Application of High-Resolution Melting and DNA Barcoding for Discrimination and Taxonomy Definition of Rocket Salad (Diplotaxis spp.) Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. DNA Barcode Primer Design
2.3. Amplification and Sequencing
2.4. High-Resolution Melting Analysis
2.5. Phylogenetic Tree Data Analysis
2.6. Genetic Diversity and Population Structure
3. Results
3.1. Sequence Comparisons
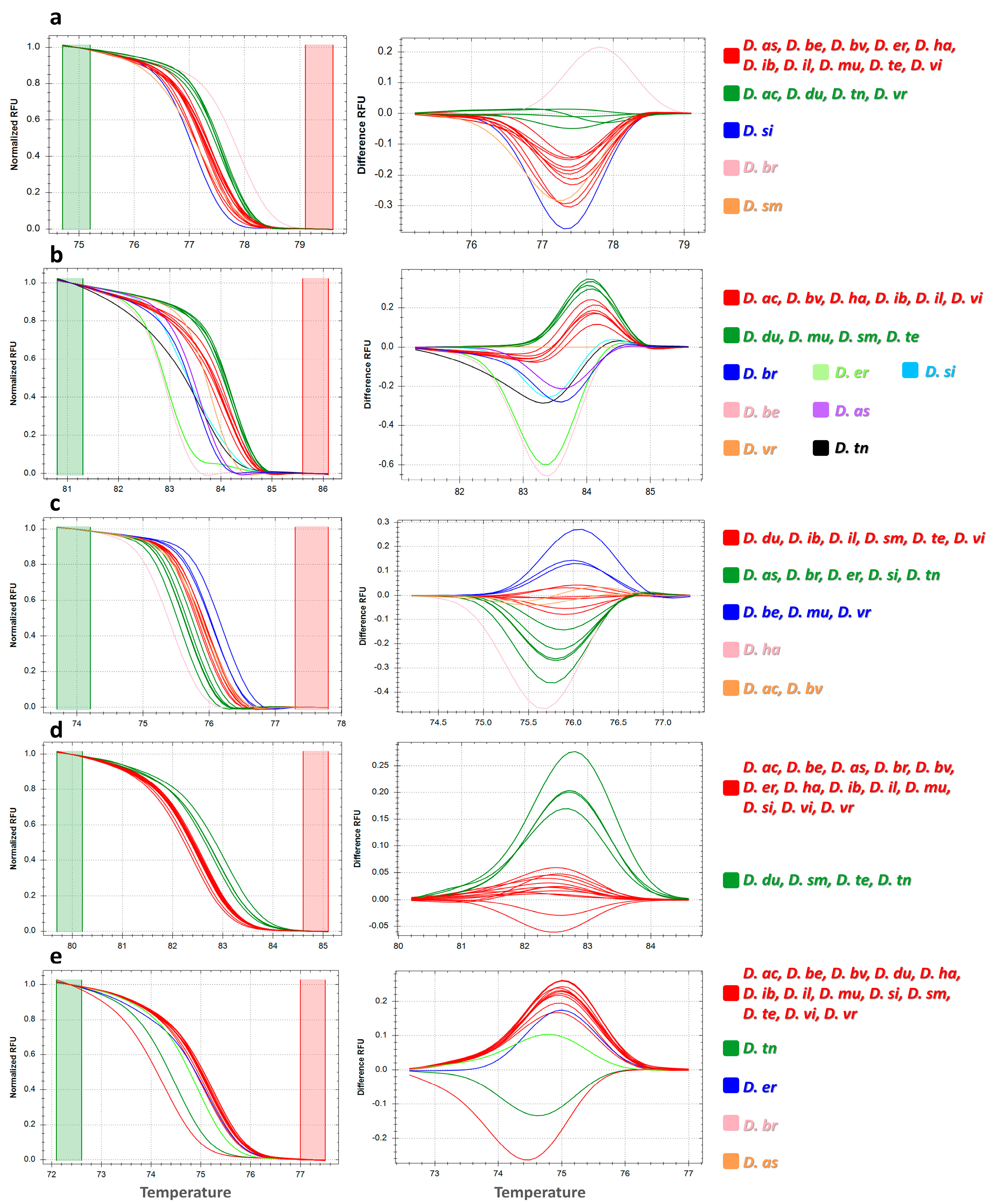
3.2. High-Resolution Melting Profiles
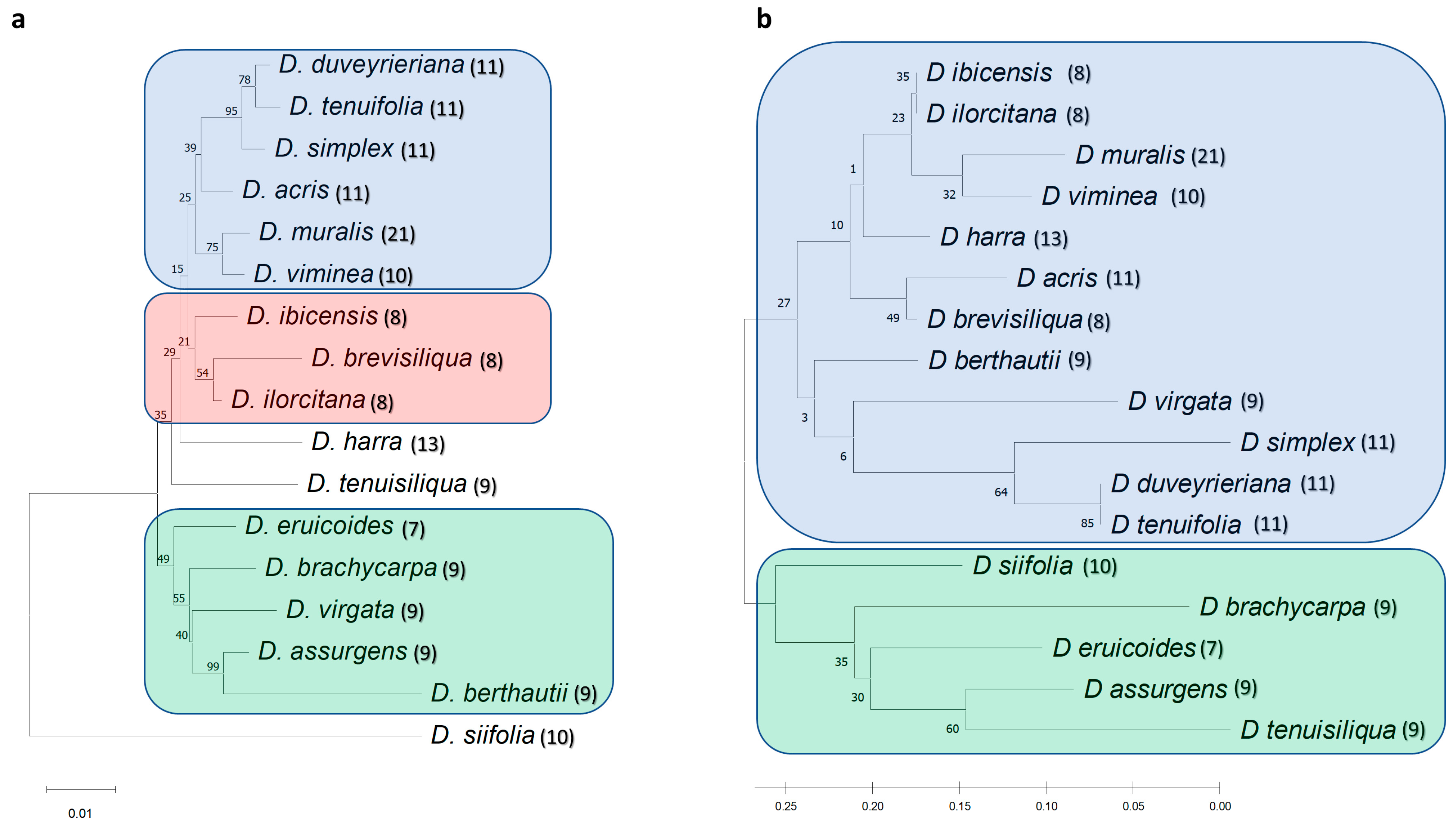
3.3. Phylogenetic Analysis
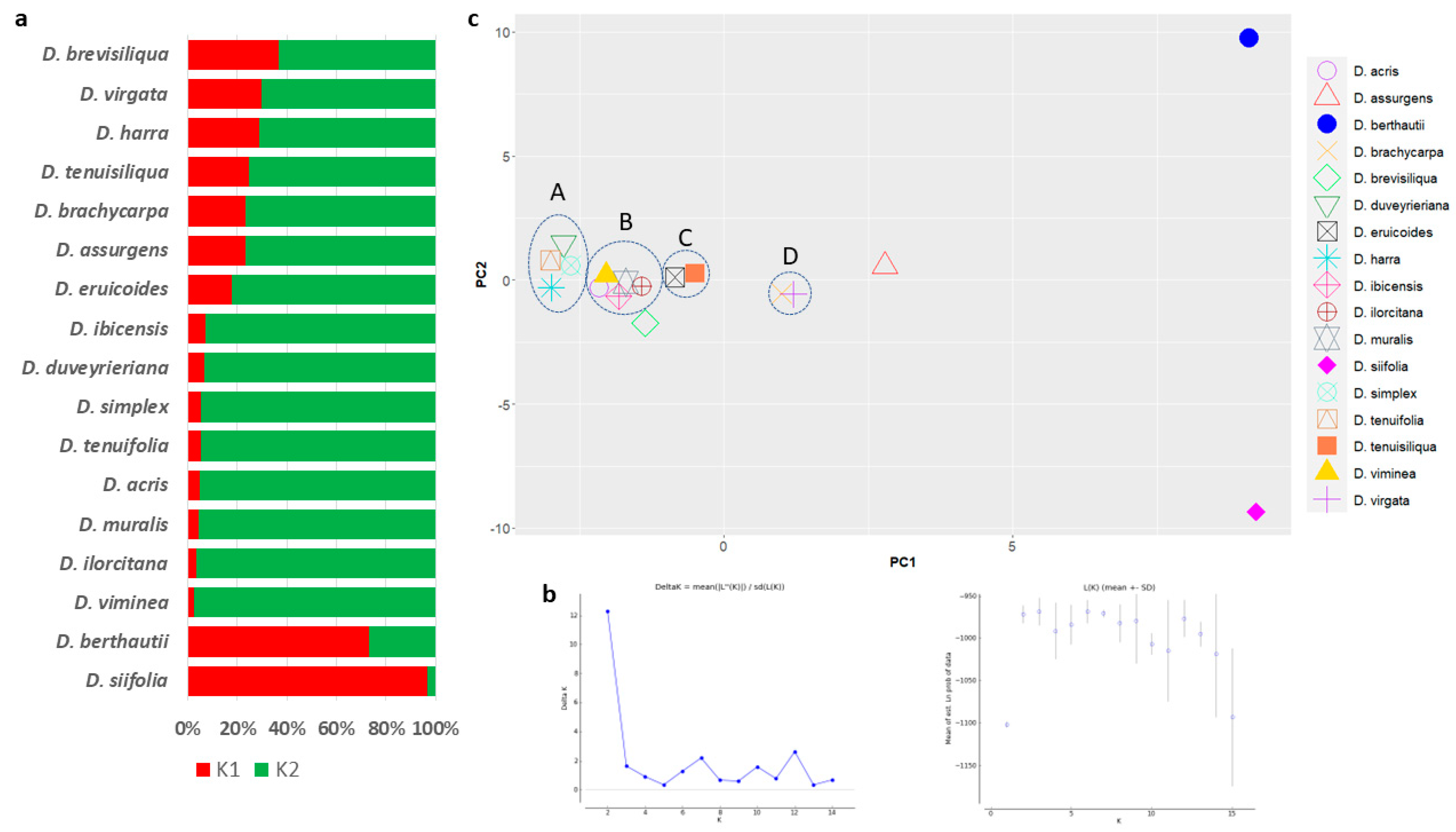
3.4. Population Structure
4. Discussion
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Warwick, S.I. Brassicaceae in Agriculture. In Genetics and Genomics of the Brassicaceae; Warwick, S.I., Ed.; Springer: New York, NY, USA, 2010; pp. 33–65. [Google Scholar]
- Tripodi, P.; Coelho, P.S.; Guijarro-Real, C. Breeding advances and prospects in rocket salad (Eruca vesicaria ssp. sativa Mill.) cultivation. In Advances in Plant Breeding Strategies: Vegetable Crops; Springer International Publishing: Cham, Switzerland, 2021; pp. 95–133. [Google Scholar]
- Gómez-Campo, C. Taxonomy. In Biology of Brassica Coenospecies; Elsevier: Amsterdam, The Netherlands, 1999; pp. 3–32. [Google Scholar]
- Martín, J.P.; Sánchez-Yélamo, M.D. Genetic relationships among species of the genus Diplotaxis (Brassicaceae) using inter-simple sequence repeat markers. Theor. Appl. Genet. 2000, 101, 1234–1241. [Google Scholar] [CrossRef]
- Harberd, D.J.; McArthur, E.D. The chromosome constitution of Diplotaxis muralis (L.) DC. Watsonia 1972, 9, 131–135. [Google Scholar]
