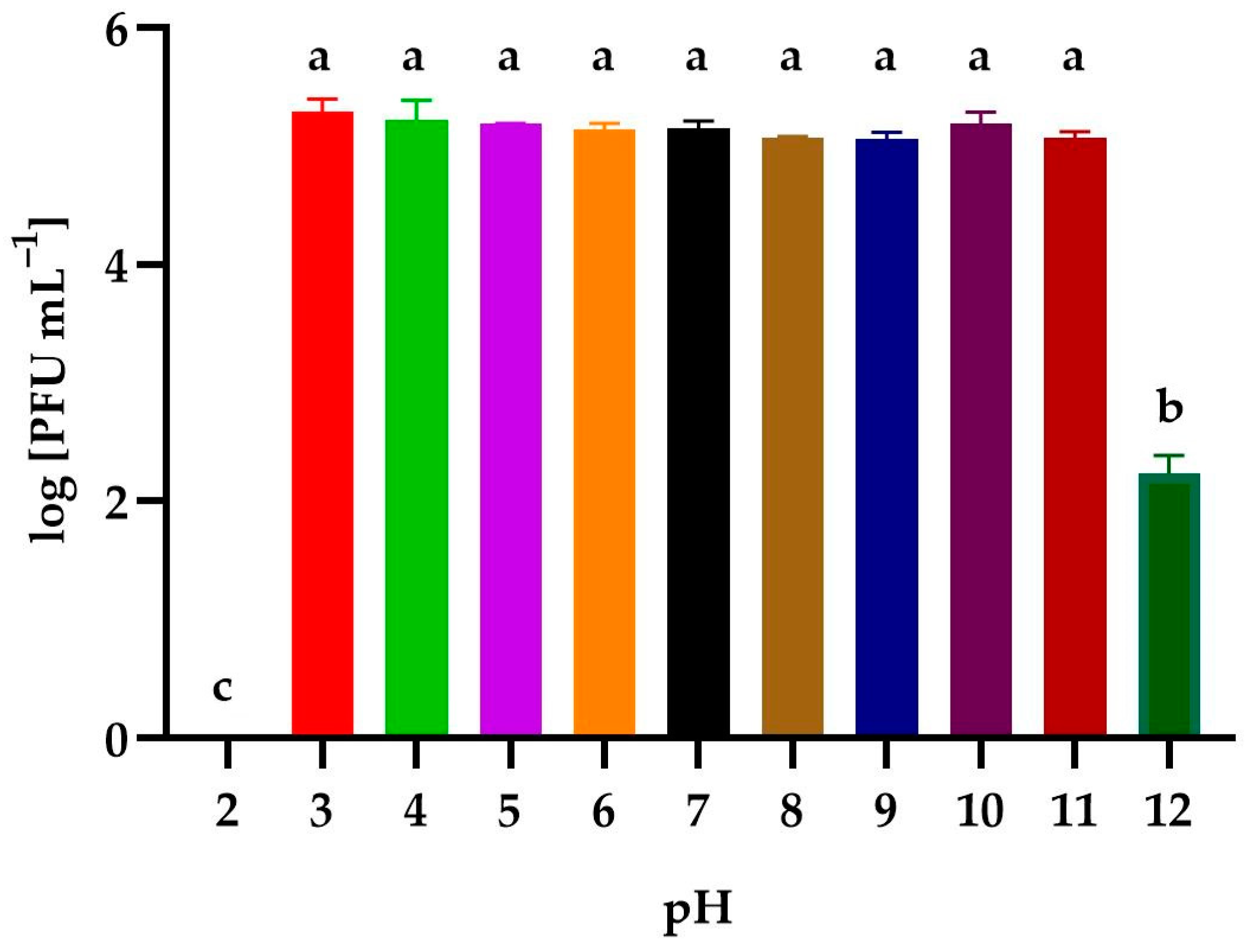
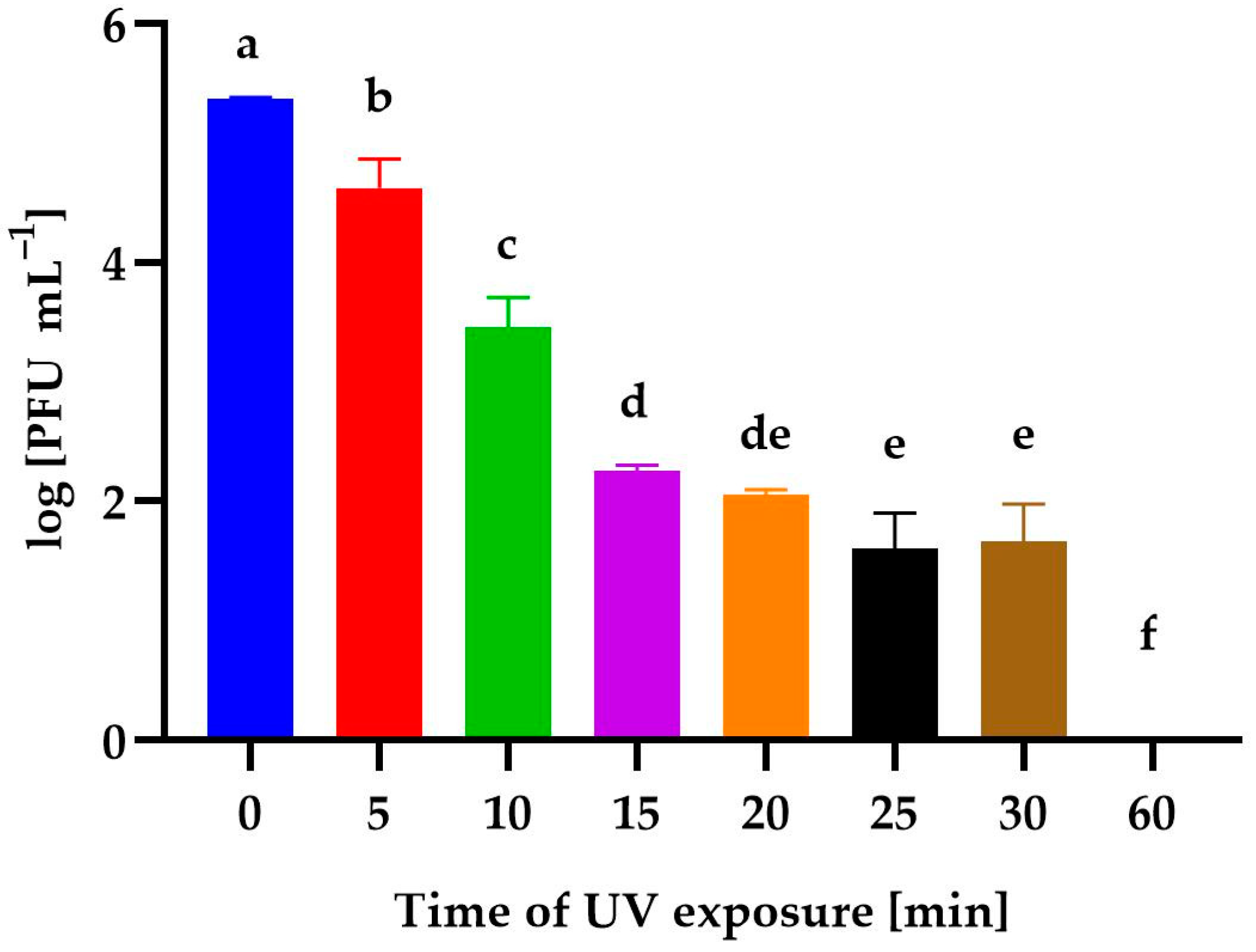
3.4. Effect of Environmental Conditions on the Phage Activity
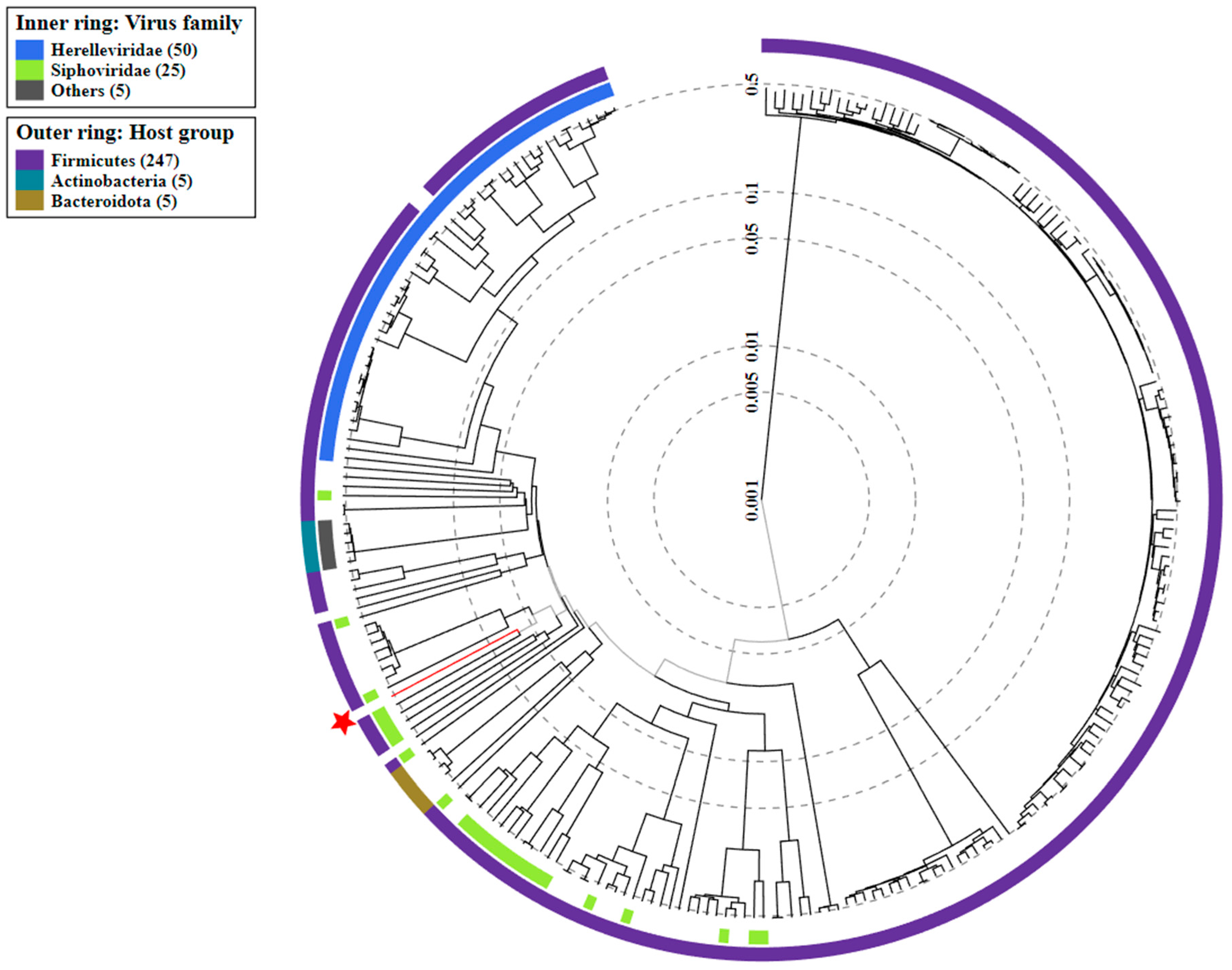
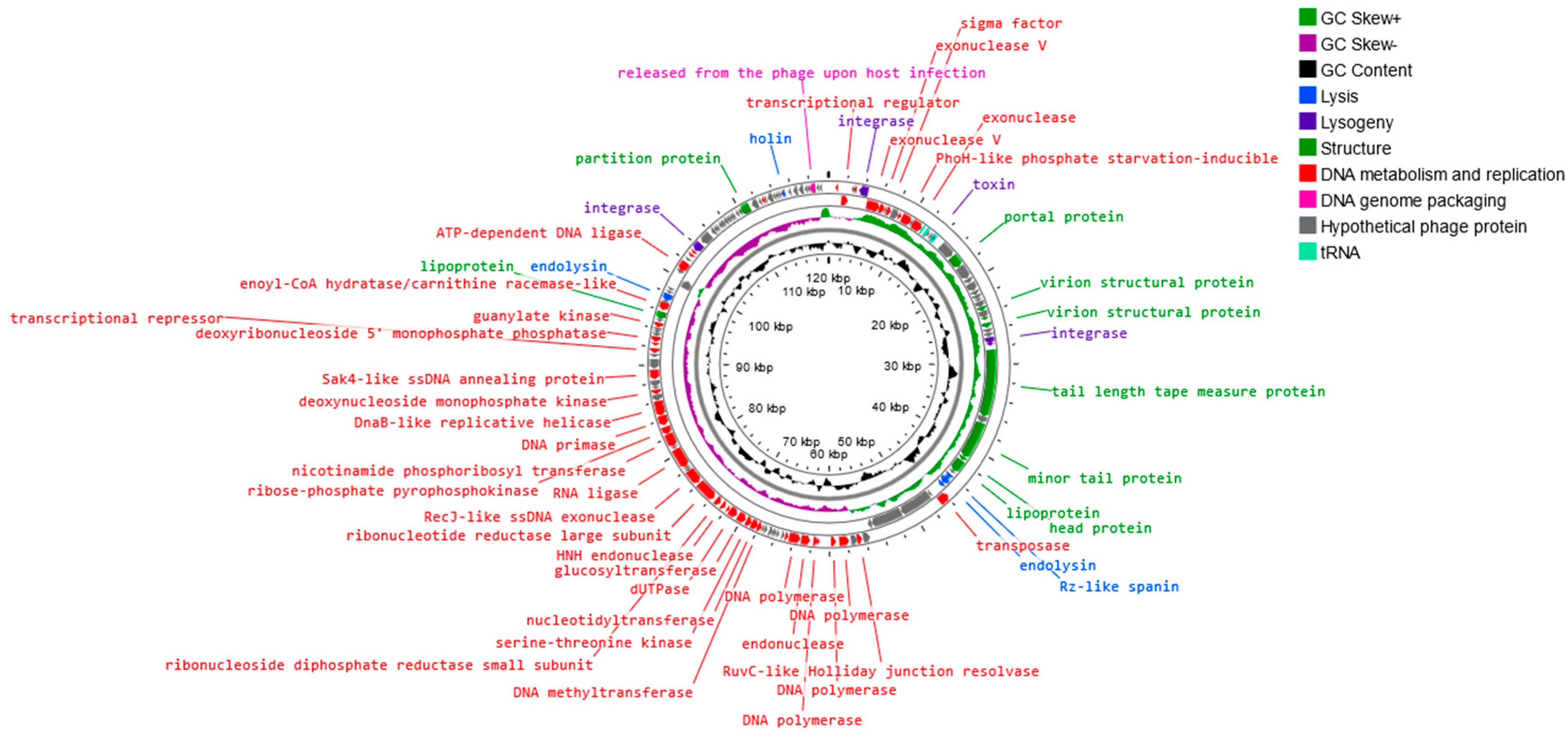
3.5. Analysis of Phage Genome
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhardwaj, R.L.; Nandal, U.; Pal, A.; Jain, S. Bioactive Compounds and Medicinal Properties of Fruit Juices. Fruits 2014, 69, 5. [Google Scholar] [CrossRef]
- Fernandes, F.A.N.; Rodrigues, S. Cold Plasma Processing on Fruits and Fruit Juices: A Review on the Effects of Plasma on Nutritional Quality. Processes 2021, 9, 2098. [Google Scholar] [CrossRef]
- Nowak, D.; Gośliński, M.; Kłębukowska, L. Antioxidant and Antimicrobial Properties of Selected Fruit Juices. Plant Foods Hum. Nutr. 2022, 77, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhang, M.; Mujumdar, A.S.; Liu, Y. Application Advantages of New Non-Thermal Technology in Juice Browning Control: A Comprehensive Review. Food Rev. Int. 2022, 1–22. [Google Scholar] [CrossRef]
- Santarelli, G.A.; Migliorati, G.; Pomilio, F.; Marfoglia, C.; Centorame, P.; D’Agostino, A.; D’Aurelio, R.; Scarpone, R.; Battistelli, N.; Di Simone, F.; et al. Assessment of Pesticide Residues and Microbial Contamination in Raw Leafy Green Vegetables Marketed in Italy. Food Control 2018, 85, 350–358. [Google Scholar] [CrossRef]
- Pérez-Cacho, P.R.; Galan-Soldevilla, H.; Mahattanatawee, K.; Elston, A.; Rouseff, R.L. Sensory lexicon for fresh squeezed and processed orange juices. Food Sci. Technol. Int. 2008, 14, 131–141. [Google Scholar] [CrossRef]
- Mphahlele, R.R.; Fawole, O.A.; Mokwena, L.M.; Opara, U.L. Effect of Extraction Method on Chemical, Volatile Composition and Antioxidant Properties of Pomegranate Juice. S. Afr. J. Bot. 2016, 103, 135–144. [Google Scholar] [CrossRef]
- World Health Organization. Measuring the Intake of Fruit and Vegetables. Available online: http://www.who.int/dietphysicalactivity/fruit/en/ (accessed on 1 March 2023).
- Aneja, K.R.; Dhiman, R.; Aggarwal, N.K.; Kumar, V.; Kaur, M. Microbes Associated with Freshly Prepared Juices of Citrus and Carrots. Int. J. Food Sci. 2014, 2014, 408085. [Google Scholar] [CrossRef]
- Kincal, D.; Hill, W.S.; Balaban, M.O.; Portier, K.M.; Wei, C.I.; Marshall, M.R. A Continuous High Pressure Carbon Dioxide System for Microbial Reduction in Orange Juice. J. Food Sci. 2005, 70, M249–M254. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, Q.; Ma, X.; Zhang, Y.; Zhang, X.; Zhang, W. A Novel Developed Method Based on Single Primer Isothermal Amplification for Rapid Detection of Alicyclobacillus acidoterrestris in Apple Juice. Food Control 2017, 75, 187–195. [Google Scholar] [CrossRef]
- Silva, L.P.; Gonzales-Barron, U.; Cadavez, V.; Sant’Ana, A.S. Modeling the Effects of Temperature and PH on the Resistance of Alicyclobacillus acidoterrestris in Conventional Heat-Treated Fruit Beverages through a Meta-Analysis Approach. Food Microbiol. 2015, 46, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Han, A.; Jo, S.; Cheon, H.; Song, H.; Jang, A.R.; Kim, D.; Lee, S.Y. Microbiological Quality and Safety of Commercial Fresh Fruit and Vegetable Juices in Korea. LWT 2021, 152, 112432. [Google Scholar] [CrossRef]
- Dewanti-Hariyadi, R.; Maria, A.; Sandoval, P. Microbiological Quality and Safety of Fruit Juices. Food Rev. Int. 2013, 1, 54–57. [Google Scholar]
- Yeni, F.; Yavaş, S.; Alpas, H.; Soyer, Y. Most Common Foodborne Pathogens and Mycotoxins on Fresh Produce: A Review of Recent Outbreaks. Crit. Rev. Food Sci. Nutr. 2016, 56, 1532–1544. [Google Scholar] [CrossRef]
- Balali, G.I.; Yar, D.D.; Afua Dela, V.G.; Adjei-Kusi, P. Microbial Contamination, an Increasing Threat to the Consumption of Fresh Fruits and Vegetables in Today’s World. Int. J. Microbiol. 2020, 2020, 3029295. [Google Scholar] [CrossRef]
