Blood Transcriptomic Analyses Reveal Functional Pathways Associated with Thermotolerance in Pregnant Ewes Exposed to Environmental Heat Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Management
2.2. Climatic Data and Thermo-Tolerance Indicator
2.3. RNA Isolation and Sequencing
2.4. Raw Data Reads and Alignment
2.5. Differential Gene Expression Analyses
2.6. Network and Functional Analysis
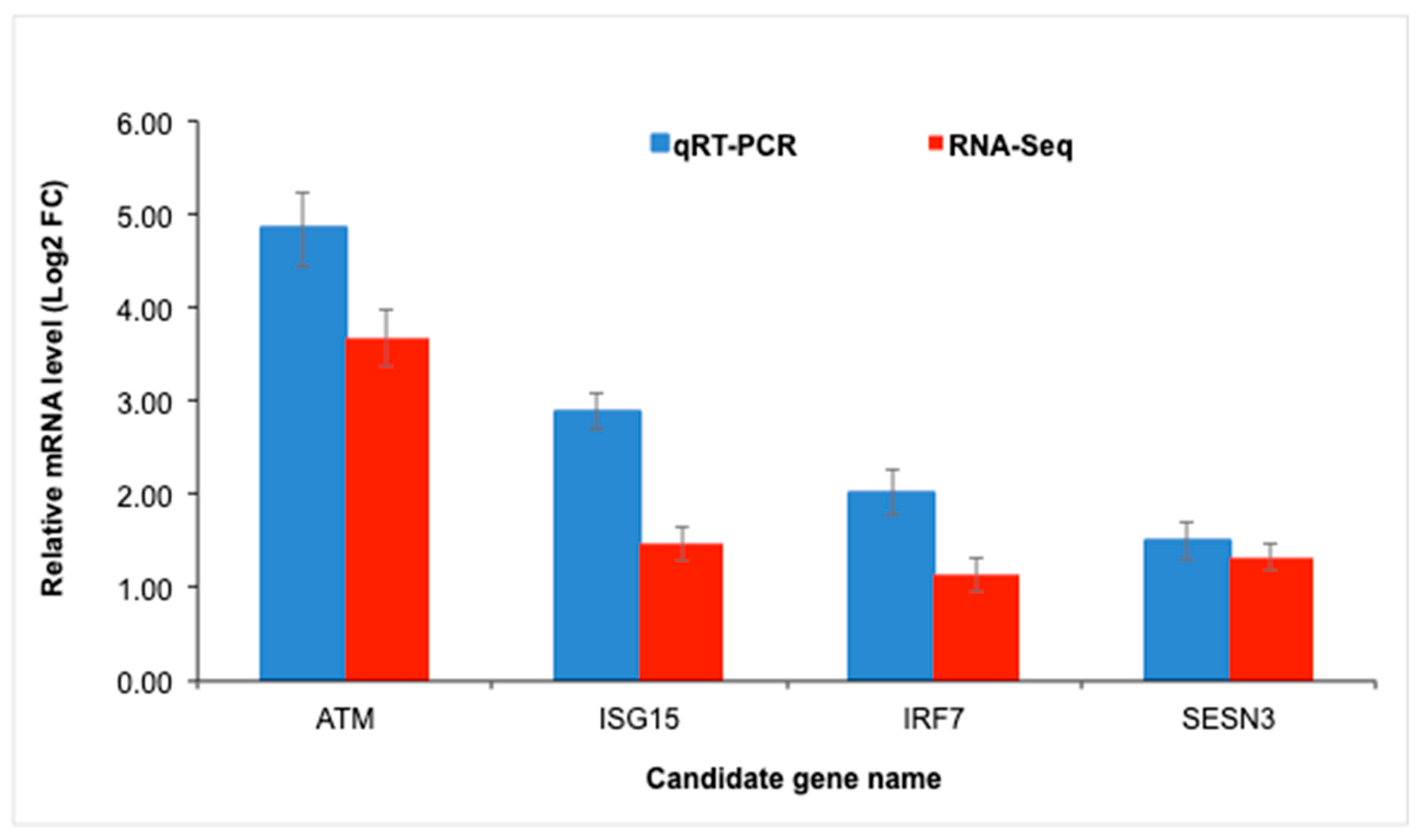
2.7. Validation of RNA-Seq Data Using Quantitative RT-PCR
2.8. Hematological Parameters
3. Results
3.1. Thermotolerance Indicator
3.2. Blood Cell Parameters
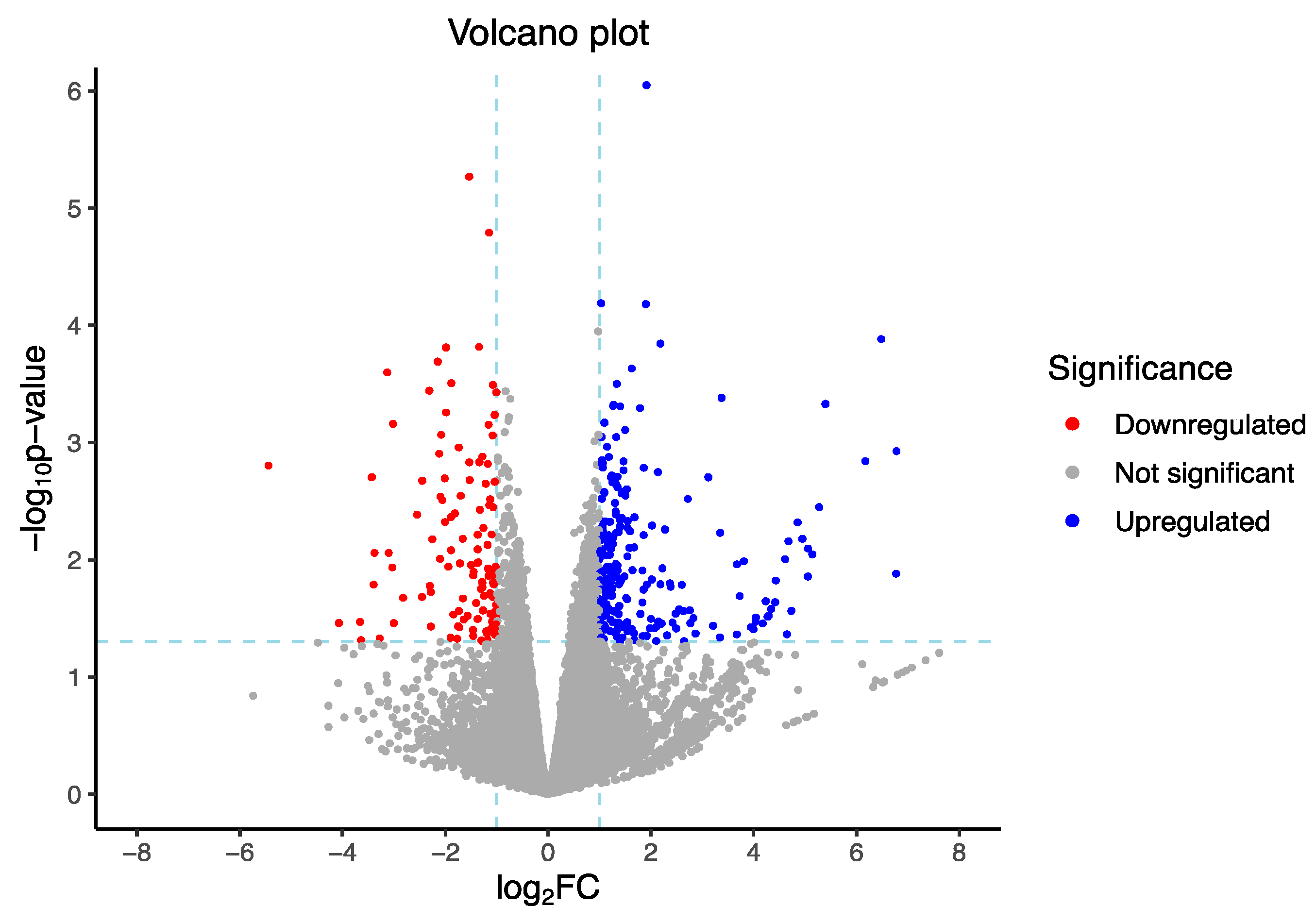
3.3. Read Mapping and Gene Expression
3.4. GO Enrichment and Functional Analyses
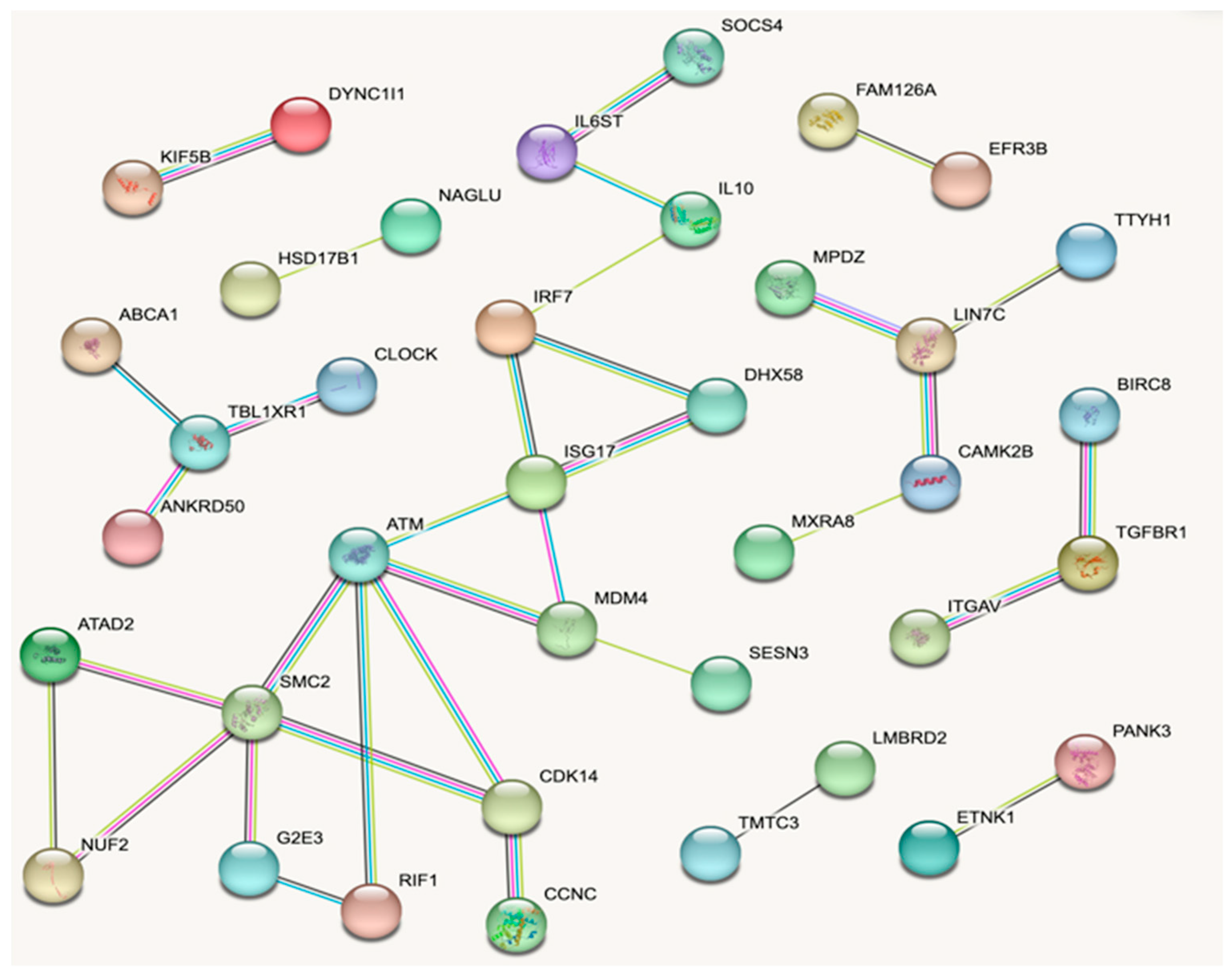
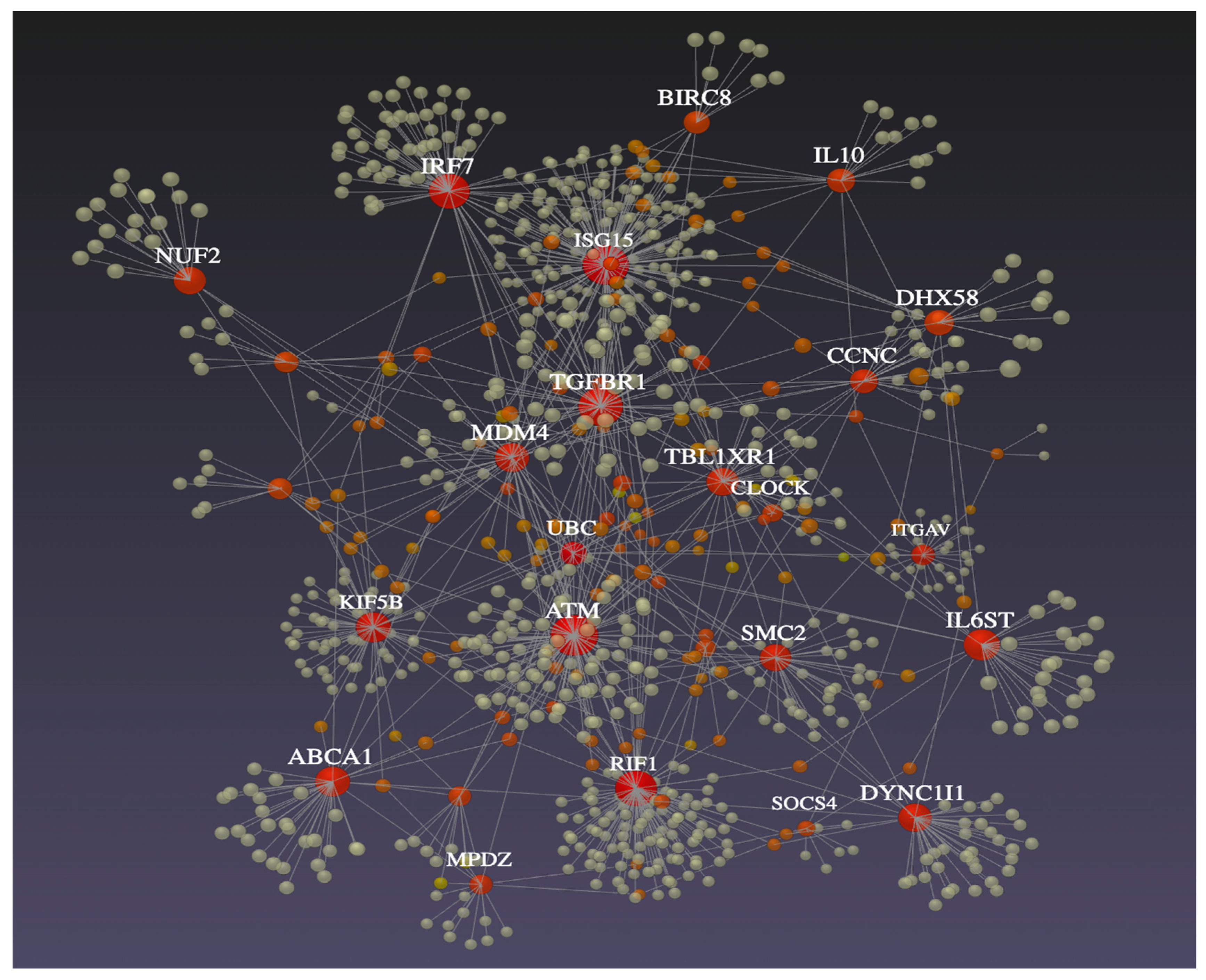
3.5. Network Analyses for DEGs
3.6. RNA-Seq Validation
4. Discussion
4.1. Apoptosis and Cell Signaling
4.2. Immune Response
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rajaud, A.; Noblet-Ducoudré, N. Tropical semi-arid regions expanding over temperate latitudes under climate change. Clim. Chang. 2017, 144, 703–719. [Google Scholar] [CrossRef]
- Henry, B.; Eckard, R.; Beauchemin, K. Adaptation of ruminant livestock production systems to climate changes. Animal 2018, 12, s445–s456. [Google Scholar] [CrossRef]
- Dikmen, S.; Wang, X.Z.; Ortega, M.S.; Cole, J.B.; Null, D.J.; Hansen, P.J. Single nucleotide polymorphisms associated with thermoregulation in lactating dairy cows exposed to heat stress. J. Anim. Breed. Genet. 2015, 132, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Sarlo Davila, K.M.; Hamblen, H.; Hansen, P.J.; Dikmen, S.; Oltenacu, P.A.; Mateescu, R.G. Genetic parameters for hair characteristics and core body temperature in a multibreed Brahman-Angus herd1. J. Anim. Sci. 2019, 97, 3246–3252. [Google Scholar] [CrossRef] [PubMed]
- Sejian, V.; Bhatta, R.; Gaughan, J.B.; Dunshea, F.R.; Lacetra, N. Review: Adaptation of animals to heat stress. Animal 2018, 12, 431–444. [Google Scholar] [CrossRef]
- Joy, A.; Dunshea, F.R.; Leury, B.J.; Clarke, I.J.; DiGiacomo, K.; Chauhan, S.S. Resilience of small ruminants to climate change and increased environmental temperature. A review. Animal 2020, 10, 867. [Google Scholar] [CrossRef]
- Carabaño, M.J.; Ramón, M.; Menéndez-Buxadera, A.; Molina, A.; Díaz, C. Selecting for heat tolerance. Anim. Front. 2019, 9, 62–68. [Google Scholar] [CrossRef]
- Liu, Z.; Ji, Z.; Wang, G.; Chao, T.; Hou, L.; Wang, J. Genome-wide analysis reveals signatures of selection for important traits in domestic sheep from different ecoregions. BMC Genom. 2016, 17, 863. [Google Scholar] [CrossRef]
- Luna-Nevarez, G.; Kelly, A.C.; Camacho, L.E.; Limesand, S.W.; Reyna-Granados, J.R.; Luna-Nevarez, P. Discovery and validation of candidate SNP markers associated to heat stress response in pregnant ewes managed inside a climate-controlled chamber. Trop. Anim. Health Prod. 2020, 52, 3457–3466. [Google Scholar] [CrossRef]