- Tripodi, P.; Francese, G.; Mennella, G. Rocket Salad: Crop Description, Bioactive Compounds and Breeding Perspectives. Adv. Hortic. Sci. 2017, 31, 107–113. [Google Scholar]
- Gómez-Campo, C. Morphology and morphotaxonomy of the Tribe Brassiceae. In Brassica Crops and Wild Allies; Tsunoda, S., Hinata, K., Gómez-Campo, C., Eds.; Japan Scientific Societies Press: Tokyo, Japan, 1980; pp. 3–31. [Google Scholar]
- Gómez-Campo, C.; Martínez-Laborde, J.B. Reajustes taxonómicos y nomenclaturales en la tribu Brassiceae (Cruciferae). An. Jardín Bot. Madrid 1998, 56, 379–381. [Google Scholar]
- D’antuono, L.F.; Elementi, S.; Neri, R. Glucosinolates in Diplotaxis and Eruca leaves: Diversity, taxonomic relations and applied aspects. Phytochemistry 2008, 69, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Yélamo, M.D. A chemosystematic survey of flavonoids in the Brassicinae: Diplotaxis. Bot. J. Linn. Soc. 1994, 115, 9–18. [Google Scholar] [CrossRef]
- Eschmann-Grupe, G.; Hurka, H.; Neuffer, B. Species relationships within Diplotaxis (Brassicaceae) and the phylogenetic origin of D. muralis. Plant. Syst. Evol. 2004, 243, 13–29. [Google Scholar] [CrossRef]
- Warwick, S.I.; Black, L.D.; Aguinagalde, I. Molecular systematics of Brassica and allied genera (subtribe Brassicinae, Brassiceae)—Chloroplast DNA variation in the genus Diplotaxis. Theorl Appl. Genet. 1992, 83, 839–850. [Google Scholar] [CrossRef]
- Warwick, S.I.; Sauder, C.A. Phylogeny of tribe Brassiceae (Brassicaceae) based on chloroplast restriction site polymorphisms and nuclear ribosomal internal transcribed spacer and chloroplast trnL intron sequences. Can. J. Bot. 2005, 83, 467–483. [Google Scholar]
