Dynamics of Amino Acid Metabolism, Gene Expression, and Circulomics in a Recombinant Chinese Hamster Ovary Cell Line Adapted to Moderate and High Levels of Extracellular Lactate
Abstract
:1. Introduction
2. Materials and Methods
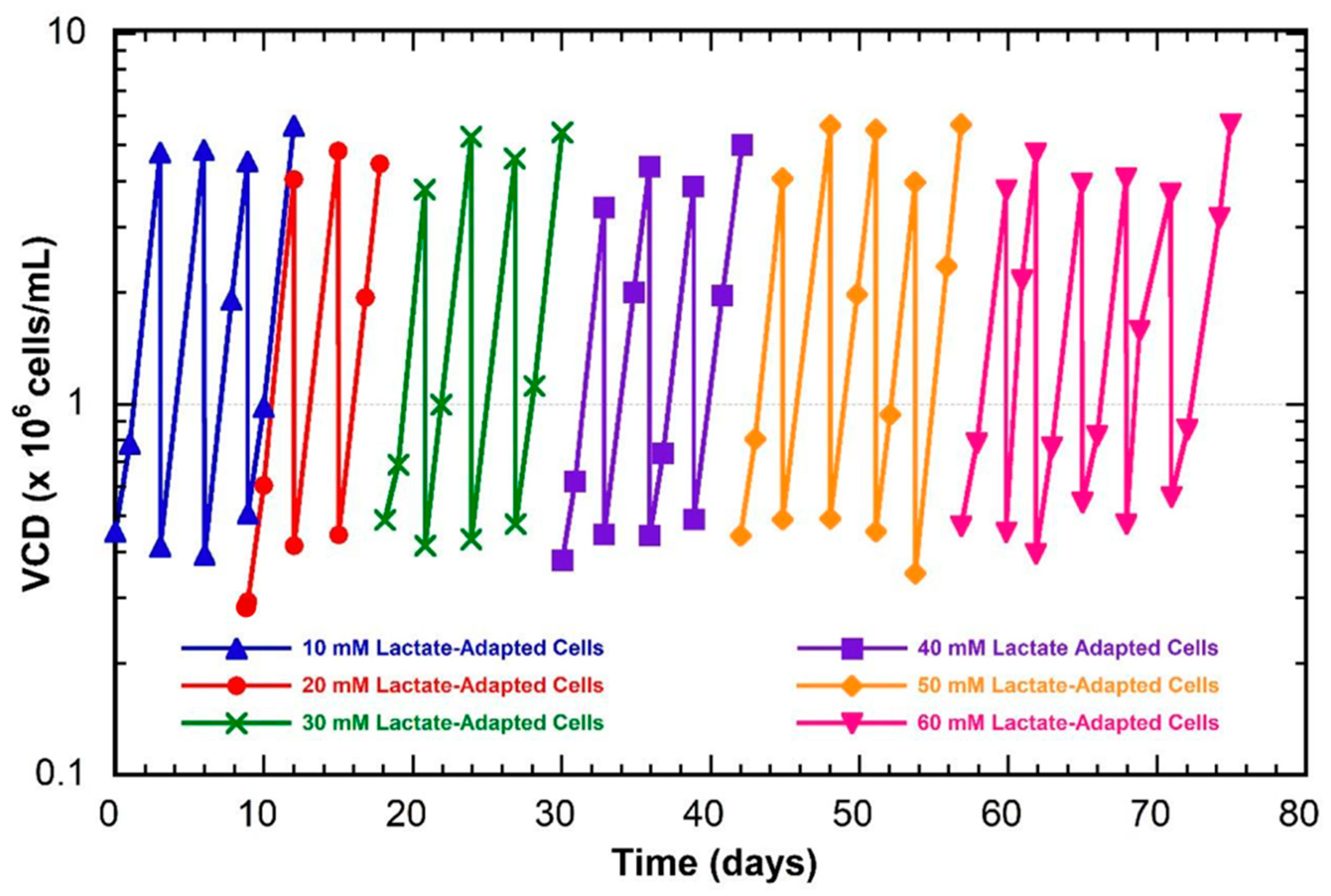
2.1. Cell Lines and Batch Culture Adaptation
2.2. EccDNA Enrichment and Sequence Library Preparation
2.3. Bioinformatic Pipeline
2.4. EccDNA Structure and Identification of Genomic Origins
3. Results
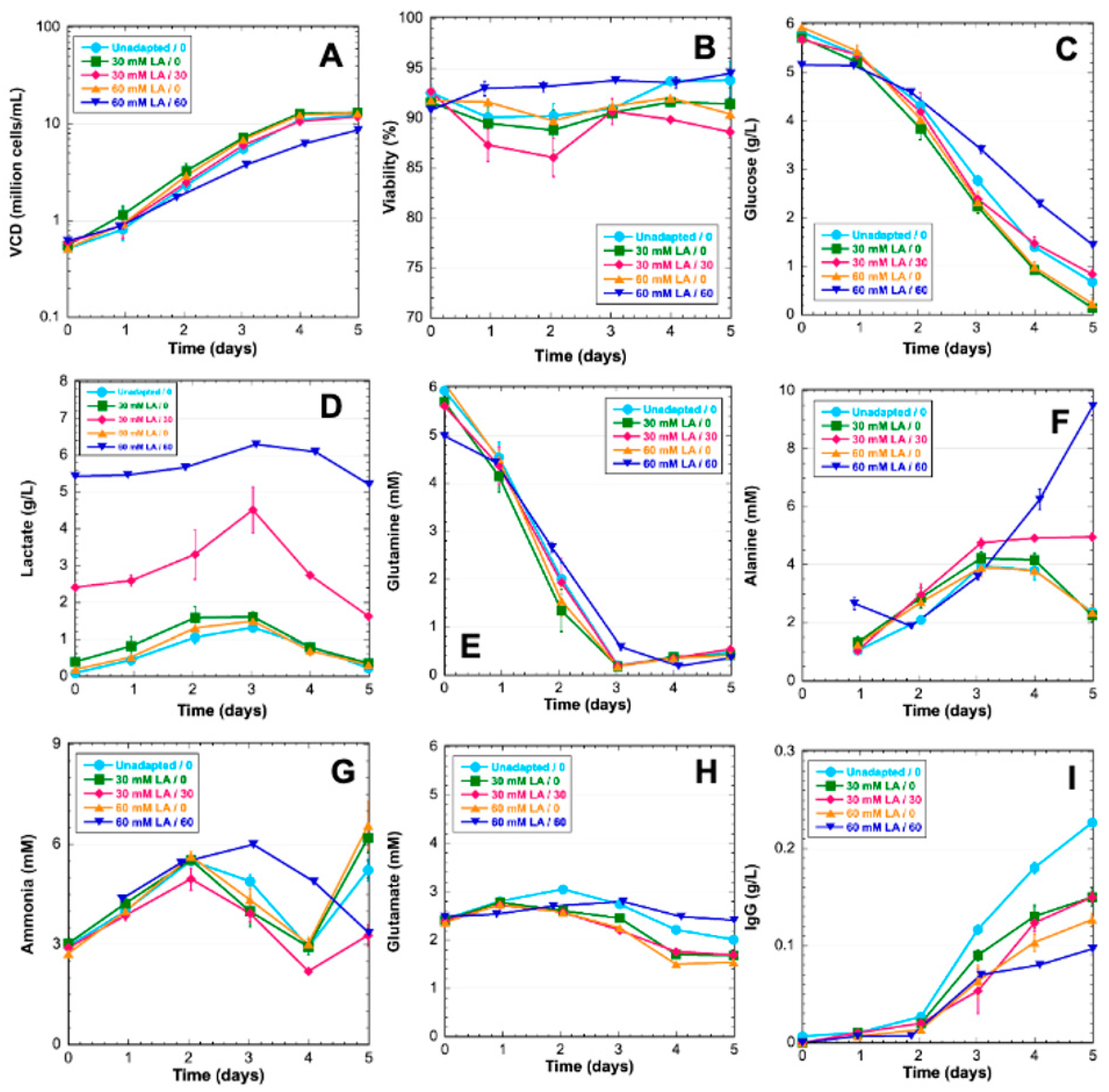
3.1. Phenotypic Cell Culture Data
3.2. EccDNA Sequence Composition
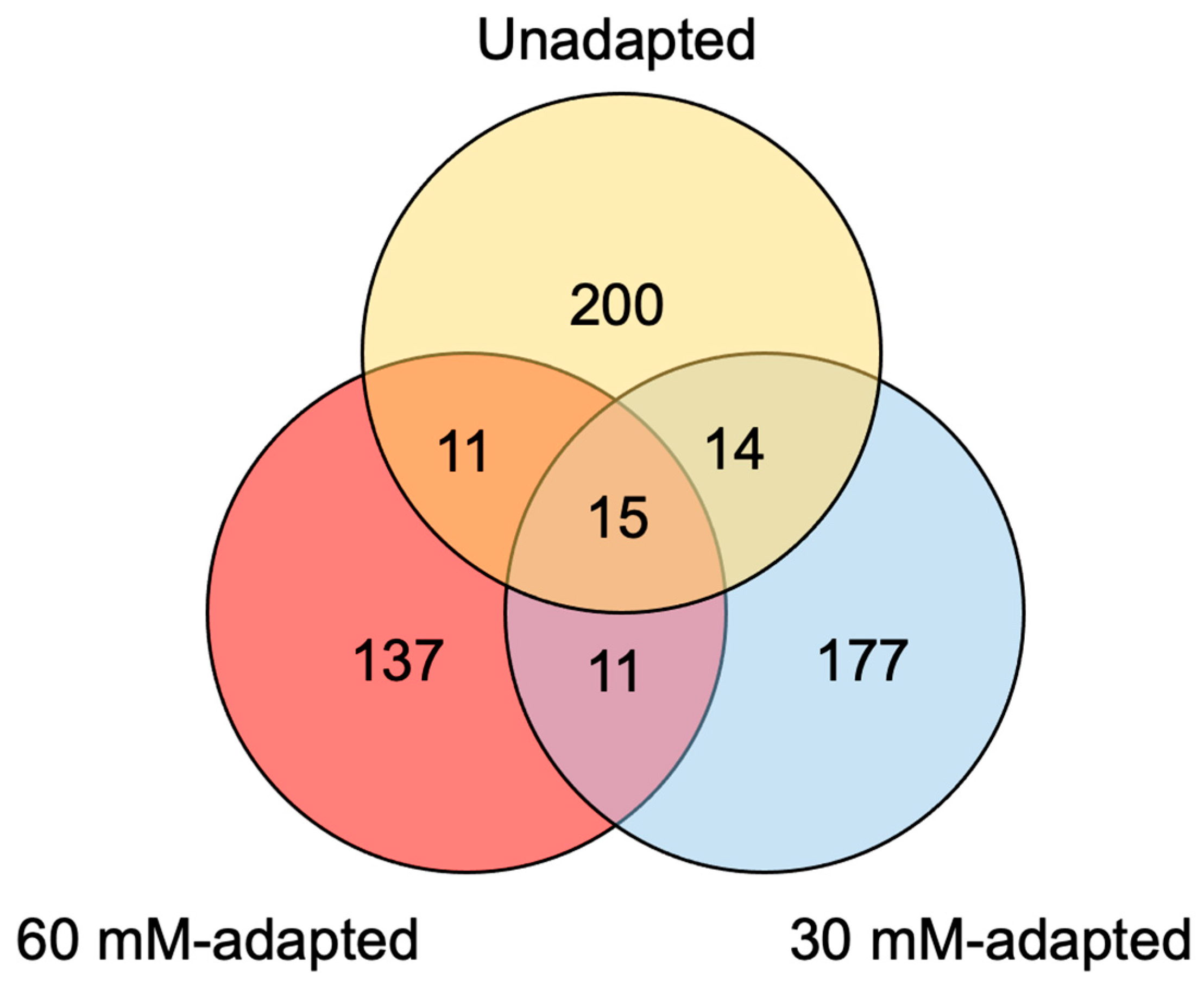
3.3. EccDNA Sequence Origins
3.4. Transcriptome Analysis
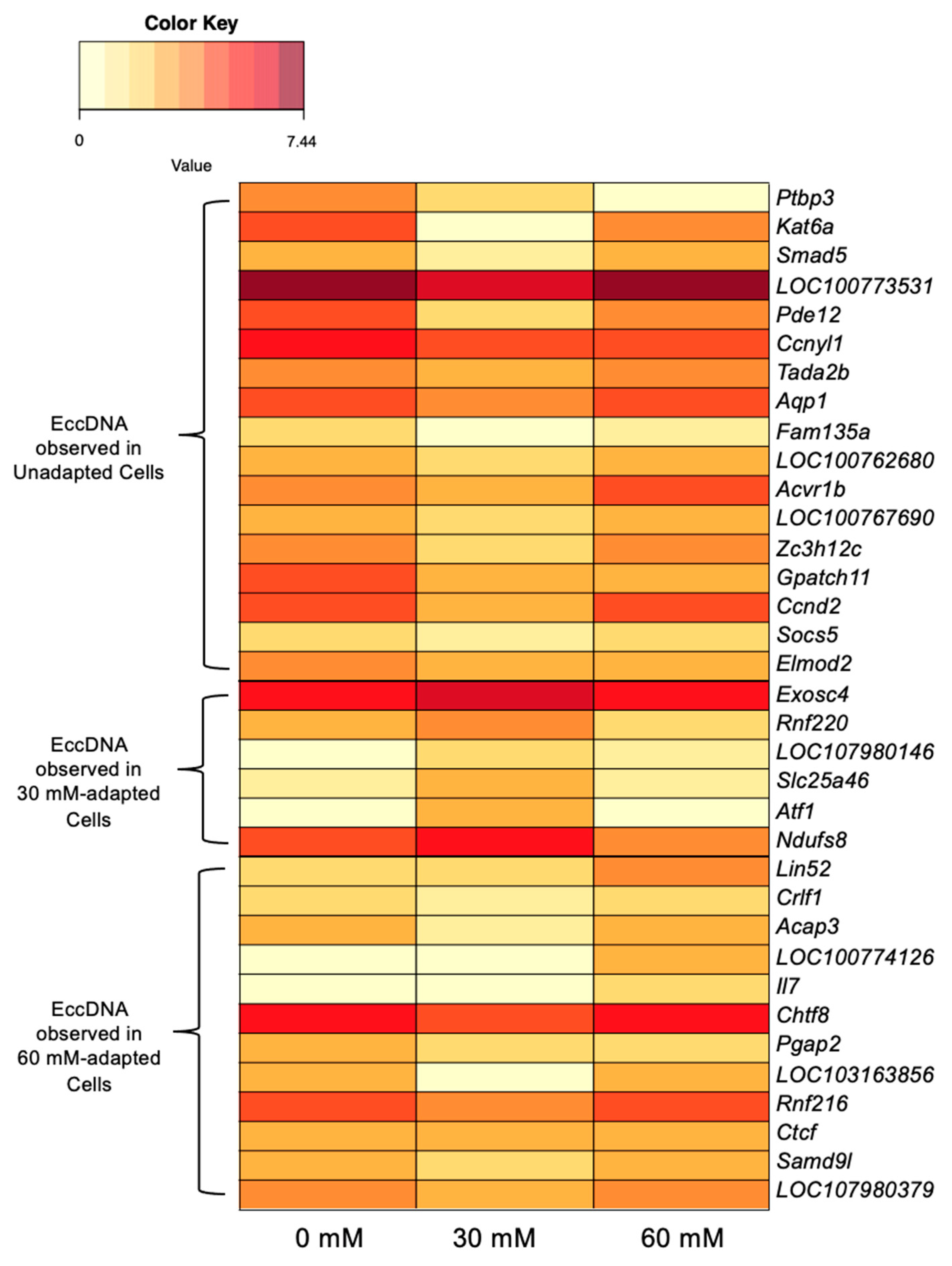
3.4.1. EccDNA-Encoded Genes
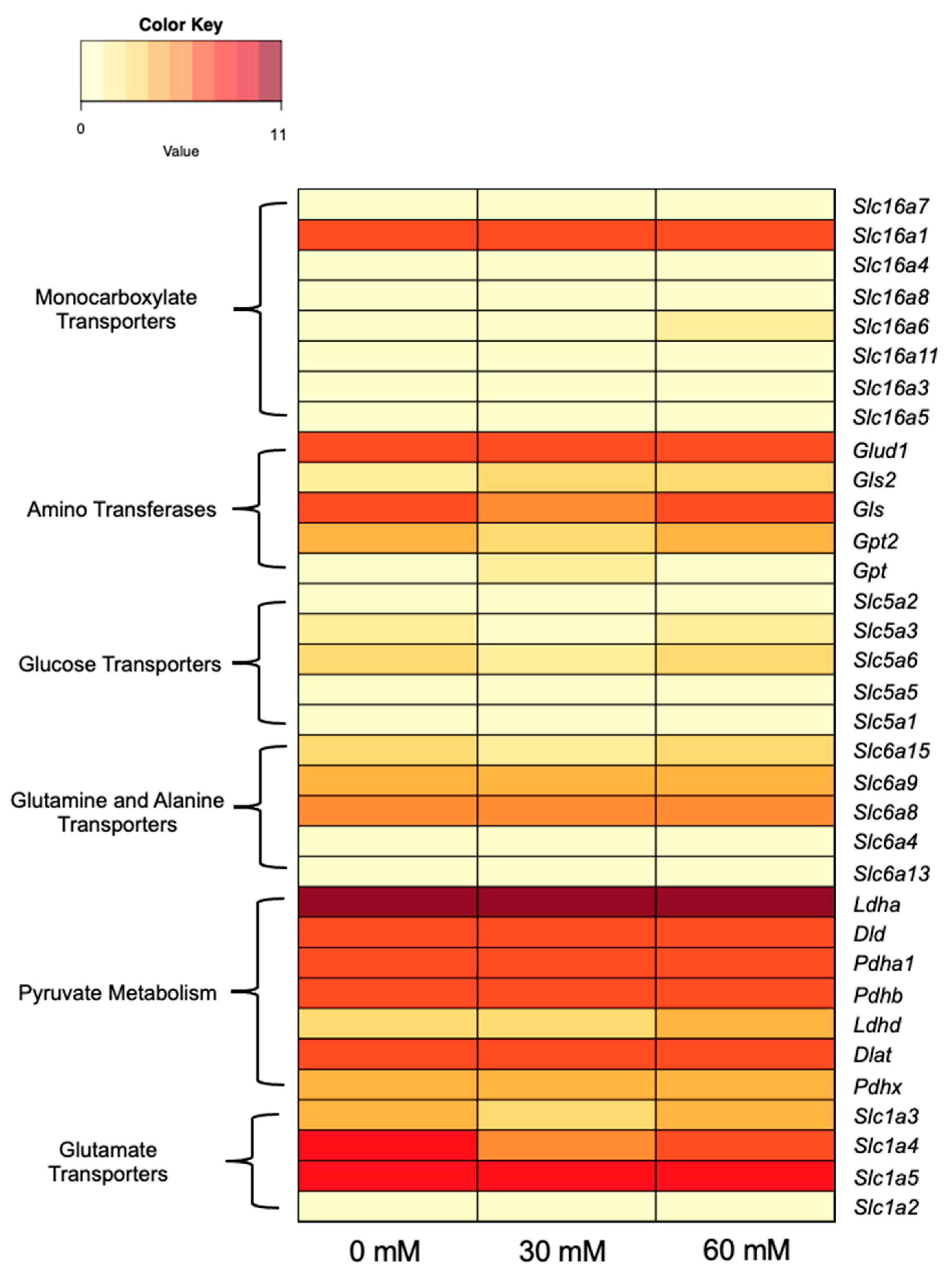
3.4.2. Metabolism-Linked Genes
3.4.3. Differentially Expressed Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lao, M.S.; Toth, D. Effects of ammonium and lactate on growth and metabolism of a recombinant Chinese hamster ovary cell culture. Biotechnol. Prog. 1997, 13, 688–691. [Google Scholar] [CrossRef]
- Ma, N.; Ellet, J.; Okediadi, C.; Hermes, P.; McCormick, E.; Casnocha, S. A single nutrient feed supports both chemically defined NS0 and CHO fed-batch processes: Improved productivity and lactate metabolism. Biotechnol. Prog. 2009, 25, 1353–1363. [Google Scholar] [CrossRef]
- Pereira, S.; Kildegaard, H.F.; Andersen, M.R. Impact of CHO Metabolism on Cell Growth and Protein Production: An Overview of Toxic and Inhibiting Metabolites and Nutrients. Biotechnol. J. 2018, 13, e1700499. [Google Scholar] [CrossRef] [Green Version]
- Ozturk, S.S.; Riley, M.R.; Palsson, B.O. Effects of ammonia and lactate on hybridoma growth, metabolism, and antibody production. Biotechnol. Bioeng. 1992, 39, 418–431. [Google Scholar] [CrossRef] [Green Version]
- Gagnon, M.; Hiller, G.; Luan, Y.T.; Kittredge, A.; DeFelice, J.; Drapeau, D. High-end pH-controlled delivery of glucose effectively suppresses lactate accumulation in CHO fed-batch cultures. Biotechnol. Bioeng. 2011, 108, 1328–1337. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, N.; Martinez, V.S.; Gerdtzen, Z.P. Engineering CHO cells galactose metabolism to reduce lactate synthesis. Biotechnol. Lett. 2019, 41, 779–788. [Google Scholar] [CrossRef]
- Ley, D.; Pereira, S.; Pedersen, L.E.; Arnsdorf, J.; Hefzi, H.; Davy, A.M.; Ha, T.K.; Wulff, T.; Kildegaard, H.F.; Andersen, M.R. Reprogramming AA catabolism in CHO cells with CRISPR/Cas9 genome editing improves cell growth and reduces byproduct secretion. Metab. Eng. 2019, 56, 120–129. [Google Scholar] [CrossRef]
- Li, J.; Wong, C.L.; Vijayasankaran, N.; Hudson, T.; Amanullah, A. Feeding lactate for CHO cell culture processes: Impact on culture metabolism and performance. Biotechnol. Bioeng. 2012, 109, 1173–1186. [Google Scholar] [CrossRef] [PubMed]
- Hauser, H.; Wagner, R. (Eds.) Mammalian Cell Biotechnology in Protein Production; Walter de Gruyter: Berlin, Germany; New York, NY, USA, 1997; p. xix. 491p. [Google Scholar]
- Hartley, F.; Walker, T.; Chung, V.; Morten, K. Mechanisms driving the lactate switch in Chinese hamster ovary cells. Biotechnol. Bioeng. 2018, 115, 1890–1903. [Google Scholar] [CrossRef]
- Mulukutla, B.C.; Gramer, M.; Hu, W.S. On metabolic shift to lactate consumption in fed-batch culture of mammalian cells. Metab. Eng. 2012, 14, 138–149. [Google Scholar] [CrossRef] [PubMed]
- Zagari, F.; Jordan, M.; Stettler, M.; Broly, H.; Wurm, F.M. Lactate metabolism shift in CHO cell culture: The role of mitochondrial oxidative activity. New Biotechnol. 2013, 30, 238–245. [Google Scholar] [CrossRef]
- Altamirano, C.; Illanes, A.; Becerra, S.; Cairo, J.J.; Godia, F. Considerations on the lactate consumption by CHO cells in the presence of galactose. J. Biotechnol. 2006, 125, 547–556. [Google Scholar] [CrossRef]
- Altamirano, C.; Paredes, C.; Illanes, A.; Cairo, J.J.; Godia, F. Strategies for fed-batch cultivation of t-PA producing CHO cells: Substitution of glucose and glutamine and rational design of culture medium. J. Biotechnol. 2004, 110, 171–179. [Google Scholar] [CrossRef]
- Ghorbaniaghdam, A.; Chen, J.; Henry, O.; Jolicoeur, M. Analyzing clonal variation of monoclonal antibody-producing CHO cell lines using an in silico metabolomic platform. PLoS ONE 2014, 9, e90832. [Google Scholar] [CrossRef] [Green Version]