- Castillo-Salas, C.A.; Luna-Nevárez, G.; Reyna-Granados, J.R.; Luna-Ramirez, R.I.; Limesand, S.W.; Luna-Nevárez, P. Molecular markers for thermo-tolerance are associated with reproductive and physiological traits in Pelibuey ewes raised in a semiarid environment. J. Therm. Biol. 2023, 112, 103475. [Google Scholar] [CrossRef]
- Li, Y.X.; Feng, X.P.; Wang, H.L.; Meng, C.H.; Zhang, J.; Qian, Y.; Zhong, J.F.; Cao, S.X. Transcriptome analysis reveals corresponding genes and key pathways involved in heat stress in Hu sheep. Cell Stress Chaperones 2019, 24, 1045–1054. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kong, L.; Deng, M.; Lian, Z.; Han, Y.; Sun, B.; Guo, Y.; Liu, G.; Liu, D. Heat stress-responsive transcriptome analysis in the liver tissue of Hu sheep. Genes 2019, 100, 395. [Google Scholar] [CrossRef]
- Lu, Z.; Chu, M.; Li, Q.; Jin, M.; Fei, X.; Ma, L.; Zhang, L.; Wei, C. Transcriptomic analysis provides novel insights into heat stress responses in sheep. Animals 2019, 9, 387. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Yang, Y.L.; Li, G.Q.; Yu, Q.; Yang, J. Identifications of immune-responsive genes for adaptative traits by comparative transcriptome analysis of spleen tissue from Kazakh and Suffolk sheep. Sci. Rep. 2021, 11, 3157. [Google Scholar] [CrossRef] [PubMed]
- Haire, A.; Bai, J.; Zhao, X.; Song, Y.; Zhao, G.; Dilixiati, A.; Li, J.; Sun, W.; Wan, P.; Fu, X.; et al. Identifying the heat resistant genes by multi-tissue transcriptome sequencing analysis in Turpan Black sheep. Theriogenology 2022, 179, 78–86. [Google Scholar] [CrossRef]
- Jiao, D.; Ji, K.; Wang, W.; Liu, H.; Zhou, J.; Degen, A.A.; Zhang, Y.; Zhou, P.; Yang, G. Transcriptome profiles of the liver in two cold-exposed sheep breeds revealed different mechanisms and candidate genes for thermogenesis. Genet. Res. 2021, 10, 5510297. [Google Scholar] [CrossRef] [PubMed]
- Ji, K.; Jiao, D.; Yang, G.; Degen, A.A.; Zhou, J.; Liu, H.; Wang, W.; Cong, H. Transcriptome analysis revealed potential genes involved in thermogenesis in muscle tissue in cold-exposed lambs. Front. Genet. 2022, 13, 1017458. [Google Scholar] [CrossRef]
- Sohn, E. Diagnosis: Frontiers in blood testing. Nature 2017, 549, S16. [Google Scholar] [CrossRef]
- Schmidt, M.; Hopp, L.; Arakelyan, A.; Kirsten, H.; Engel, C.; Wirkner, K.; Krohn, K.; Burkhardt, R.; Thiery, J.; Loeffler, M.; et al. The human blood transcriptome in a large population cohort and its relation to aging and health. Front. Big Data 2020, 3, 548873. [Google Scholar] [CrossRef]
- Hanash, S.M.; Baik, C.S.; Kallioniemi, O. Emerging molecular biomarkers—Blood-based strategies to detect and monitor cancer. Nat. Rev. Clin. Oncol. 2011, 8, 142–150. [Google Scholar] [CrossRef]
- Baird, A.E.; Soper, S.A.; Pullagurla, S.R.; Adamski, M.G. Recent and near-future advances in nucleic acid-based diagnosis of stroke. Expert Rev. Mol. Diagn. 2015, 15, 665–679. [Google Scholar]