- Bhilwadikar, T.; Pounraj, S.; Manivannan, S.; Rastogi, N.K.; Negi, P.S. Decontamination of Microorganisms and Pesticides from Fresh Fruits and Vegetables: A Comprehensive Review from Common Household Processes to Modern Techniques. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1003–1038. [Google Scholar] [CrossRef]
- Lykov, I.N.; Kakharova, M.A.; Kureber, V.S.; Yurova, A.E. Research of Antibiotic Resistance of Microorganisms Isolated from Fruits and Vegetables. IOP Conf. Ser. Earth Environ. Sci. 2021, 839, 042003. [Google Scholar] [CrossRef]
- Misra, N.N. The Contribution of Non-Thermal and Advanced Oxidation Technologies towards Dissipation of Pesticide Residues. Trends Food Sci. Technol. 2015, 45, 229–244. [Google Scholar] [CrossRef]
- Silva, F.V.M.; Tan, E.K.; Farid, M. Bacterial Spore Inactivation at 45–65 °C Using High Pressure Processing: Study of Alicyclobacillus acidoterrestris in Orange Juice. Food Microbiol. 2012, 32, 206–211. [Google Scholar] [CrossRef]
- Nasiłowska, J.; Sokołowska, B.; Fonberg-Broczek, M. Long term storage of vegetable juices treated by high hydrostatic pressure–assurance of the microbial safety. BioMed Res. Int. 2018, 12, 7389381. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Mittal, G.S. Effects of High-Pressure Processing (HPP) on Bacterial Spores: An Overview. Food Rev. Int. 2008, 24, 330–351. [Google Scholar] [CrossRef]
- Sokołowska, B.; Skąpska, S.; Fonberg-Broczek, M.; Niezgoda, J.; Chotkiewicz, M.; Dekowska, A.; Rzoska, S.J. Factors influencing the inactivation of Alicyclobacillus acidoterrestris spores exposed to high hydrostatic pressure in apple juice. High Press. Res. 2013, 33, 73–82. [Google Scholar] [CrossRef]
- Sokołowska, B.; Połaska, M.; Dekowska, A. Alicyclobacillus–still current issues in the beverage industry. In Safety Issues in Beverage Production; Academic Press: Cambridge, MA, USA, 2020; Volume 18, pp. 105–146. [Google Scholar]
- Molva, C.; Baysal, A.H. Evaluation of Bioactivity of Pomegranate Fruit Extract against Alicyclobacillus acidoterrestris DSM 3922 Vegetative Cells and Spores in Apple Juice. LWT 2015, 62, 989–995. [Google Scholar] [CrossRef]
- Sen Chang, S.; Kang, D.H. Alicyclobacillus spp. in the Fruit Juice Industry: History, Characteristics, and Current Isolation/Detection Procedures. Crit. Rev. Microbiol. 2004, 30, 55–74. [Google Scholar] [CrossRef] [PubMed]
- Smit, Y.; Cameron, M.; Venter, P.; Witthuhn, R.C. Alicyclobacillus Spoilage and Isolation-A Review. Food Microbiol. 2011, 28, 331–349. [Google Scholar] [CrossRef] [PubMed]
- Uchida, R.; Silva, F.V.M. Alicyclobacillus acidoterrestris Spore Inactivation by High Pressure Combined with Mild Heat: Modeling the Effects of Temperature and Soluble Solids. Food Control 2017, 73, 426–432. [Google Scholar] [CrossRef]
- Durak, M.Z.; Churey, J.J.; Danyluk, M.D.; Worobo, R.W. Identification and Haplotype Distribution of Alicyclobacillus Spp. from Different Juices and Beverages. Int. J. Food Microbiol. 2010, 142, 286–291. [Google Scholar] [CrossRef]
- Gutiérrez, D.; Rodríguez-Rubio, L.; Martínez, B.; Rodríguez, A.; García, P. Bacteriophages as Weapons against Bacterial Biofilms in the Food Industry. Front. Microbiol. 2016, 7, 825. [Google Scholar] [CrossRef]
- Li, J.; Zhao, F.; Zhan, W.; Li, Z.; Zou, L.; Zhao, Q. Challenges for the Application of Bacteriophages as Effective Antibacterial Agents in the Food Industry. J. Sci. Food Agric. 2022, 102, 461–471. [Google Scholar] [CrossRef]
- Połaska, M.; Sokołowska, B. Review Bacteriophages—A New Hope or a Huge Problem in the Food Industry. AIMS Microbiol. 2019, 5, 324. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, X. The Application and Research Progress of Bacteriophages in Food Safety. J. Appl. Microbiol. 2022, 133, 2137–2147. [Google Scholar] [CrossRef]
- Osei, E.K.; Mahony, J.; Kenny, J.G. From Farm to Fork: Streptococcus suis as a Model for the Development of Novel Phage-Based Biocontrol Agents. Viruses 2022, 14, 1996. [Google Scholar] [CrossRef] [PubMed]