- Martinez, V.S.; Dietmair, S.; Quek, L.E.; Hodson, M.P.; Gray, P.; Nielsen, L.K. Flux balance analysis of CHO cells before and after a metabolic switch from lactate production to consumption. Biotechnol. Bioeng. 2013, 110, 660–666. [Google Scholar] [CrossRef]
- Wahrheit, J.; Nicolae, A.; Heinzle, E. Dynamics of growth and metabolism controlled by glutamine availability in Chinese hamster ovary cells. Appl. Microbiol. Biotechnol. 2014, 98, 1771–1783. [Google Scholar] [CrossRef] [PubMed]
- Liste-Calleja, L.; Lecina, M.; Lopez-Repullo, J.; Albiol, J.; Sola, C.; Cairo, J.J. Lactate and glucose concomitant consumption as a self-regulated pH detoxification mechanism in HEK293 cell cultures. Appl. Microbiol. Biotechnol. 2015, 99, 9951–9960. [Google Scholar] [CrossRef]
- Zalai, D.; Koczka, K.; Parta, L.; Wechselberger, P.; Klein, T.; Herwig, C. Combining mechanistic and data-driven approaches to gain process knowledge on the control of the metabolic shift to lactate uptake in a fed-batch CHO process. Biotechnol. Prog. 2015, 31, 1657–1668. [Google Scholar] [CrossRef]
- Luo, J.; Vijayasankaran, N.; Autsen, J.; Santuray, R.; Hudson, T.; Amanullah, A.; Li, F. Comparative metabolite analysis to understand lactate metabolism shift in Chinese hamster ovary cell culture process. Biotechnol. Bioeng. 2012, 109, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Converti, A.; Sommariva, C.; Del Borghi, M.; Ferraiolo, G. The effects of mixing on bioprocesses. Concentration distributions and mechanical shear stress. Bioprocess Eng. 1993, 9, 183–189. [Google Scholar] [CrossRef]
- Fomina-Yadlin, D.; Gosink, J.J.; McCoy, R.; Follstad, B.; Morris, A.; Russell, C.B.; McGrew, J.T. Cellular responses to individual amino-acid depletion in antibody-expressing and parental CHO cell lines. Biotechnol. Bioeng. 2014, 111, 965–979. [Google Scholar] [CrossRef]
- Synoground, B.F.; McGraw, C.E.; Elliott, K.S.; Leuze, C.; Roth, J.R.; Harcum, S.W.; Sandoval, N.R. Transient ammonia stress on Chinese hamster ovary (CHO) cells yield alterations to alanine metabolism and IgG glycosylation profiles. Biotechnol. J. 2021, 16, e2100098. [Google Scholar] [CrossRef]
- Dorner, A.J.; Wasley, L.C.; Raney, P.; Haugejorden, S.; Green, M.; Kaufman, R.J. The stress response in Chinese hamster ovary cells. Regulation of ERp72 and protein disulfide isomerase expression and secretion. J. Biol. Chem. 1990, 265, 22029–22034. [Google Scholar] [CrossRef]
- Hernandez, I.; Dhiman, H.; Klanert, G.; Jadhav, V.; Auer, N.; Hanscho, M.; Baumann, M.; Esteve-Codina, A.; Dabad, M.; Gomez, J.; et al. Epigenetic regulation of gene expression in Chinese Hamster Ovary cells in response to the changing environment of a batch culture. Biotechnol. Bioeng. 2019, 116, 677–692. [Google Scholar] [CrossRef] [Green Version]
- Molin, W.T.; Yaguchi, A.; Blenner, M.; Saski, C.A. The EccDNA Replicon: A Heritable, Extranuclear Vehicle That Enables Gene Amplification and Glyphosate Resistance in Amaranthus palmeri. Plant Cell 2020, 32, 2132–2140. [Google Scholar] [CrossRef] [Green Version]
- Spier Camposano, H.; Molin, W.T.; Saski, C.A. Sequence characterization of eccDNA content in glyphosate sensitive and resistant Palmer amaranth from geographically distant populations. PLoS ONE 2022, 17, e0260906. [Google Scholar] [CrossRef]
- Wang, M.; Chen, X.; Yu, F.; Ding, H.; Zhang, Y.; Wang, K. Extrachromosomal Circular DNAs: Origin, formation and emerging function in Cancer. Int. J. Biol. Sci. 2021, 17, 1010–1025. [Google Scholar] [CrossRef]
- Qiu, H.; Shao, Z.Y.; Wen, X.; Zhang, L.Z. New insights of extrachromosomal DNA in tumorigenesis and therapeutic resistance of cancer. Am. J. Cancer Res. 2020, 10, 4056–4065. [Google Scholar]
- Verhaak, R.G.W.; Bafna, V.; Mischel, P.S. Extrachromosomal oncogene amplification in tumour pathogenesis and evolution. Nat. Rev. Cancer 2019, 19, 283–288. [Google Scholar] [CrossRef]
- Yan, Y.; Guo, G.; Huang, J.; Gao, M.; Zhu, Q.; Zeng, S.; Gong, Z.; Xu, Z. Current understanding of extrachromosomal circular DNA in cancer pathogenesis and therapeutic resistance. J. Hematol. Oncol. 2020, 13, 124. [Google Scholar] [CrossRef]