- Chaussabel, D. Assessment of immune status using blood transcriptomics and potential implications for global health. Semin. Immunol. 2015, 27, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Johannsen, N.M.; Priest, E.L.; Dixit, V.D.; Earnest, C.P.; Blair, S.N.; Church, T.S. Association of white blood cell subfraction concentration with fitness and fatness. Br. J. Sports Med. 2010, 44, 588–593. [Google Scholar] [CrossRef] [PubMed]
- Burton, K.J.; Pimentel, G.; Zangger, N.; Vionnet, N.; Drai, J.; McTernan, P.G. Modulation of the peripheral blood transcriptome by the ingestion of probiotic yoghurt and acidified milk in healthy, young men. PLoS ONE 2018, 13, e0192947. [Google Scholar] [CrossRef]
- Pendleton, A.L.; Antolic, A.T.; Kelly, A.C.; Davis, M.A.; Camacho, L.E.; Doubleday, K.; Anderson, M.J.; Langlais, P.R.; Lynch, R.M.; Limesand, S.W. Lower oxygen consumption and Complex I activity in mitochondria isolated from skeletal muscle of fetal sheep with intrauterine growth restriction. Am. J. Physiol. Endocrinol. Metab. 2020, 319, E67–E80. [Google Scholar] [CrossRef]
- Chen, X.; Fahy, A.L.; Green, A.; Anderson, M.J.; Rhoads, R.P.; Limesand, S.W. β2-adrenergic receptor desensitization in perirenal adipose tissue in fetuses and lambs with placental insufficiency-induced intrauterine growth restriction. J. Physiol. 2010, 588, 3539–3549. [Google Scholar] [CrossRef]
- Macko, A.R.; Yates, D.T.; Chen, X.; Green, A.S.; Kelly, A.C.; Brown, L.D.; Limesand, S.W. Elevated plasma norepinephrine inhibits insulin secretion, but adrenergic blockade reveals enhanced β-cell responsiveness in an ovine model of placental insufficiency at 0.7 of gestation. J. Dev. Orig. Health Dis. 2013, 4, 402–410. [Google Scholar] [CrossRef]
- Yates, D.T.; Camacho, L.E.; Kelly, A.C.; Steyn, L.V.; Davis, M.A.; Antolic, A.T.; Anderson, M.J.; Goyal, R.; Allen, R.E.; Papas, K.K.; et al. Postnatal β2 adrenergic treatment improves insulin sensitivity in lambs with IUGR but not persistent defects in pancreatic islets or skeletal muscle. J. Physiol. 2019, 597, 5835–5858. [Google Scholar] [CrossRef]
- Pendleton, A.L.; Humphreys, L.R.; Davis, M.A.; Camacho, L.E.; Anderson, M.J.; Limesand, S.W. Increased pyruvate dehydrogenase activity in skeletal muscle of growth-restricted ovine fetuses. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2019, 317, R513–R520. [Google Scholar] [CrossRef]
- MacKay, H.; Scott, C.A.; Duryea, J.D. DNA methylation in AgRP neurons regulates voluntary exercise behavior in mice. Nat. Commun. 2019, 10, 5364. [Google Scholar] [CrossRef]
- Park, W.; Srikanth, K.; Lim, D.; Park, M.; Hur, T.; Kemp, S.; Dessie, T.; Kim, M.S.; Lee, S.R.; Te Pas, M.F.W.; et al. Comparative transcriptome analysis of Ethiopian indigenous chickens from low and high altitudes under heat stress condition reveals differential immune response. Anim. Genet. 2019, 50, 42–53. [Google Scholar] [CrossRef]