- Moye, Z.D.; Woolston, J.; Sulakvelidze, A. Bacteriophage application for food production and processing. Viruses 2018, 10, 205. [Google Scholar] [CrossRef] [PubMed]
- Fernández, L.; Gutiérrez, D.; Rodríguez, A.; García, P. Application of Bacteriophages in the Agro-Food Sector: A Long Way toward Approval. Front. Cell. Infect Microbiol. 2018, 8, 296. [Google Scholar] [CrossRef] [PubMed]
- Ye, M.; Sun, M.; Huang, D.; Zhang, Z.; Zhang, H.; Zhang, S.; Hu, F.; Jiang, X.; Jiao, W. A Review of Bacteriophage Therapy for Pathogenic Bacteria Inactivation in the Soil Environment. Environ. Int. 2019, 129, 488–496. [Google Scholar] [CrossRef]
- Harada, L.K.; Silva, E.C.; Campos, W.F.; Del Fiol, F.S.; Vila, M.; Dąbrowska, K.; Krylov, V.N.; Balcão, V.M. Biotechnological Applications of Bacteriophages: State of the Art. Microbiol. Res. 2018, 212, 38–58. [Google Scholar] [CrossRef] [PubMed]
- Butala, M.; Dragoš, A. Unique Relationships between Phages and Endospore-Forming Hosts. Trends Microbiol. 2022, 31, 498–510. [Google Scholar] [CrossRef]
- Mok, J.H.; Sun, Y.; Pyatkovskyy, T.; Hu, X.; Sastry, S.K. Mechanisms of Bacillus subtilis Spore Inactivation by Single- and Multi-Pulse High Hydrostatic Pressure (MP-HHP). Innov. Food Sci. Emerg. Technol. 2022, 81, 103147. [Google Scholar] [CrossRef]
- Porębska, I.; Sokołowska, B.; Skąpska, S.; Rzoska, S.J. Treatment with High Hydrostatic Pressure and Supercritical Carbon Dioxide to Control Alicyclobacillus acidoterrestris Spores in Apple Juice. Food Control 2017, 73, 24–30. [Google Scholar] [CrossRef]
- Sarker, M.R.; Akhtar, S.; Torres, J.A.; Paredes-Sabja, D. High Hydrostatic Pressure-Induced Inactivation of Bacterial Spores. Crit. Rev. Microbiol. 2015, 41, 18–26. [Google Scholar] [CrossRef]
- Mirzaei, M.K.; Nilsson, A.S. Isolation of Phages for Phage Therapy: A Comparison of Spot Tests and Efficiency of Plating Analyses for Determination of Host Range and Efficacy. PLoS ONE 2015, 10, e0118557. [Google Scholar] [CrossRef] [PubMed]
- Dekowska, A.; Niezgoda, J.; Sokołowska, B. Genetic heterogeneity of Alicyclobacillus strains revealed by RFLP analysis of vdc region and rpoB gene. Bio Med. Res. Intern. 2018, 2018, 9608756. [Google Scholar]
- Połaska, M.; Dekowska, A.; Sokołowska, B. Isolation and identification of guaiacol-producing Alicyclobacillus fastidiosus strains from orchards in Poland. Acta Bioch. Pol. 2021, 68, 301–307. [Google Scholar]
- Alves, D.; Cerqueira, M.A.; Pastrana, L.M.; Sillankorva, S. Entrapment of a Phage Cocktail and Cinnamaldehyde on Sodium Alginate Emulsion-Based Films to Fight Food Contamination by Escherichia coli and Salmonella Enteritidis. Food Res. Int. 2020, 128, 108791. [Google Scholar] [CrossRef] [PubMed]
- Echeverría-Vega, A.; Morales-Vicencio, P.; Saez-Saavedra, C.; Gordillo-Fuenzalida, F.; Araya, R. A Rapid and Simple Protocol for the Isolation of Bacteriophages from Coastal Organisms. Methods X 2019, 6, 2614–2619. [Google Scholar] [CrossRef]
- Jamal, M.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Das, C.R. Isolation and Characterization of a Bacteriophage and Its Utilization against Multi-Drug Resistant Pseudomonas aeruginosa-2995. Life Sci. 2017, 190, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, H.W. 5500 Phages examined in the electron microscope. Arch. Virol. 2007, 151, 227–243. [Google Scholar] [CrossRef]
- Mahmoud, M.; Askora, A.; Barakat, A.B.; Rabie, O.E.F.; Hassan, S.E. Isolation and Characterization of Polyvalent Bacteriophages Infecting Multi Drug Resistant Salmonella Serovars Isolated from Broilers in Egypt. Int. J. Food Microbiol. 2018, 266, 8–13. [Google Scholar] [CrossRef]
- Amarillas, L.; Lightbourn-Rojas, L.; Angulo-Gaxiola, A.K.; Basilio Heredia, J.; González-Robles, A.; León-Félix, J. The Antibacterial Effect of Chitosan-Based Edible Coating Incorporated with a Lytic Bacteriophage against Escherichia coli O157:H7 on the Surface of Tomatoes. J. Food Saf. 2018, 38, e12571. [Google Scholar] [CrossRef]