- van Loon, N.; Miller, D.; Murnane, J.P. Formation of extrachromosomal circular DNA in HeLa cells by nonhomologous recombination. Nucleic Acids Res. 1994, 22, 2447–2452. [Google Scholar] [CrossRef]
- Paulsen, T.; Malapati, P.; Eki, R.; Abbas, T.; Dutta, A. EccDNA formation is dependent on MMEJ, repressed by c-NHEJ pathway, and stimulated by DNA double-strand break. bioRxiv 2020, 410480. [Google Scholar] [CrossRef]
- Cohen, S.; Yacobi, K.; Segal, D. Extrachromosomal circular DNA of tandemly repeated genomic sequences in Drosophila. Genome Res. 2003, 13, 1133–1145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hull, R.M.; King, M.; Pizza, G.; Krueger, F.; Vergara, X.; Houseley, J. Transcription-induced formation of extrachromosomal DNA during yeast ageing. PLoS Biol. 2019, 17, e3000471. [Google Scholar] [CrossRef]
- Tandon, I.; Pal, R.; Pal, J.K.; Sharma, N.K. Extrachromosomal circular DNAs: An extra piece of evidence to depict tumor heterogeneity. Future Sci. OA 2019, 5, FSO390. [Google Scholar] [CrossRef] [Green Version]
- Mehta, D.; Cornet, L.; Hirsch-Hoffmann, M.; Zaidi, S.S.; Vanderschuren, H. Full-length sequencing of circular DNA viruses and extrachromosomal circular DNA using CIDER-Seq. Nat. Protoc. 2020, 15, 1673–1689. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: Improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Korf, I.; Robb, S.M.; Parra, G.; Ross, E.; Moore, B.; Holt, C.; Sanchez Alvarado, A.; Yandell, M. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008, 18, 188–196. [Google Scholar] [CrossRef] [Green Version]
- Hilliard, W.; MacDonald, M.L.; Lee, K.H. Chromosome-scale scaffolds for the Chinese hamster reference genome assembly to facilitate the study of the CHO epigenome. Biotechnol. Bioeng. 2020, 117, 2331–2339. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [Green Version]
- Hao, Z.; Lv, D.; Ge, Y.; Shi, J.; Weijers, D.; Yu, G.; Chen, J. RIdeogram: Drawing SVG graphics to visualize and map genome-wide data on the idiograms. PeerJ Comput. Sci. 2020, 6, e251. [Google Scholar] [CrossRef] [Green Version]
- Chitwood, D.G.; Wang, Q.; Klaubert, S.R.; Green, K.; Wu, C.H.; Harcum, S.W.; Saski, C.A. Microevolutionary dynamics of eccDNA in Chinese hamster ovary cells grown in fed-batch cultures under control and lactate-stressed conditions. Sci. Rep. 2023, 13, 1200. [Google Scholar] [CrossRef]
- Rorbach, J.; Nicholls, T.J.; Minczuk, M. PDE12 removes mitochondrial RNA poly(A) tails and controls translation in human mitochondria. Nucleic Acids Res. 2011, 39, 7750–7763. [Google Scholar] [CrossRef] [Green Version]
- Poulsen, J.B.; Andersen, K.R.; Kjaer, K.H.; Durand, F.; Faou, P.; Vestergaard, A.L.; Talbo, G.H.; Hoogenraad, N.; Brodersen, D.E.; Justesen, J.; et al. Human 2′-phosphodiesterase localizes to the mitochondrial matrix with a putative function in mitochondrial RNA turnover. Nucleic Acids Res. 2011, 39, 3754–3770. [Google Scholar] [CrossRef]
- Ohta, S.; Bukowski-Wills, J.C.; Sanchez-Pulido, L.; Alves Fde, L.; Wood, L.; Chen, Z.A.; Platani, M.; Fischer, L.; Hudson, D.F.; Ponting, C.P.; et al. The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell 2010, 142, 810–821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, C.; Wu, K.; Gu, S.; Wang, W.; Xie, S.; Zhou, Y. PTBP3 mediates TGF-β-induced EMT and metastasis of lung adenocarcinoma. Cell Cycle 2022, 21, 1406–1421. [Google Scholar] [CrossRef]
- Hou, P.; Chen, F.; Yong, H.; Lin, T.; Li, J.; Pan, Y.; Jiang, T.; Li, M.; Chen, Y.; Song, J.; et al. PTBP3 contributes to colorectal cancer growth and metastasis via translational activation of HIF-1alpha. J. Exp. Clin. Cancer Res. 2019, 38, 301. [Google Scholar] [CrossRef] [Green Version]
- de Sury, R.; Martinez, P.; Procaccio, V.; Lunardi, J.; Issartel, J.P. Genomic structure of the human NDUFS8 gene coding for the iron-sulfur TYKY subunit of the mitochondrial NADH:ubiquinone oxidoreductase. Gene 1998, 215, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Taniue, K.; Tanu, T.; Shimoura, Y.; Mitsutomi, S.; Han, H.; Kakisaka, R.; Ono, Y.; Tamamura, N.; Takahashi, K.; Wada, Y.; et al. RNA Exosome Component EXOSC4 Amplified in Multiple Cancer Types Is Required for the Cancer Cell Survival. Int. J. Mol. Sci. 2022, 23, 496. [Google Scholar] [CrossRef]