- Yan, M.; Xiong, Y.; Liu, R.; Deng, M.; Song, J. The application and limitation of universal chloroplast markers in discriminating east Asian evergreen oaks. Front. Plant. Sci. 2018, 9, 569. [Google Scholar] [CrossRef]
- Hollingsworth, P.M.; Graham, S.W.; Little, D.P. Choosing and Using a Plant DNA Barcode. PLoS ONE 2011, 6, e19254. [Google Scholar] [CrossRef]
- Dunning, L.T.; Savolainen, V. Broad-scale amplification of matK for DNA barcoding plants, a technical note. Bot. J. Linn. Soc. 2010, 164, 1–9. [Google Scholar] [CrossRef]
- Bafeel, S.O.; Arif, I.A.; Bakir, M.A.; Al Homaidan, A.A.; Al Farhan, A.H.; Khan, H.A. DNA barcoding of arid wild plants using rbcL gene sequences. Genet. Mol. Res. 2012, 11, 1934–1941. [Google Scholar] [CrossRef] [PubMed]
- China Plant BOL Group; Li, D.Z.; Gao, L.M.; Li, H.T.; Wang, H.; Ge, X.J.; Liu, J.Q.; Chen, Z.D.; Zhou, S.L.; Chen, S.L.; et al. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Natl. Acad. Sci. USA 2011, 108, 19641–19646. [Google Scholar] [PubMed]
- Hollingsworth, P.M.; Forrest, L.L.; Spouge, J.L.; Hajibabaei, M.; Ratnasingham, S.; van der Bank, M.; Chase, M.W.; Cowan, R.S.; Erickson, D.L.; Fazekas, A.J.; et al. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar]
- Kress, W.J. Plant DNA Barcodes: Applications Today and in the Future. Front. Plant. Syst. Evol. 2017, 55, 291–307. [Google Scholar] [CrossRef]
- Müller, K.F.; Borsch, T.; Hilu, K.W. Phylogenetic utility of rapidly evolving DNA at high taxonomical levels: Contrasting matK, trnT-F, and rbcL in basal angiosperms. Mol. Phylogen. Evol. 2006, 41, 99–117. [Google Scholar] [CrossRef]