- Strathdee, S.A.; Hatfull, G.F.; Mutalik, V.K.; Schooley, R.T. Phage Therapy: From Biological Mechanisms to Future Directions. Cell 2023, 186, 17–31. [Google Scholar] [CrossRef]
- Wójcicki, M.; Świder, O.; Średnicka, P.; Shymialevich, D.; Ilczuk, T.; Koperski, Ł.; Cieślak, H.; Sokołowska, B.; Juszczuk-Kubiak, E. Newly Isolated Virulent Salmophages for Biocontrol of Multidrug-Resistant Salmonella in Ready-to-Eat Plant-Based Food. Int. J. Mol. Sci. 2023, 24, 10134. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of Long, Error-Prone Reads Using Repeat Graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Mcnair, K.; Zhou, C.; Dinsdale, E.A.; Souza, B.; Edwards, R.A. PHANOTATE: A Novel Approach to Gene Identification in Phage Genomes. Bioinformatics 2019, 35, 4537–4542. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, Q.; Li, M.; Xu, J.; Wang, C.; Zhang, J.; Xiao, M.; Bin, Y.; Xia, J. PhaGAA: An Integrated Web Server Platform for Phage Genome Annotation and Analysis. Bioinformatics 2023, 39, btad120. [Google Scholar] [CrossRef]
- Proksee Software. Available online: https://proksee.ca/ (accessed on 2 March 2023).
- Mihara, T.; Nishimura, Y.; Shimizu, Y.; Nishiyama, H.; Yoshikawa, G.; Uehara, H.; Hingamp, P.; Goto, S.; Ogata, H. Linking Virus Genomes with Host Taxonomy. Viruses 2016, 8, 66. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.; Yoshida, T.; Kuronishi, M.; Uehara, H.; Ogata, H.; Goto, S. ViPTree: The Viral Proteomic Tree Server. Bioinformatics 2017, 33, 2379–2380. [Google Scholar] [CrossRef]
- Tynecki, P.; Guziński, A.; Kazimierczak, J.; Jadczuk, M.; Dastych, J.; Onisko, A. PhageAI-Bacteriophage Life Cycle Recognition with Machine Learning and Natural Language Processing. bioRxiv 2020, 198606. [Google Scholar]
- Abedon, S.T.; Yin, J. Bacteriophage Plaques: Theory and Analysis. Methods Mol. Biol. 2009, 501, 161–174. [Google Scholar]
- Zheng, X.-F.; Yang, Z.; Zhangl, H.; Jinl, W.-X.; Xul, C.-W.; Gaol, L.; Raol, S.-Q.; Jiao, X. Isolation of virulent phages infecting dominant mesophilic aerobic bacteria in cucumber pickle fermentation. Food Microbiol. 2020, 86, 103330. [Google Scholar] [CrossRef]
- Cornelissen, A.; Ceyssens, P.J.; T’Syen, J.; van Praet, H.; Noben, J.P.; Shaburova, O.V.; Krylov, V.N.; Volckaert, G.; Lavigne, R. The T7-Related Pseudomonas Putida Phage Φ15 Displays Virion-Associated Biofilm Degradation Properties. PLoS ONE 2011, 6, e18597. [Google Scholar] [CrossRef]
- Krone, S.M.; Abedon, S.T. Modeling phage plaque growth. Bacteriophage Ecol. 2008, 15, 415–438. [Google Scholar]
- Glonti, T.; Pirnay, J.P. In Vitro Techniques and Measurements of Phage Characteristics That Are Important for Phage Therapy Success. Viruses 2022, 14, 1490. [Google Scholar] [CrossRef] [PubMed]
- Jaschke, P.R.; Dotson, G.A.; Hung, K.S.; Liu, D.; Endy, D. Definitive Demonstration by Synthesis of Genome Annotation Completeness. Proc. Natl. Acad. Sci. USA 2019, 116, 24206–24213. [Google Scholar] [CrossRef]
- Wright, B.W.; Ruan, J.; Molloy, M.P.; Jaschke, P.R. Genome Modularization Reveals Overlapped Gene Topology Is Necessary for Efficient Viral Reproduction. ACS Synth. Biol. 2020, 9, 3079–3090. [Google Scholar] [CrossRef]
- Hyman, P. Phages for Phage Therapy: Isolation, Characterization, and Host Range Breadth. Pharmaceuticals 2019, 12, 3079–3090. [Google Scholar] [CrossRef]
- Leibovici, L.; Shraga, I.; Drucker, M.; Konigsberger, H.; Samra, Z.; Pitlik, S.D. The Benefit of Appropriate Empirical Antibiotic Treatment in Patients with Bloodstream Infection. J. Intern. Med. 1998, 244, 379–386. [Google Scholar] [CrossRef]
- Ross, A.; Ward, S.; Hyman, P. More Is Better: Selecting for Broad Host Range Bacteriophages. Front. Microbiol. 2016, 7, 1352. [Google Scholar] [CrossRef] [PubMed]