- Iness, A.N.; Felthousen, J.; Ananthapadmanabhan, V.; Sesay, F.; Saini, S.; Guiley, K.Z.; Rubin, S.M.; Dozmorov, M.; Litovchick, L. The cell cycle regulatory DREAM complex is disrupted by high expression of oncogenic B-Myb. Oncogene 2019, 38, 1080–1092. [Google Scholar] [CrossRef]
- Kaufman, R.J.; Sharp, P.A.; Latt, S.A. Evolution of chromosomal regions containing transfected and amplified dihydrofolate reductase sequences. Mol. Cell Biol. 1983, 3, 699–711. [Google Scholar] [CrossRef]
- Halestrap, A.P.; Meredith, D. The SLC16 gene family-from monocarboxylate transporters (MCTs) to aromatic amino acid transporters and beyond. Pflügers Arch. 2004, 447, 619–628. [Google Scholar] [CrossRef]
- Adeva, M.; Gonzalez-Lucan, M.; Seco, M.; Donapetry, C. Enzymes involved in l-lactate metabolism in humans. Mitochondrion 2013, 13, 615–629. [Google Scholar] [CrossRef]
- Wright, E.M.; Turk, E. The sodium/glucose cotransport family SLC5. Pflügers Arch. 2004, 447, 510–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, N.H.; Reith, M.E.; Quick, M.W. Synaptic uptake and beyond: The sodium- and chloride-dependent neurotransmitter transporter family SLC6. Pflügers Arch. 2004, 447, 519–531. [Google Scholar] [CrossRef]
- Grewer, C.; Gameiro, A.; Rauen, T. SLC1 glutamate transporters. Pflügers Arch. 2014, 466, 3–24. [Google Scholar] [CrossRef] [Green Version]
- Rincon-Arano, H.; Rosales, R.; Mora, N.; Rodriguez-Castaneda, A.; Rosales, C. R-Ras promotes tumor growth of cervical epithelial cells. Cancer 2003, 97, 575–585. [Google Scholar] [CrossRef]
- Szanto, I. NADPH Oxidase 4 (NOX4) in Cancer: Linking Redox Signals to Oncogenic Metabolic Adaptation. Int. J. Mol. Sci. 2022, 23, 2702. [Google Scholar] [CrossRef]
- Kontorovich, T.; Cohen, Y.; Nir, U.; Friedman, E. Promoter methylation patterns of ATM, ATR, BRCA1, BRCA2 and p53 as putative cancer risk modifiers in Jewish BRCA1/BRCA2 mutation carriers. Breast Cancer Res. Treat. 2009, 116, 195–200. [Google Scholar] [CrossRef]
- Wu, Y.; Zhao, W.; Liu, Y.; Tan, X.; Li, X.; Zou, Q.; Xiao, Z.; Xu, H.; Wang, Y.; Yang, X. Function of HNRNPC in breast cancer cells by controlling the dsRNA-induced interferon response. EMBO J. 2018, 37, e99017. [Google Scholar] [CrossRef]
- Xia, N.; Yang, N.; Shan, Q.; Wang, Z.; Liu, X.; Chen, Y.; Lu, J.; Huang, W.; Wang, Z. HNRNPC regulates RhoA to induce DNA damage repair and cancer-associated fibroblast activation causing radiation resistance in pancreatic cancer. J. Cell Mol. Med. 2022, 26, 2322–2336. [Google Scholar] [CrossRef]
- Glacken, M.W.; Fleischaker, R.J.; Sinskey, A.J. Reduction of waste product excretion via nutrient control: Possible strategies for maximizing product and cell yields on serum in cultures of mammalian cells. Biotechnol. Bioeng. 1986, 28, 1376–1389. [Google Scholar] [CrossRef]
- Chang, R.S.; Geyer, R.P. Propagation of conjunctival and HeLa cells in various carbohydrate media. Proc. Soc. Exp. Biol. Med. 1957, 96, 336–340. [Google Scholar] [CrossRef]
- Altamirano, C.; Paredes, C.; Cairo, J.J.; Godia, F. Improvement of CHO cell culture medium formulation: Simultaneous substitution of glucose and glutamine. Biotechnol. Prog. 2000, 16, 69–75. [Google Scholar] [CrossRef]
- Europa, A.F.; Gambhir, A.; Fu, P.C.; Hu, W.S. Multiple steady states with distinct cellular metabolism in continuous culture of mammalian cells. Biotechnol. Bioeng. 2000, 67, 25–34. [Google Scholar] [CrossRef]
- Zhou, W.; Rehm, J.; Hu, W.S. High viable cell concentration fed-batch cultures of hybridoma cells through on-line nutrient feeding. Biotechnol. Bioeng. 1995, 46, 579–587. [Google Scholar] [CrossRef]
- Yoon, S.K.; Choi, S.L.; Song, J.Y.; Lee, G.M. Effect of culture pH on erythropoietin production by Chinese hamster ovary cells grown in suspension at 32.5 and 37.0 degrees C. Biotechnol. Bioeng. 2005, 89, 345–356. [Google Scholar] [CrossRef]
- Langheinrich, C.; Nienow, A.W. Control of pH in large-scale, free suspension animal cell bioreactors: Alkali addition and pH excursions. Biotechnol. Bioeng. 1999, 66, 171–179. [Google Scholar] [CrossRef]
- Li, F.; Vijayasankaran, N.; Shen, A.Y.; Kiss, R.; Amanullah, A. Cell culture processes for monoclonal antibody production. MAbs 2010, 2, 466–479. [Google Scholar] [CrossRef] [Green Version]