- Howard, C.; Lockie-Williams, C.; Slater, A. Applied Barcoding: The Practicalities of DNA Testing for Herbals. Plants 2020, 9, 1150. [Google Scholar] [CrossRef] [PubMed]
- Simko, I. High-resolution DNA melting analysis in plant research. Trends Plant. Sci. 2016, 21, 528–537. [Google Scholar] [CrossRef]
- Jaakola, L.; Suokas, M.; Häggman, H. Novel Approaches Based on DNA Barcoding and High-Resolution Melting of Amplicons for Authenticity Analyses of Berry Species. Food Chem. 2010, 123, 494–500. [Google Scholar] [CrossRef]
- Chen, W.; Chen, X.; Xu, J.; Cai, J.; Wang, X. Identification of Dendrobium officinale using DNA barcoding method combined with HRM and qPCR technology. Food Anal. Meth. 2022, 15, 1310–1320. [Google Scholar] [CrossRef]
- Song, M.; Li, J.; Xiong, C.; Liu, H.; Liang, J. Applying high-resolution melting (HRM) technology to identify five commonly used Artemisia species. Sci. Rep. 2016, 6, 34133. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Li, J.; Xiong, C.; Zhao, B.; Chen, S. The Potential Power of Bar-HRM Technology in Herbal Medicine Identification. Front. Plant. Sci. 2016, 7, 367. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Wu, X.; Liu, C.; Newmaster, S.; Ragupathy, S.; Kress, W.J. Progress in the Use of DNA Barcodes in the Identification and Classification of Medicinal Plants. Ecotoxicol. Environ. Saf. 2021, 208, 111691. [Google Scholar] [CrossRef]