- Klohonatz, K.M.; Coleman, S.J.; Cameron, A.D.; Hess, A.M.; Reed, K.J.; Canovas, A.; Medrano, J.F.; Islas-Trejo, A.D.; Kalbfleisch, T.; Bouma, G.J.; et al. Non-coding RNA sequencing of equine endometrium during maternal recognition of pregnancy. Genes 2019, 10, 821. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Benner, M.J.; Hancock, R.E. NetworkAnalyst—Integrative approaches for protein-protein interaction network analysis and visual exploration. Nucleic Acids Res. 2014, 42, W167–W174. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Soufan, O.; Ewald, J.; Hancock, R.E.W.; Basu, N.; Xia, J. NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019, 47, W234–W241. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Tang, S.; Li, M.; Sun, Y.; Liao, Y.; Wu, X.; Zhong, R.; Chen, L.; Zhang, H. Effects of chronic heat stress on the immunophenotyping of lymphocytes in immune organs of growing pigs. J. Anim. Sci. 2022, 100, skac317. [Google Scholar] [CrossRef]
- Hoekstra, S.P.; Wright, A.K.A.; Bishop, N.C.; Leicht, C.A. The effect of temperature and heat shock protein 72 on the ex vivo acute inflammatory response in monocytes. Cell Stress Chaperones 2019, 4, 461–467. [Google Scholar] [CrossRef]
- Honda, B.T.; Calefi, A.S.; Costola-de-Souza, C.; Quinteiro-Filho, W.M.; da Silva Fonseca, J.G.; de Paula, V.F.; Palermo-Neto, J. Effects of heat stress on peripheral T and B lymphocyte profiles and IgG and IgM serum levels in broiler chickens vaccinated for Newcastle disease virus. Poult. Sci. 2015, 94, 2375–2381. [Google Scholar] [CrossRef]
- Matsuki, S.; Iuchi, Y.; Ikeda, Y.; Sasagawa, I.; Tomita, Y.; Fujii, J. Suppression of cytochrome c release and apoptosis in testes with heat stress by minocycline. Biochem. Biophys. Res. Commun. 2003, 312, 843–849. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.T.; Wang, H.; Li, L.; Liu, Y.S.; Deng, X.B.; Huo, S.F.; Yuan, F.F.; Liu, Z.F.; Tong, H.S.; Su, L. Heat stress induces apoptosis through transcription-independent p53-mediated mitochondrial pathways in human umbilical vein endothelial cell. Sci. Rep. 2014, 26, 4469. [Google Scholar] [CrossRef]
- Lee, S.J.; Yang, E.S.; Kim, S.Y.; Kim, S.Y.; Shin, S.W.; Park, J.W. Regulation of heat shock-induced apoptosis by sensitive to apoptosis gene protein. Free Radic. Biol. Med. 2008, 45, 167–176. [Google Scholar] [CrossRef]
- Gong, L.; Zhang, Q.; Pan, X.; Chen, S.; Yang, L.; Liu, B.; Yang, W.; Yu, L.; Xiao, Z.X.; Feng, X.H.; et al. p53 protects cells from death at the heatstroke threshold temperature. Cell Rep. 2019, 29, 3693–3707. [Google Scholar] [CrossRef] [PubMed]
- Shaltiel, I.A.; Krenning, L.; Bruinsma, W.; Medema, R.H. The same, only different—DNA damage checkpoints and their reversal throughout the cell cycle. J. Cell Sci. 2015, 128, 607–620. [Google Scholar] [CrossRef] [PubMed]
- Dikomey, E.; Franzke, J. Effect of heat on induction and repair of DNA strand breaks in X-irradiated CHO cells. Int. J. Radiat. Biol. 1992, 61, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Phan, L.M.; Rezaeian, A.H. ATM: Main features, signaling pathways, and its diverse roles in dna damage response, tumor suppression, and cancer development. Genes 2021, 12, 845. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Huang, T.; Yu, Z.; Yu, Q.; Wang, Y.; Hu, J.; Shi, J.; Yang, G. The functions and roles of sestrins in regulating human diseases. Cell. Mol. Biol. Lett. 2022, 27, 2. [Google Scholar] [CrossRef]