- Le, H.; Kabbur, S.; Pollastrini, L.; Sun, Z.; Mills, K.; Johnson, K.; Karypis, G.; Hu, W.S. Multivariate analysis of cell culture bioprocess data--lactate consumption as process indicator. J. Biotechnol. 2012, 162, 210–223. [Google Scholar] [CrossRef]
- Toussaint, C.; Henry, O.; Durocher, Y. Metabolic engineering of CHO cells to alter lactate metabolism during fed-batch cultures. J. Biotechnol. 2016, 217, 122–131. [Google Scholar] [CrossRef]
- Gupta, S.K.; Srivastava, S.K.; Sharma, A.; Nalage, V.H.H.; Salvi, D.; Kushwaha, H.; Chitnis, N.B.; Shukla, P. Metabolic engineering of CHO cells for the development of a robust protein production platform. PLoS ONE 2017, 12, e0181455. [Google Scholar] [CrossRef] [Green Version]
- Zhou, M.; Crawford, Y.; Ng, D.; Tung, J.; Pynn, A.F.; Meier, A.; Yuk, I.H.; Vijayasankaran, N.; Leach, K.; Joly, J.; et al. Decreasing lactate level and increasing antibody production in Chinese Hamster Ovary cells (CHO) by reducing the expression of lactate dehydrogenase and pyruvate dehydrogenase kinases. J. Biotechnol. 2011, 153, 27–34. [Google Scholar] [CrossRef]
- Kim, S.H.; Lee, G.M. Down-regulation of lactate dehydrogenase-A by siRNAs for reduced lactic acid formation of Chinese hamster ovary cells producing thrombopoietin. Appl. Microbiol. Biotechnol. 2007, 74, 152–159. [Google Scholar] [CrossRef]
- Kim, S.H.; Lee, G.M. Functional expression of human pyruvate carboxylase for reduced lactic acid formation of Chinese hamster ovary cells (DG44). Appl. Microbiol. Biotechnol. 2007, 76, 659–665. [Google Scholar] [CrossRef] [PubMed]
- Noh, S.M.; Park, J.H.; Lim, M.S.; Kim, J.W.; Lee, G.M. Reduction of ammonia and lactate through the coupling of glutamine synthetase selection and downregulation of lactate dehydrogenase-A in CHO cells. Appl. Microbiol. Biotechnol. 2017, 101, 1035–1045. [Google Scholar] [CrossRef] [PubMed]
- Freund, N.W.; Croughan, M.S. A Simple Method to Reduce both Lactic Acid and Ammonium Production in Industrial Animal Cell Culture. Int. J. Mol. Sci. 2018, 19, 385. [Google Scholar] [CrossRef] [Green Version]
- Pickett, H.A.; Cesare, A.J.; Johnston, R.L.; Neumann, A.A.; Reddel, R.R. Control of telomere length by a trimming mechanism that involves generation of t-circles. EMBO J. 2009, 28, 799–809. [Google Scholar] [CrossRef] [Green Version]
- Tomaska, L.; Nosek, J.; Kramara, J.; Griffith, J.D. Telomeric circles: Universal players in telomere maintenance? Nat. Struct. Mol. Biol. 2009, 16, 1010–1015. [Google Scholar] [CrossRef]
- Molin, W.T.; Yaguchi, A.; Blenner, M.; Saski, C.A. Autonomous replication sequences from the Amaranthus palmeri eccDNA replicon enable replication in yeast. BMC Res. Notes 2020, 13, 330. [Google Scholar] [CrossRef]
- Von Hoff, D.D.; Needham-VanDevanter, D.R.; Yucel, J.; Windle, B.E.; Wahl, G.M. Amplified human MYC oncogenes localized to replicating submicroscopic circular DNA molecules. Proc. Natl. Acad. Sci. USA 1988, 85, 4804–4808. [Google Scholar] [CrossRef]
- Kim, H.; Nguyen, N.P.; Turner, K.; Wu, S.; Gujar, A.D.; Luebeck, J.; Liu, J.; Deshpande, V.; Rajkumar, U.; Namburi, S.; et al. Extrachromosomal DNA is associated with oncogene amplification and poor outcome across multiple cancers. Nat. Genet. 2020, 52, 891–897. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, H.; Zhou, Y.; Shi, J. Extrachromosomal circular DNA: A new potential role in cancer progression. J. Transl. Med. 2021, 19, 257. [Google Scholar] [CrossRef]
- Sadasivam, S.; DeCaprio, J.A. The DREAM complex: Master coordinator of cell cycle-dependent gene expression. Nat. Rev. Cancer 2013, 13, 585–595. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Weng, L.; Jia, Y.; Liu, B.; Wu, S.; Xue, L.; Yin, X.; Mao, A.; Wang, Z.; Shang, M. PTBP3 promotes malignancy and hypoxia-induced chemoresistance in pancreatic cancer cells by ATG12 up-regulation. J. Cell Mol. Med. 2020, 24, 2917–2930. [Google Scholar] [CrossRef]
- Xie, C.; Long, F.; Li, L.; Li, X.; Ma, M.; Lu, Z.; Wu, R.; Zhang, Y.; Huang, L.; Chou, J.; et al. PTBP3 modulates P53 expression and promotes colorectal cancer cell proliferation by maintaining UBE4A mRNA stability. Cell Death Dis. 2022, 13, 128. [Google Scholar] [CrossRef]