- Gientka, I.; Wójcicki, M.; Żuwalski, A.W.; Błażejak, S. Use of phage cocktail for improving the overall microbiological quality of sprouts—Two methods of application. Appl. Microbiol. 2021, 1, 21. [Google Scholar] [CrossRef]
- Lu, Z.; Breidt, F. Escherichia coli O157:H7 Bacteriophage F241 Isolated from an Industrial Cucumber Fermentation at High Acidity and Salinity. Front. Microbiol. 2015, 6, 67. [Google Scholar] [CrossRef]
- Kazantseva, O.A.; Piligrimova, E.G.; Shadrin, A.M. Novel Bacillus-Infecting Bacteriophage B13—The Founding Member of the Proposed New Genus Bunatrivirus. Viruses 2022, 14, 2300. [Google Scholar] [CrossRef]
- Christensen, L.L.; Josephsen, J. The methyltransferase from the LlaDII restriction-modification system influences the level of expression of its own gene. J. Bacteriol. 2004, 186, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Egido, J.E.; Costa, A.R.; Aparicio-Maldonado, C.; Haas, P.J.; Brouns, S.J. Mechanisms and clinical importance of bacteriophage resistance. FEMS Microbiol. Rev. 2022, 46, fuab048. [Google Scholar] [CrossRef] [PubMed]
- Mills, S.; McAuliffe, O.E.; Coffey, A.; Fitzgerald, G.F.; Ross, R.P. Plasmids of lactococci–genetic accessories or genetic necessities? FEMS Microbiol. Rev. 2006, 30, 243–273. [Google Scholar] [CrossRef]
- Dupuis, M.È.; Villion, M.; Magadán, A.H.; Moineau, S. CRISPR-Cas and restriction–modification systems are compatible and increase phage resistance. Nat. Commun. 2013, 4, 2087. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Dutta, V.; Elhanafi, D.; Lee, S.; Osborne, J.A.; Kathariou, S. A novel restriction-modification system is responsible for temperature-dependent phage resistance in Listeria monocytogenes ECII. Appl. Environ. Microbiol. 2012, 78, 1995–2004. [Google Scholar] [CrossRef]
- Watson, B.N.; Steens, J.A.; Staals, R.H.; Westra, E.R.; van Houte, S. Coevolution between bacterial CRISPR-Cas systems and their bacteriophages. Cell Host Microbe 2021, 29, 715–725. [Google Scholar] [CrossRef]
- Pilosof, S.; Alcala-Corona, S.A.; Wang, T.; Kim, T.; Maslov, S.; Whitaker, R.; Pascual, M. The network structure and eco-evolutionary dynamics of CRISPR-induced immune diversification. Nat. Commun. 2020, 4, 1650–1660. [Google Scholar] [CrossRef]
- Gabiatti, N.; Yu, P.; Mathieu, J.; Lu, G.W.; Wang, X.; Zhang, H.; Soares, H.M.; Alvarez, P.J.J. Bacterial Endospores as Phage Genome Carriers and Protective Shells. Appl. Environ. Microbiol. 2018, 84, e01186-18. [Google Scholar] [CrossRef]
- Pires, D.P.; Melo, L.D.R.; Vilas Boas, D.; Sillankorva, S.; Azeredo, J. Phage Therapy as an Alternative or Complementary Strategy to Prevent and Control Biofilm-Related Infections. Curr. Opin. Microbiol. 2017, 39, 48–56. [Google Scholar] [CrossRef]
- Oechslin, F.; Piccardi, P.; Mancini, S.; Gabard, J.; Moreillon, P.; Entenza, J.M.; Resch, G.; Que, Y.A. Synergistic Interaction between Phage Therapy and Antibiotics Clears Pseudomonas aeruginosa Infection in Endocarditis and Reduces Virulence. J. Infect. Dis. 2017, 215, 703–712. [Google Scholar] [CrossRef]
- Morozova, V.; Bokovaya, O.; Kozlova, Y.; Kurilshikov, A.; Babkin, I.; Tupikin, A.; Bondar, A.; Ryabchikova, E.; Brayanskaya, A.; Peltek, S.; et al. A Novel Thermophilic Aeribacillus Bacteriophage AP45 Isolated from the Valley of Geysers, Kamchatka: Genome Analysis Suggests the Existence of a New Genus within the Siphoviridae Family. Extremophiles 2019, 23, 599–612. [Google Scholar] [CrossRef]
- Thung, T.Y.; Lee, E.; Mahyudin, N.A.; Wan Mohamed Radzi, C.W.J.; Mazlan, N.; Tan, C.W.; Radu, S. Partial Characterization and in Vitro Evaluation of a Lytic Bacteriophage for Biocontrol of Campylobacter jejuni in Mutton and Chicken Meat. J. Food Saf. 2020, 40, e12770. [Google Scholar] [CrossRef]
- Capra, M.L.; Quiberoni, A.; Reinheimer, J. Phages of Lactobacillus casei/paracasei: Response to Environmental Factors and Interaction with Collection and Commercial Strains. J. Appl. Microbiol. 2006, 100, 334–342. [Google Scholar] [CrossRef]