- Rai, N.; Dey, S. Protective response of Sestrin under stressful conditions in aging. Ageing Res. Rev. 2020, 64, 101186. [Google Scholar] [CrossRef]
- Huang, M.; Kim, H.G.; Zhong, X.; Dong, C.; Zhang, B.; Fang, Z.; Zhang, Y.; Lu, X.; Saxena, R.; Liu, Y.; et al. Sestrin 3 protects against diet-induced nonalcoholic steatohepatitis in mice through suppression of transforming growth factor β signal transduction. Hepatology 2020, 71, 76–92. [Google Scholar] [CrossRef]
- Matos-Perdomo, E.; Machín, F. TORC1, stress and the nucleolus. Aging 2018, 10, 857–858. [Google Scholar] [CrossRef]
- Guo, Z.; Gao, S.; Ouyang, J.; Ma, L.; Bu, D. Impacts of heat stress-induced oxidative stress on the milk protein biosynthesis of dairy cows. Animals 2021, 11, 726. [Google Scholar] [CrossRef]
- Gonzalez, S.; Rallis, C. The TOR signaling pathway in spatial and temporal control of cell size and growth. Front. Cell. Dev. Biol. 2017, 5, 61. [Google Scholar] [CrossRef]
- Yue, S.; Wang, Z.; Wang, L.; Peng, Q.; Xue, B. Transcriptome Functional Analysis of Mammary Gland of Cows in Heat Stress and Thermoneutral Condition. Animals 2020, 10, 1015. [Google Scholar] [CrossRef]
- Di Conza, G.; Mancini, F.; Buttarelli, M.; Pontecorvi, A.; Trimarchi, F.; Moretti, F. MDM4 enhances p53 stability by promoting an active conformation of the protein upon DNA damage. Cell Cycle 2012, 11, 670–749. [Google Scholar] [CrossRef][Green Version]
- Hu, W.; Feng, Z.; Levine, A.J. The regulation of multiple p53 stress responses is mediated through MDM2. Genes Cancer 2012, 3, 199–208. [Google Scholar] [CrossRef]
- Sammad, A.; Luo, H.; Hu, L.; Zhu, H.; Wang, Y. Transcriptome Reveals Granulosa Cells Coping through Redox, Inflammatory and Metabolic Mechanisms under Acute Heat Stress. Cells 2022, 11, 1443. [Google Scholar] [CrossRef]
- Zhang, X.; Tang, N.; Hadden, T.J.; Rishi, A.K. Akt, FoxO and regulation of apoptosis. Biochim. Biophys. Acta 2011, 1813, 1978–1986. [Google Scholar] [CrossRef]
- Yoshihara, T.; Kobayashi, H.; Kakigi, R.; Sugiura, T.; Naito, H. Heat stress-induced phosphorylation of FoxO3a signalling in rat skeletal muscle. Acta Physiol. 2016, 218, 178–187. [Google Scholar] [CrossRef]
- Zhang, Y.; Alexander, P.B.; Wang, X.F. TGF-β family signaling in the control of cell proliferation and survival. Cold Spring Harb. Perspect. Biol. 2017, 9, a022145. [Google Scholar] [CrossRef]
- Dooley, S.; Ten Dijke, P. TGF-β in progression of liver disease. Cell Tissue Res. 2012, 347, 245–256. [Google Scholar] [CrossRef]