- Liang, X.; Shi, H.; Yang, L.; Qiu, C.; Lin, S.; Qi, Y.; Li, J.; Zhao, A.; Liu, J. Inhibition of polypyrimidine tract-binding protein 3 induces apoptosis and cell cycle arrest, and enhances the cytotoxicity of 5- fluorouracil in gastric cancer cells. Br. J. Cancer 2017, 116, 903–911. [Google Scholar] [CrossRef] [Green Version]
- de Nadal, E.; Ammerer, G.; Posas, F. Controlling gene expression in response to stress. Nat. Rev. Genet. 2011, 12, 833–845. [Google Scholar] [CrossRef]
- Lopez-Maury, L.; Marguerat, S.; Bahler, J. Tuning gene expression to changing environments: From rapid responses to evolutionary adaptation. Nat. Rev. Genet. 2008, 9, 583–593. [Google Scholar] [CrossRef]
- Macia, J.; Regot, S.; Peeters, T.; Conde, N.; Sole, R.; Posas, F. Dynamic signaling in the Hog1 MAPK pathway relies on high basal signal transduction. Sci. Signal 2009, 2, ra13. [Google Scholar] [CrossRef] [Green Version]
- Acar, M.; Mettetal, J.T.; van Oudenaarden, A. Stochastic switching as a survival strategy in fluctuating environments. Nat. Genet. 2008, 40, 471–475. [Google Scholar] [CrossRef]
- Swindell, W.R.; Huebner, M.; Weber, A.P. Plastic and adaptive gene expression patterns associated with temperature stress in Arabidopsis thaliana. Heredity 2007, 99, 143–150. [Google Scholar] [CrossRef] [Green Version]
- McClintock, B. The significance of responses of the genome to challenge. Science 1984, 226, 792–801. [Google Scholar] [CrossRef] [Green Version]

| Condition | Sequences | Sequences Clustered | Max Length (bp) | Average Length (bp) | Repeat bases masked | GC (%) | eccDNA with genes | eccDNA with rDNA | eccDNA with tRNA | ORI (>95%) |
|---|---|---|---|---|---|---|---|---|---|---|
| Unadapted | 8202 | 8109 | 24,436 | 4905 | 15,159,445 bp (37.9%) | 40.6% | 295 (3.6%) | 6 | 800 (9.9%) | 3 |
| 30 mM Adapted | 6476 | 6419 | 27,326 | 4976 | 12,132,410 bp (37.9%) | 40.5% | 255 (4.0%) | 4 | 618 (9.6%) | 3 |
| 60 mM Adapted | 4563 | 4511 | 24,598 | 5515 | 9,671,540 bp (38.9%) | 40.3% | 233 (5.2%) | 4 | 484 (10.7%) | 4 |
| Unadapted | 30 mM Adapted | 60 mM Adapted | ||||||||
| Repeat Structure | Subcategory | Number of Sequences | Base pairs (bp) | Percent of total bases | Number of Sequences | Base pairs (bp) | Percent of total bases | Number of Sequences | Base pairs (bp) | Percent of total bases |
| SINEs: | 23,088 | 2,994,270 | 7.49 | 17,928 | 2,322,980 | 7.25 | 13,529 | 1,750,013 | 7.03 | |
| Alu/B1 | 9915 | 1,172,210 | 2.93 | 7715 | 910,743 | 2.84 | 6008 | 712,601 | 2.86 | |
| MIRs | 1263 | 142,664 | 0.36 | 972 | 108,753 | 0.34 | 688 | 77,202 | 0.31 | |
| LINEs: | 12,326 | 5,818,281 | 14.6 | 9816 | 4,729,537 | 14.8 | 7735 | 3,856,083 | 15.5 | |
| LINE1 | 11,416 | 5,678,568 | 14.2 | 9140 | 4,623,778 | 14.4 | 7284 | 3,781,556 | 15.2 | |
| LINE2 | 725 | 111,589 | 0.28 | 564 | 88,862 | 0.28 | 359 | 62,787 | 0.25 | |
| L3/CR1 | 136 | 20,991 | 0.05 | 81 | 11,098 | 0.03 | 68 | 8645 | 0.03 | |
| RTE | 45 | 6525 | 0.02 | 26 | 5024 | 0.02 | 22 | 2683 | 0.01 | |
| LTR elements: | 13,196 | 3,972,926 | 9.94 | 10,381 | 3,175,847 | 9.91 | 8222 | 2,552,906 | 10.3 | |
| ERVL | 1223 | 349,001 | 0.87 | 888 | 267,321 | 0.83 | 795 | 217,908 | 0.88 | |
| ERVL-MaLRs | 5550 | 1,498,301 | 3.75 | 4444 | 1,207,001 | 3.77 | 3434 | 927,879 | 3.73 | |
| ERR_class I | 1259 | 262,964 | 0.66 | 972 | 189,106 | 0.59 | 799 | 172,688 | 0.69 | |
| ERV_class II | 5043 | 1,804,631 | 4.52 | 3991 | 1,464,909 | 4.57 | 3115 | 1,182,229 | 4.75 | |
| DNA elements: | 2334 | 431,383 | 1.08 | 1899 | 358,406 | 1.12 | 1363 | 250,375 | 1.01 | |
| hAT-Charlie | 1415 | 247,910 | 0.62 | 1173 | 205,506 | 0.64 | 832 | 147,135 | 0.59 | |
| TcMar-Tigger | 598 | 123,340 | 0.31 | 456 | 98,914 | 0.31 | 326 | 69,009 | 0.28 | |
| Unclassified: | 626 | 244,548 | 0.61 | 464 | 197,627 | 0.62 | 389 | 141,496 | 0.57 | |
| Total Interspaced Repeats: | - | 13,461,408 | 33.7 | - | 10,784,397 | 33.7 | - | 8,550,873 | 34.4 | |
| Small RNA: | 581 | 41,990 | 0.11 | 467 | 34,506 | 0.11 | 412 | 31,344 | 0.13 | |
| Satellites | 1694 | 727,205 | 1.82 | 1338 | 579,860 | 1.81 | 1112 | 491,622 | 1.98 | |
| Simple Repeats | 16,679 | 836,271 | 2.09 | 13,098 | 665,925 | 2.08 | 10,325 | 541,927 | 2.18 | |
| Low Complexity | 2008 | 105,297 | 0.26 | 1514 | 77,218 | 0.24 | 1231 | 62,244 | 0.25 | |
| Unadapted | ||||||