- Krasowska, A.; Biegalska, A.; Augustyniak, D.; Łoś, M.; Richert, M.; Łukaszewicz, M. Isolation and Characterization of Phages Infecting Bacillus subtilis. Biomed. Res. Int. 2015, 2015, 179597. [Google Scholar] [CrossRef] [PubMed]
- Hazem, A. Effects of Temperatures, Ph-Values, Ultra-Violet Light, Ethanol and Chloroform on the Growth of Isolated Thermophilic Bacillus Phages. New Microbiol. 2002, 25, 469–476. [Google Scholar]
- Kim, S.; Kim, S.H.; Rahman, M.; Kim, J. Characterization of a Salmonella Enteritidis Bacteriophage Showing Broad Lytic Activity against Gram-Negative Enteric Bacteria. J. Microbiol. 2018, 56, 917–925. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, Y.; Wang, S.; Duan, X.; Zhang, F.; Guo, A.; Qian, P. Exploring the benefits of metal ions in phage cocktail for the treatment of methicillin-resistant Staphylococcus aureus (MRSA) infection. Infect. Drug Resist. 2022, 15, 2689–2702. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.H. The stability of bacterial viruses in solutions of salts. J. Gen. Physiol. 1949, 32, 579–594. [Google Scholar] [CrossRef] [PubMed]
- Carey-Smith, G.V.; Billington, C.; Cornelius, A.J.; Hudson, J.A.; Heinemann, J.A. Isolation and characterization of bacteriophages infecting Salmonella spp. FEMS Microbiol. Lett. 2006, 258, 182–186. [Google Scholar] [CrossRef]
- Chhibber, S.; Kaur, T.; Kaur, S. Essential role of calcium in the infection process of broad-spectrum methicillin-resistant Staphylococcus aureus bacteriophage. J. Microbiol. 2014, 54, 775–780. [Google Scholar]
- Grabowski, Ł.; Łepek, K.; Stasiłojć, M.; Kosznik-Kwaśnicka, K.; Zdrojewska, K.; Maciąg-Dorszyńska, M.; Węgrzyn, G.; Węgrzyn, A. Bacteriophage-Encoded Enzymes Destroying Bacterial Cell Membranes and Walls, and Their Potential Use as Antimicrobial Agents. Microbiol. Res. 2021, 248, 126746. [Google Scholar] [CrossRef]
- Bujak, K.; Decewicz, P.; Kamiński, J.; Radlińska, M. Identification, characterization, and genomic analysis of novel Serratia temperate phages from a gold mine. Int. J. Mol. Sci. 2020, 21, 6709. [Google Scholar] [CrossRef]
- Jamal, M.; Bukhari, S.M.A.U.S.; Andleeb, S.; Ali, M.; Raza, S.; Nawaz, M.A.; Hussain, T.; Rahman, S.U.; Shah, S.S.A. Bacteriophages: An overview of the control strategies against multiple bacterial infections in different fields. J. Basic Microbiol. 2019, 59, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Chamakura, K.; Young, R. Phage Single-Gene Lysis: Finding the Weak Spot in the Bacterial Cell Wall. J. Biolog. Chem. 2019, 294, 3350–3358. [Google Scholar] [CrossRef]
- Wang, I.N. Lysis Timing and Bacteriophage Fitness. Genetics 2006, 172, 17–26. [Google Scholar] [CrossRef]
- Berry, J.D.; Rajaure, M.; Young, R.Y. Spanin Function Requires Subunit Homodimerization through Intermolecular Disulfide Bonds. Mol. Microbiol. 2013, 88, 35–47. [Google Scholar] [CrossRef]
- Holt, A.; Cahill, J.; Ramsey, J.; Martin, C.; O’Leary, C.; Moreland, R.; Maddox, L.T.; Galbadage, T.; Sharan, R.; Sule, P.; et al. Phage-Encoded Cationic Antimicrobial Peptide Required for Lysis. J. Bacter. 2022, 204, e00214–21. [Google Scholar] [CrossRef] [PubMed]
- Canchaya, C.; Fournous, G.; Brüssow, H. The impact of prophages on bacterial chromosomes. Mol. Microbiol. 2004, 53, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Bailly-Bechet, M.; Vergassola, M.; Rocha, E. Causes for the intriguing presence of tRNAs in phages. Genome Res. 2007, 17, 1486–1495. [Google Scholar] [CrossRef] [PubMed]
- Cieślik, M.; Bagińska, N.; Jończyk-Matysiak, E.; Węgrzyn, A.; Węgrzyn, G.; Górski, A. Temperate Bacteriophages—The Powerful Indirect Modulators of Eukaryotic Cells and Immune Functions. Viruses 2021, 13, 1013. [Google Scholar] [CrossRef]