- Saadeldin, I.M.; Abdel-Aziz Swelum, A.; Zakri, A.M.; Tukur, H.A.; Allowaimer, A.N. Effects of acute hyperthermia on the thermotolerance of cow and sheep skin-derived fibroblasts. Animals 2020, 10, 545. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Fernandez, M.R.; Ochoa-Ibarrola, L.; Quintana-Gallardo, L.; Valuesta, J.M. HSP70—A master regulator in protein degradation. FEBS Lett. 2017, 591, 2648–2660. [Google Scholar] [CrossRef] [PubMed]
- Amaral, C.D.S.; Correa, G.R.E.; Serrano Mujica, L.K.; Fiorenza, M.F.; Rosa, S.G.; Nogueira, C.W.; Portela, V.M.; Comim, F.V.; Schoenau, W.; Smirnova, N.P.; et al. Heat stress modulates polymorphonuclear cell response in early pregnancy cows: I. interferon pathway and oxidative stress. PLoS ONE 2021, 16, e0257418. [Google Scholar] [CrossRef] [PubMed]
- Rehwinkel, J.; Gack, M.U. RIG-I-like receptors: Their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020, 20, 537–551. [Google Scholar] [CrossRef]
- Albert, M.; Bécares, M.; Falqui, M.; Fernández-Lozano, C.; Guerra, S. ISG15, a small molecule with huge implications: Regulation of mitochondrial homeostasis. Viruses 2018, 10, 629. [Google Scholar] [CrossRef]
- Zhang, M.; Li, J.; Yan, H.; Huang, J.; Wang, F.; Liu, T.; Zeng, L.; Zhou, F. ISGylation in innate antiviral immunity and pathogen defense responses: A review. Front. Cell. Dev. Biol. 2021, 9, 788410. [Google Scholar] [CrossRef]
- Dos Santos, P.F.; Mansur, D.S. Beyond ISGlylation: Functions of free intracellular and extracellular ISG15. J. Interferon Cytokine Res. 2017, 37, 246–253. [Google Scholar] [CrossRef]
- Munnur, D.; Banducci-Karp, A.; Sanyal, S. ISG15 driven cellular responses to virus infection. Biochem. Soc. Trans. 2022, 50, 1837–1846. [Google Scholar] [CrossRef]
- Sulistio, Y.A.; Heese, K. The Ubiquitin-Proteasome System and molecular chaperone deregulation in Alzheimer’s disease. Mol. Neurobiol. 2016, 53, 905–931. [Google Scholar] [CrossRef]
- Lee, K.J.; Lee, H.; Joo, C.H. Negative regulation of IKKε-mediated IRF7 phosphorylation by HSP70. J. Immunol. 2020, 204, 2562–2574. [Google Scholar] [CrossRef]
- Liang, Q.; Deng, H.; Sun, C.W.; Townes, T.M.; Zhu, F. Negative regulation of IRF7 activation by activating transcription factor 4 suggests a cross-regulation between the IFN responses and the cellular integrated stress responses. J. Immunol. 2011, 186, 1001–1010. [Google Scholar] [CrossRef] [PubMed]
- Palacios, C.; Abecia, J.A. Meteorological variables affect fertility rate after intrauterine artificial insemination in sheep in a seasonal-dependent manner: A 7-year study. Int. J. Biometeorol. 2015, 59, 585–592. [Google Scholar] [CrossRef] [PubMed]
- van Wettere, W.H.E.J.; Kind, K.L.; Gatford, K.L.; Swinbourne, A.M.; Leu, S.T.; Hayman, P.T.; Kelly, J.M.; Weaver, A.C.; Kleemann, D.O.; Walker, S.K. Review of the impact of heat stress on reproductive performance of sheep. J. Anim. Sci. Biotechnol. 2021, 12, 26. [Google Scholar] [CrossRef] [PubMed]
- Limesand, S.W.; Camacho, L.E.; Kelly, A.C.; Antolic, A.T. Impact of thermal stress on placental function and fetal physiology. Anim. Reprod. 2018, 15 (Suppl. 1), 886–898. [Google Scholar] [CrossRef]