- Qiao, J.; Zhang, X.; Chen, B.; Huang, F.; Xu, K.; Huang, Q.; Huang, Y.; Hu, Q.; Wu, X. Comparison of the cytoplastic genomes by resequencing: Insights into the genetic diversity and the phylogeny of the agriculturally important genus Brassica. BMC Genom. 2020, 21, 480. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol. Ecol. Not. 2007, 7, 574–578. [Google Scholar] [CrossRef]
- Earl, D.A.; VonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Cons. Genet. Res. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Wickham, H. Programming with Ggplot2. In ggplot2; Springer: Berling/Heidelberg, Germany, 2016; pp. 241–253. [Google Scholar]
- Adhikari, S.; Saha, S.; Biswas, A.; Rana, T.S.; Bandyopadhyay, T.K.; Ghosh, P. Application of molecular markers in plant genome analysis: A review. Nucleus 2017, 60, 283–297. [Google Scholar] [CrossRef]
- Mehmood, F.; Abdullah; Ubaid, Z.; Bao, Y.; Poczai, P.; Mirza, B. Comparative plastomics of Ashwagandha (Withania, Solanaceae) and identification of mutational hotspots for barcoding medicinal plant. Plants 2020, 9, 752. [Google Scholar] [CrossRef]
- Hellberg, R.S.; Isaacs, R.B.; Hernandez, E.L. Identification of shark species in commercial products using DNA barcoding. Fish. Res. 2019, 210, 81–88. [Google Scholar] [CrossRef]
- Wurzbacher, C.; Larsson, E.; Bengtsson-Palme, J.; Van den Wyngaert, S.; Svantesson, S.; Kristiansson, E.; Kagami, M.; Nilsson, R.H. Introducing ribosomal tandem repeat barcoding for fungi. Mol. Ecol. Resour. 2019, 19, 118–127. [Google Scholar] [CrossRef]
- Choudhary, P.; Singh, B.N.; Chakdar, H.; Saxena, A.K. DNA Barcoding of Phytopathogens for Disease Diagnostics and Bio-Surveillance. World J. Microbiol. Biotechnol. 2021, 37, 54. [Google Scholar] [CrossRef]