- Howard-Varona, C.; Hargreaves, K.R.; Abedon, S.T.; Sullivan, M.B. Lysogeny in nature: Mechanisms, impact and ecology of temperate phages. ISME J. 2017, 11, 1511–1520. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, L.; Yang, D.; Song, J.; Wang, C.; Sun, E.; Gu, C.; Chen, H.; Tong, Y.; Tao, P.; et al. Specific Integration of Temperate Phage Decreases the Pathogenicity of Host Bacteria. Front. Cell. Infect. Microbiol. 2020, 10, 14. [Google Scholar] [CrossRef] [PubMed]
- Krger, A.; Lucchesi, P.M.A. Shiga toxins and stx phages: Highly diverse entities. Microbiology 2015, 161, 451–462. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.M.; Waldor, M.K. Mobile Genetic Elements and Bacterial Pathogenesis; Craig, N.L., Craigie, R., Gellert, M., Lambowitz, A.M., Eds.; Mobile DNA II; ASM Press: Washington, DC, USA, 2007; pp. 1040–1059. [Google Scholar]
- Boyd, E.F.; Brüssow, H. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 2002, 10, 521–529. [Google Scholar] [CrossRef]
- Knowles, B.; Silveira, C.B.; Bailey, B.A.; Barott, K.; Cantu, V.A.; Cobián-Güemes, A.G.; Coutinho, F.H.; Dinsdale, E.A.; Felts, B.; Furby, K.A.; et al. Erratum: Corrigendum: Lytic to temperate switching of viral communities. Nat. Cell Biol. 2016, 539, 123. [Google Scholar]
| Bacterial Host Strain (n = 53) | Year of Isolation | Source of Isolation | Guaiacol Production | GenBank Accession Number | Infect with Phage |
|---|---|---|---|---|---|
| Strains from the Culture Collection of Industrial Microorganisms—Microbiological Resources Center, IAFB (n = 46) | |||||
| A. acidoterrestris strain KKP 377 | 2011 | Concentrated cherry juice | + | OQ285871 | ++ |
| A. acidoterrestris strain KKP 381 | 2005 | Concentrated apple juice | + | OQ284561 | − |
| A. acidoterrestris strain KKP 382 | 2005 | Concentrated apple juice | + | OQ284562 | − |
| A. acidoterrestris strain KKP 383 | 2006 | Concentrated apple juice | + | OQ285847 | − |
| A. acidoterrestris strain KKP 394 | 2005 | Concentrated apple juice | + | OQ284103 | − |
| A. acidoterrestris strain KKP 395 | 2002 | Concentrated apple juice | + | OQ284104 | + |
| A. acidoterrestris strain KKP 400 | 2002 | Concentrated apple juice | + | OQ285872 | − |
| A. acidoterrestris strain KKP 404 | 2012 | Concentrated black currant juice | + | OQ284075 | − |
| A. acidoterrestris strain KKP 564 | 2006 | Concentrated apple juice | + | OQ285856 | − |
| A. acidoterrestris strain KKP 596 | 2006 | Concentrated apple juice | + | OQ285855 | − |
| A. acidoterrestris strain KKP 609 | 2013 | Concentrated strawberry juice | + | OQ285862 | − |
| A. acidoterrestris strain KKP 610 | 2006 | Concentrated apple juice | + | OQ285860 | − |
| A. acidoterrestris strain KKP 611 | 2006 | Concentrated apple juice | − | OQ285857 | − |
| A. acidoterrestris strain KKP 613 | 2006 | Concentrated apple juice | + | OQ318516 | + |
| Alicyclobacillus fastidiosus strain KKP 3000 | 2019 | Soil from a pear orchard | + | KY088044 | − |
| A. fastidiosus strain KKP 3001 | 2019 | Soil from a pear orchard | + | KY088045 | − |
| A. fastidiosus strain KKP 3002 | 2002 | Soil from a pear orchard | + | KY088046 | + |
| A. acidoterrestris strain KKP 3133 | 2002 | Concentrated apple juice | + | OQ261766 | ++ |
| A. acidoterrestris strain KKP 3134 | 2004 | Carbonated ingredient | + | OQ263350 | − |
| Alicyclobacillus acidocaldarius strain KKP 3135 | 2004 | Beverage powder | − | OQ285905 | ++ |
| A. acidoterrestris strain KKP 3136 | 2006 | Spoiled orange drink | + | OQ263355 | − |
| A. acidoterrestris strain KKP 3137 | 2006 | Concentrated apple juice | + | OQ263363 | − |
| A. acidoterrestris strain KKP 3138 | 2006 | Banana nectar | + | OQ262932 | − |
| A. acidoterrestris strain KKP 3139 | 2007 | Concentrated apple juice | + | OQ263354 | − |
| A. acidoterrestris strain KKP 3140 | 2007 | Concentrated apple juice | + | OQ262861 | − |
| A. acidoterrestris strain KKP 3141 | 2008 | Concentrated blackcurrant juice | + | KX371237 | ++ |