- Luna-Nevárez, G.; Pendleton, A.L.; Luna-Ramirez, R.I.; Limesand, S.W.; Reyna-Granados, J.R.; Luna-Nevárez, P. Genome-wide association study of a thermo-tolerance indicator in pregnant ewes exposed to an artificial heat-stressed environment. J. Therm. Biol. 2021, 101, 103095. [Google Scholar] [CrossRef]
- Hori, T.S.; Gamperl, A.K.; Booman, M. A moderate increase in ambient temperature modulates the Atlantic cod (Gadus morhua) spleen transcriptome response to intraperitoneal viral mimic injection. BMC Genomics 2012, 13, 431. [Google Scholar] [CrossRef]
- Suen, W.W.; Imoda, M.; Thomas, A.W.; Nasir, N.N.B.M.; Tearnsing, N.; Wang, W.; Bielefeldt-Ohmann, H. An acute stress model in New Zealand white rabbits exhibits altered immune response to infection with West Nile virus. Pathogens 2019, 8, 195. [Google Scholar] [CrossRef]


| Heat Tolerant (HT) Ewes | Non-Heat Tolerant (NHT) Ewes | ||||||
|---|---|---|---|---|---|---|---|
| ID 1 | β1 2 | R2 3 | p-value 4 | ID 1 | β1 2 | R2 3 | p-Value 4 |
| Y18 | 0.8902 | 0.5922 | <0.0001 | 5 | −0.8194 | 0.4095 | 0.0216 |
| A44 | 0.7759 | 0.5012 | 0.0002 | 844 | −0.5273 | 0.5476 | 0.0002 |
| R40 | 0.4918 | 0.3595 | 0.0485 | 831 | −0.4058 | 0.5569 | <0.0001 |
| 845 | 0.3710 | 0.5712 | 0.0005 | 1 | −0.3939 | 0.4340 | 0.0065 |
| 810 | 0.3606 | 0.4281 | 0.0092 | 826 | −0.3595 | 0.5227 | 0.0004 |
| 815 | 0.3160 | 0.4931 | 0.0013 | 816 | −0.3136 | 0.4816 | 0.0018 |
| 858 | 0.2291 | 0.4911 | 0.0002 | 822 | −0.2857 | 0.5670 | 0.0003 |
| 817 | 0.1522 | 0.4576 | 0.0076 | 819 | −0.1066 | 0.4133 | 0.0314 |
| Heat Stressed Groups | ||||
|---|---|---|---|---|
| Blood Trait 1 | Normal Range 2 | HT 3 | NHT 4 | p-Value 5 |
| WBC | 4.0–12.0 × 103/μL | 6.04 a | 7.81 b | 0.024 |
| RBC | 9.0–15.0 × 106/μL | 11.16 a | 9.65 b | 0.021 |
| HGB | 9.0–18.0 g/dL | 11.18 | 10.64 | 0.632 |
| HCT | 27.0–45.0% | 35.38 | 32.73 | 0.131 |
| MCV | 28–40 fL | 33.10 | 32.30 | 0.489 |
| MCH | 8.0–12.0 pg | 10.49 | 10.65 | 0.642 |
| MCHC | 31–34 g/dL | 31.68 | 32.74 | 0.391 |
| Platelets | 240–700 × 103/μL | 511.75 | 504.63 | 0.920 |
| Neutrophils | 700–6000/μL | 2890.75 | 2742.25 | 0.819 |
| Lymphocytes | 2000–10,000/μL | 2615.75 a | 4388.35 b | <0.001 |
| Monocytes | 100–300/μL | 134.21 a | 203.63 b | 0.026 |
| Eosinophil | 0–1000/μL | 200.38 | 278.88 | 0.099 |
| Basophils | 0/μL | 22.88 | 3.75 | 0.187 |
| Gene Ontology (GO) Term 1 | GO Group 2 | No. DEGs 3 | p-Value 4 |
|---|---|---|---|
| Regulation of TORC1 signaling | BP | 15 | 5.3 × 10−4 |
| Regulation of cell communication | BP | 15 | 6.4 × 10−4 |
| Type I interferon production | BP | 11 | 6.6 × 10−4 |
| Regulation of signal transduction | BP | 14 | 6.9 × 10−4 |
| Regulation of signaling | BP | 12 | 1.7 × 10−3 |
| Regulation of immune system process | BP | 10 | 2.9 × 10−3 |
| Lipoprotein metabolic process | BP | 11 | 3.7 × 10−3 |
| Regulation of cytokine production | BP | 10 | 4.2 × 10−3 |
| Cellular metabolic process | BP | 10 | 6.4 × 10−3 |
| Regulation of response to stress | BP | 9 | 1.7 × 10−2 |