| Chr | Start | End | Binned EccDNAs | Binned EccDNAs with Genes (#) | Binned EccDNAs with Genes (%) | Z-score |
| 9 | 15,000,000 | 15,500,000 | 217 | 10 | 4.6 | 42.66 |
| 9 | 14,500,000 | 15,000,000 | 113 | 22 | 19.5 | 21.98 |
| 9 | 14,000,000 | 14,500,000 | 89 | 7 | 7.9 | 17.2 |
| 9 | 15,500,000 | 16,000,000 | 88 | 3 | 3.4 | 17 |
| 9 | 13,500,000 | 14,000,000 | 84 | 0 | 0 | 16.21 |
| 9 | 13,000,000 | 13,500,000 | 51 | 1 | 2 | 9.64 |
| 10 | 25,500,000 | 26,000,000 | 47 | 0 | 0 | 8.85 |
| 9 | 16,500,000 | 17,000,000 | 36 | 1 | 2.8 | 6.66 |
| 9 | 16,000,000 | 16,500,000 | 35 | 2 | 5.7 | 6.46 |
| 9 | 17,500,000 | 18,000,000 | 35 | 0 | 0 | 6.46 |
| 9 | 25,500,000 | 26,000,000 | 32 | 2 | 6.3 | 5.87 |
| 4 | 131,500,000 | 132,000,000 | 29 | 0 | 0 | 5.27 |
| 7 | 134,000,000 | 134,359,064 | 27 | 0 | 0 | 4.87 |
| 9 | 17,000,000 | 17,500,000 | 27 | 2 | 7.4 | 4.87 |
| 1_2 | 195,000,00 | 195,500,000 | 23 | 0 | 0 | 4.08 |
| 30 mM-adapted | ||||||
| Chr | Start | End | Binned EccDNAs | Binned EccDNAs with Genes (#) | Binned EccDNAs with Genes (%) | Z-score |
| 9 | 15,000,000 | 15,500,000 | 185 | 8 | 4.3 | 41.41 |
| 9 | 14,500,000 | 15,000,000 | 117 | 19 | 16.2 | 26.03 |
| 9 | 14,000,000 | 14,500,000 | 84 | 3 | 3.6 | 18.56 |
| 9 | 13,500,000 | 14,000,000 | 76 | 3 | 3.9 | 16.75 |
| 9 | 15,500,000 | 16,000,000 | 61 | 0 | 0 | 13.36 |
| 9 | 16,000,000 | 16,500,000 | 55 | 6 | 10.9 | 12 |
| 9 | 12,500,000 | 13,000,000 | 47 | 1 | 2.1 | 10.19 |
| 9 | 17,500,000 | 18,000,000 | 41 | 4 | 9.8 | 8.83 |
| 9 | 13,000,000 | 13,500,000 | 36 | 3 | 8.3 | 7.7 |
| 1_2 | 196,000,000 | 196,500,000 | 34 | 1 | 2.9 | 7.25 |
| 9 | 16,500,000 | 17,000,000 | 29 | 2 | 6.9 | 6.12 |
| 2 | 34,500,000 | 35,000,000 | 21 | 4 | 19 | 4.31 |
| 4 | 72,500,000 | 73,000,000 | 20 | 0 | 0 | 4.08 |
| 7 | 134,000,000 | 134,359,064 | 19 | 0 | 0 | 3.86 |
| 9 | 25,500,000 | 26,000,000 | 19 | 3 | 15.8 | 3.86 |
| 60 mM-adapted | ||||||
| Chr | Start | End | Binned EccDNAs | Binned EccDNAs with Genes (#) | Binned EccDNAs with Genes (%) | Z-score |
| 9 | 15,000,000 | 15,500,000 | 141 | 7 | 5 | 35.89 |
| 9 | 14,500,000 | 15,000,000 | 98 | 23 | 23.5 | 24.84 |
| 9 | 14,000,000 | 14,500,000 | 85 | 5 | 5.9 | 21.49 |
| 9 | 13,500,000 | 14,000,000 | 77 | 1 | 1.3 | 19.44 |
| 9 | 15,500,000 | 16,000,000 | 53 | 0 | 0 | 13.26 |
| 7 | 134,000,000 | 134,359,064 | 41 | 0 | 0 | 10.18 |
| X | 106,500,000 | 107,000,000 | 38 | 0 | 0 | 9.41 |
| 9 | 12,500,000 | 13,000,000 | 35 | 2 | 5.7 | 8.64 |
| 9 | 16,000,000 | 16,500,000 | 33 | 10 | 30.3 | 8.12 |
| 2 | 41,000,000 | 41,500,000 | 32 | 0 | 0 | 7.86 |
| 7 | 88,000,000 | 88,500,000 | 32 | 2 | 6.3 | 7.86 |
| 9 | 17,500,000 | 18,000,000 | 32 | 2 | 6.3 | 7.86 |
| 2 | 34,500,000 | 35,000,000 | 29 | 9 | 31 | 7.09 |
| 4 | 5,000,000 | 5,500,000 | 29 | 0 | 0 | 7.09 |
| 9 | 17,000,000 | 17,500,000 | 28 | 1 | 3.6 | 6.83 |
| RefSeq ID | Gene | log2FC | RefSeq ID | Gene | log2FC | RefSeq ID | Gene | log2FC |
|---|---|---|---|---|---|---|---|---|
| XP_027259257.1 | Pnisr | 5.08 | XP_027263110.1 | Mms19 | 2.3 | XP_027262108.1 | C3H16orf87 | −2.43 |
| XP_027271342.1 | LOC107977511 | 4.72 | XP_027268884.1 | Yeats2 | 2.29 | XP_035302467.1 | Asip | −2.43 |
| XP_035300595.1 | Camk2n2 | 4.07 | XP_027266487.1 | Banp | 2.28 | XP_027250346.1 | Oard1 | −2.44 |
| XP_027260962.1 | Usp6nl | 4.01 | XP_027272584.1 | Trip10 | 2.26 | XP_035296751.1 | Prkag1 | −2.45 |
| XP_027243872.1 | Spred2 | 3.97 | XP_027260324.1 | Kat8 | 2.26 | XP_027246344.1 | Tinf2 | −2.45 |
| XP_035299495.1 | Vps11 | 3.81 | XP_027281260.1 | Baiap2 | 2.25 | XP_035309594.1 | LOC118237753 | −2.54 |
| XP_027270425.1 | Ccdc51 | 3.77 | XP_027281261.1 | Baiap2 | 2.25 | XP_035296374.1 | Tmem161b | −2.62 |
| XP_027265866.1 | Tmem41b | 3.64 | XP_027269839.1 | LOC100765617 | 2.2 | XP_027243712.1 | Mpv17l | −2.62 |
| XP_035309292.1 | Tdrd3 | 3.52 | XP_027279239.1 | Zswim3 | 2.19 | XP_027243713.1 | Mpv17l | −2.62 |
| XP_027284541.1 | Spr | 3.51 | XP_027256563.1 | Spata24 | 2.18 | XP_035308661.1 | Mpv17l | −2.62 |
| XP_027274795.1 | Nit1 | 3.33 | XP_027253398.1 | Asb1 | 2.18 | XP_027285208.1 | Ino80b | −2.7 |
| XP_035301857.1 | Nit1 | 3.33 | XP_035301484.1 | Rbbp5 | 2.18 | XP_027285383.1 | Tra2a | −2.71 |
| XP_027258717.1 | Ccn2 | 3.32 | XP_027264466.1 | LOC100757535 | 2.15 | XP_027283850.1 | Brca1 | −2.71 |