| A. acidoterrestris strain KKP 3142 | 2008 | Concentrated apple juice | + | KX371238 | − |
| A. acidoterrestris strain KKP 3143 | 2008 | Fresh apple | + | KX371239 | − |
| A. acidocaldarius strain KKP 3144 | 2009 | Sugar syrup | + | OQ285906 | − |
| A. acidoterrestris strain KKP 3145 | 2009 | Concentrated apple juice | + | KX371240 | − |
| A. acidoterrestris strain KKP 3146 | 2009 | Spoiled apple drink | + | OQ261717 | − |
| A. acidoterrestris strain KKP 3147 | 2009 | Concentrated apple juice | + | OQ263351 | − |
| A. acidoterrestris strain KKP 3148 | 2009 | Spoiled apple juice | − | OQ261718 | ++ |
| A. acidocaldarius strain KKP 3149 | 2010 | Concentrated strawberry juice | − | KX371241 | − |
| A. acidoterrestris strain KKP 3150 | 2010 | Concentrated red beet juice | + | KX371242 | − |
| A. acidoterrestris strain KKP 3151 | 2011 | Concentrated cherry juice | + | KX371243 | ++ |
| A. acidoterrestris strain KKP 3152 | 2011 | Concentrated cherry juice | + | KX371245 | − |
| A. acidoterrestris strain KKP 3153 | 2011 | Concentrated raspberry juice | + | KX371246 | − |
| A. acidoterrestris strain KKP 3154 | 2011 | Tomato juice | + | OQ285874 | − |
| A. acidoterrestris strain KKP 3156 | 2012 | Tomato juice | + | OQ263364 | − |
| A. acidocaldarius strain KKP 3157 | 2013 | Concentrated apple juice | − | OQ285907 | ++ |
| A. acidoterrestris strain KKP 3194 | 2020 | Soil from an apple orchard | + | KY088041 | − |
| A. acidoterrestris strain KKP 3195 | 2020 | Soil from an apple orchard | + | KY088042 | + |
| A. acidoterrestris strain KKP 3347 | 2020 | Soil from an apple orchard | + | KY088043 | − |
| A. acidoterrestris strain KKP 3348 | 2020 | Soil from an apple orchard | + | KY088047 | − |
| A. acidoterrestris strain KKP 3349 | 2020 | Soil from an apple orchard | + | MW332524 | − |
| Strains from the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures (n = 7) | |||||
| Alicyclobacillus acidocaldarius subsp. acidocaldarius strain DSM 446 | 1990 | Acid hot spring | − | CP001727 | − |
| Alicyclobacillus hesperidum strain DSM 12489 | 1998 | Solfataric soils | + | FNOJ00000000 | − |
| Alicyclobacillus herbarius strain DSM 13609 | 2000 | Dry flower of Hibiscus | + | AUMH00000000 | − |
| Alicyclobacillus acidophilus strain DSM 14558 | 2001 | Acidic beverage, which was off-flavored | + | AB076660 | − |
| A. fastidiosus strain DSM 17978 | 2003 | Apple juice | − | NR_041471 | − |
| A. acidoterrestris strain DSM 2498 | 1982 | Juice | + | AB059675 | ++ |
| A. acidoterrestris strain DSM 3922 | 1986 | Garden soil | + | AURB00000000 | − |
| Control Culture | MOI = 1.0 | MOI = 0.1 | |
|---|---|---|---|
| ΔOD | 0.066 | 0.038 | 0.007 |
| μ [h—1] | 0.064 | 0.031 | 0.032 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shymialevich, D.; Wójcicki, M.; Świder, O.; Średnicka, P.; Sokołowska, B. Characterization and Genome Study of a Newly Isolated Temperate Phage Belonging to a New Genus Targeting Alicyclobacillus acidoterrestris. Genes 2023, 14, 1303. https://doi.org/10.3390/genes14061303
Shymialevich D, Wójcicki M, Świder O, Średnicka P, Sokołowska B. Characterization and Genome Study of a Newly Isolated Temperate Phage Belonging to a New Genus Targeting Alicyclobacillus acidoterrestris. Genes. 2023; 14(6):1303. https://doi.org/10.3390/genes14061303
Chicago/Turabian StyleShymialevich, Dziyana, Michał Wójcicki, Olga Świder, Paulina Średnicka, and Barbara Sokołowska. 2023. "Characterization and Genome Study of a Newly Isolated Temperate Phage Belonging to a New Genus Targeting Alicyclobacillus acidoterrestris" Genes 14, no. 6: 1303. https://doi.org/10.3390/genes14061303
APA StyleShymialevich, D., Wójcicki, M., Świder, O., Średnicka, P., & Sokołowska, B. (2023). Characterization and Genome Study of a Newly Isolated Temperate Phage Belonging to a New Genus Targeting Alicyclobacillus acidoterrestris. Genes, 14(6), 1303. https://doi.org/10.3390/genes14061303