| Bicellular tight junction | CC | 3 | 1.8 × 10−2 |
| External side of plasma membrane | CC | 3 | 4.6 × 10−2 |
| ATP binding | MF | 10 | 4.1 × 10−4 |
| Cytokine binding | MF | 10 | 1.5 × 10−3 |
| KEGG Pathway 1 | Main Genes 2 | p-Value 3 | Corrected p-Value 4 |
|---|---|---|---|
| p53 signaling pathway | ATM, SESN3, MDM4 | 2.7 × 10−4 | 1.8 × 10−3 |
| RIG-I-like receptor signaling pathway | ISG15, IRF7, DXH58 | 7.4 × 10−4 | 2.5 × 10−3 |
| FoxO signaling pathway | TGFBR1, ATM, IL10, IL6ST | 1.1 × 10−3 | 4.3 × 10−3 |
| Dopaminergic synapse | KIF5B, CAMK2B, CLOCK | 2.8 × 10−3 | 7.6 × 10−2 |
| Jak-STAT signaling pathway | IL10, SOCS4, IL6ST | 5.1 × 10−3 | 9.7 × 10−2 |
| Viral protein interaction with cytokine | IL10, TGFBR1, IL6ST | 8.7 × 10−3 | 1.2 × 10−1 |
| Th17 cell differentiation | TGFBR1, IL6ST | 1.3 × 10−2 | 1.3 × 10−1 |
| Cytokine–cytokine receptor interaction | IL10, TGFBR1, IL6ST | 1.6 × 10−2 | 1.4 × 10−1 |
| Wnt signaling pathway | TBL1XR1, CAMK2B | 2.5 × 10−2 | 1.7 × 10−1 |
| Cellular senescence | TGFBR1, ATM | 2.6 × 10−2 | 1.8 × 10−1 |
| Circadian rhythm | CLOCK1 | 3.4 × 10−2 | 1.9 × 10−1 |
| Gene Symbol | No. Connec. 1 | Gene Symbol | No. Interac. 2 |
|---|---|---|---|
| ATM | 5 | ISG15 | 188 |
| SMC2 | 5 | RIF1 | 120 |
| ISG15 | 4 | ATM | 117 |
| IRF7 | 3 | KIF5B | 73 |
| MDM4 | 3 | IRF7 | 66 |
| CDK14 | 3 | TGFBR1 | 65 |
| TBL1XR1 | 3 | MDM4 | 48 |
| LIN7C | 3 | DYNCIII | 41 |
| DHX58 | 2 | ABCA1 | 40 |
| TGFBR1 | 2 | DHX58 | 37 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luna-Ramirez, R.I.; Limesand, S.W.; Goyal, R.; Pendleton, A.L.; Rincón, G.; Zeng, X.; Luna-Nevárez, G.; Reyna-Granados, J.R.; Luna-Nevárez, P. Blood Transcriptomic Analyses Reveal Functional Pathways Associated with Thermotolerance in Pregnant Ewes Exposed to Environmental Heat Stress. Genes 2023, 14, 1590. https://doi.org/10.3390/genes14081590
Luna-Ramirez RI, Limesand SW, Goyal R, Pendleton AL, Rincón G, Zeng X, Luna-Nevárez G, Reyna-Granados JR, Luna-Nevárez P. Blood Transcriptomic Analyses Reveal Functional Pathways Associated with Thermotolerance in Pregnant Ewes Exposed to Environmental Heat Stress. Genes. 2023; 14(8):1590. https://doi.org/10.3390/genes14081590
Chicago/Turabian StyleLuna-Ramirez, Rosa I., Sean W. Limesand, Ravi Goyal, Alexander L. Pendleton, Gonzalo Rincón, Xi Zeng, Guillermo Luna-Nevárez, Javier R. Reyna-Granados, and Pablo Luna-Nevárez. 2023. "Blood Transcriptomic Analyses Reveal Functional Pathways Associated with Thermotolerance in Pregnant Ewes Exposed to Environmental Heat Stress" Genes 14, no. 8: 1590. https://doi.org/10.3390/genes14081590
APA StyleLuna-Ramirez, R. I., Limesand, S. W., Goyal, R., Pendleton, A. L., Rincón, G., Zeng, X., Luna-Nevárez, G., Reyna-Granados, J. R., & Luna-Nevárez, P. (2023). Blood Transcriptomic Analyses Reveal Functional Pathways Associated with Thermotolerance in Pregnant Ewes Exposed to Environmental Heat Stress. Genes, 14(8), 1590. https://doi.org/10.3390/genes14081590