| XP_027278256.1 | Pex16 | 3.16 | XP_027277209.2 | Asip | 2.14 | XP_027283851.1 | Brca1 | −2.71 |
| XP_027282741.1 | CUNH17orf49 | 3.09 | XP_035296265.1 | Nhsl1 | 2.08 | XP_027283852.1 | Brca1 | −2.71 |
| XP_027276511.1 | Rras | 3.06 | XP_027258948.1 | Anp32b | 2.06 | XP_027283853.1 | Brca1 | −2.71 |
| XP_027268403.1 | Slc35a5 | 3.05 | XP_027254583.1 | Il7 | 2.06 | XP_027256348.1 | Rnf138 | −2.71 |
| XP_027263390.1 | Nox4 | 3.05 | XP_027264703.1 | Mob2 | 2.05 | XP_027274946.1 | Atxn3 | −2.75 |
| XP_027279804.1 | Coro7 | 2.98 | XP_027273144.1 | Acbd6 | 2.04 | XP_035298824.1 | Hivep1 | −2.77 |
| XP_027254088.1 | H6pd | 2.95 | XP_027281267.1 | Chmp6 | 2.03 | XP_027243022.1 | Ctbp1 | −2.82 |
| XP_027268965.1 | Klhl22 | 2.95 | XP_027266778.1 | Mthfs | 2 | XP_035298483.1 | Cpne2 | −2.83 |
| XP_027247336.1 | Agk | 2.94 | XP_027266779.1 | Mthfs | 2 | XP_027245634.1 | Fam133b | −2.87 |
| XP_027246345.1 | Tinf2 | 2.92 | XP_035307155.1 | LOC100774954 | −2.04 | XP_027271986.1 | Hip1 | −2.89 |
| XP_027286836.2 | Zc3h4 | 2.9 | XP_027274041.1 | Trmt5 | −2.05 | XP_027252162.1 | Pphln1 | −3.06 |
| XP_027252267.1 | Cacnb3 | 2.88 | XP_027250449.1 | Nr2c2ap | −2.07 | XP_027287883.1 | Mospd1 | −3.11 |
| XP_035306485.1 | Ptpn12 | 2.87 | XP_027258438.1 | Poli | −2.08 | XP_027244120.1 | Dnajb12 | −3.13 |
| XP_027278991.1 | Ptpa | 2.86 | XP_027274796.1 | Nit1 | −2.13 | XP_027244121.1 | Dnajb12 | −3.13 |
| XP_027259074.1 | Stoml2 | 2.84 | XP_027274797.1 | Nit1 | −2.13 | XP_035304186.1 | Top3a | −3.2 |
| XP_027255528.1 | Id3 | 2.83 | XP_027274798.1 | Nit1 | −2.13 | XP_027271702.1 | Pus1 | −3.2 |
| XP_027275696.1 | Slco4a1 | 2.81 | XP_027274799.1 | Nit1 | −2.13 | XP_027247306.1 | Hipk2 | −3.28 |
| XP_027243830.1 | Commd1 | 2.73 | XP_027274800.1 | Nit1 | −2.13 | XP_027258058.1 | Rtn4ip1 | −3.41 |
| XP_035295692.1 | Ift74 | 2.68 | XP_027274801.1 | Nit1 | −2.13 | XP_027262953.1 | Ldb1 | −3.47 |
| XP_027252455.1 | Eif4b | 2.6 | XP_027258090.1 | Pde4b | −2.15 | XP_027250779.1 | LOC103162709 | −3.66 |
| XP_027288617.1 | Mospd2 | 2.57 | XP_027266034.1 | Eef2k | −2.16 | XP_027255726.1 | Zbtb17 | −3.67 |
| XP_027265269.1 | Zkscan8 | 2.56 | XP_027288241.1 | Fam3a | −2.18 | XP_027270426.1 | Ccdc51 | −3.73 |
| XP_035294787.1 | LOC113833870 | 2.55 | XP_035295624.1 | Ndc1 | −2.19 | XP_027285126.1 | Atp6v1e1 | −3.87 |
| XP_027282857.1 | Vamp2 | 2.55 | XP_035307073.1 | LOC100755006 | −2.21 | XP_035303760.1 | Vamp2 | −3.91 |
| XP_027283079.1 | Top3a | 2.46 | XP_027267297.1 | Pde4a | −2.21 | XP_027266816.1 | Ube3d | −3.95 |
| XP_027283080.1 | Top3a | 2.46 | XP_027288197.1 | Taz | −2.23 | XP_035298723.1 | Usp6nl | −4.16 |
| XP_027258844.1 | Palm2akap2 | 2.42 | XP_027265636.1 | Uimc1 | −2.27 | XP_027266818.1 | Ube3d | −4.33 |
| XP_027271581.1 | Pnpla6 | 2.4 | XP_027269840.1 | LOC100765617 | −2.3 | XP_027262827.1 | Smndc1 | −4.34 |
| XP_027255969.1 | Exoc3 | 2.4 | XP_027248483.1 | Baz2a | −2.32 | XP_027250669.1 | Smim7 | −4.38 |
| XP_027278905.1 | Rapgef1 | 2.39 | XP_027248484.1 | Baz2a | −2.32 | XP_027257570.1 | Tmem167a | −5.29 |
| XP_027248513.1 | Ankrd52 | 2.34 | XP_035306520.1 | Baz2a | −2.32 | XP_027242456.1 | Hnrnpc | −5.62 |
| XP_027257344.1 | Ppwd1 | 2.34 | XP_035300748.1 | Khsrp | −2.38 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chitwood, D.G.; Uy, L.; Fu, W.; Klaubert, S.R.; Harcum, S.W.; Saski, C.A. Dynamics of Amino Acid Metabolism, Gene Expression, and Circulomics in a Recombinant Chinese Hamster Ovary Cell Line Adapted to Moderate and High Levels of Extracellular Lactate. Genes 2023, 14, 1576. https://doi.org/10.3390/genes14081576
Chitwood DG, Uy L, Fu W, Klaubert SR, Harcum SW, Saski CA. Dynamics of Amino Acid Metabolism, Gene Expression, and Circulomics in a Recombinant Chinese Hamster Ovary Cell Line Adapted to Moderate and High Levels of Extracellular Lactate. Genes. 2023; 14(8):1576. https://doi.org/10.3390/genes14081576
Chicago/Turabian StyleChitwood, Dylan G., Lisa Uy, Wanfang Fu, Stephanie R. Klaubert, Sarah W. Harcum, and Christopher A. Saski. 2023. "Dynamics of Amino Acid Metabolism, Gene Expression, and Circulomics in a Recombinant Chinese Hamster Ovary Cell Line Adapted to Moderate and High Levels of Extracellular Lactate" Genes 14, no. 8: 1576. https://doi.org/10.3390/genes14081576