- Ongchai, S.; Chokchaitaweesuk, C.; Kongdang, P.; Chomdej, S.; Buddhachat, K. In vitro chondroprotective potential of Senna alata and Senna tora in porcine cartilage explants and their species differentiation by DNA barcoding-high resolution melting (Bar-HRM) analysis. PLoS ONE 2019, 14, e0215664. [Google Scholar] [CrossRef]
- Thongkhao, K.; Tungphatthong, C.; Phadungcharoen, T.; Sukrong, S. The use of plant DNA barcoding coupled with HRM analysis to differentiate edible vegetables from poisonous plants for food safety. Food Control 2020, 109, 106896. [Google Scholar] [CrossRef]
- Prantl, K. Diplotaxis. In Die Naturlichen Pflanzenfamilien; Wilhelm Engelmann: Leipzig, Germany, 1915. [Google Scholar]
- Schulz, O.E. Diplotaxis. In Das Pflanzenreich IV; Engler, A., Ed.; Wilhelm Engelmann: Leipzig, Germany, 1919; Volume 105, pp. 149–179. [Google Scholar]
- Martínez-Laborde, J.B. Notes on the taxonomy of Diplotaxis DC. (Brassiceae). Bot. J. Linn. Soc. 1991, 106, 67–71. [Google Scholar] [CrossRef]
- Takahata, Y.; Hinata, K. Studies on cytodemes in subtribe Brassicinae (Cruciferae). Tohoku J. Agric. Res. 1983, 33, 111–124. [Google Scholar]
- Mummenhoff, K.; Eschmann-Grupe, G.; Zunk, K. Subunit polypeptide composition of Rubisco indicates Diplotaxis viminea as maternal parent species of amphiploid Diplotaxis muralis. Phytochemistry 1993, 34, 429–431. [Google Scholar] [CrossRef]
- Mader, E.E.; Ruzicka, J.; Schmiderer, C.; Novak, J. Quantitative high-resolution melting analysis for detecting adulterations. Anal. Biochem. 2011, 409, 153–155. [Google Scholar] [CrossRef]
- Jiang, K.W.; Zhang, R.; Zhang, Z.F.; Pan, B.; Tian, B. DNA barcoding and molecular phylogeny of Dumasia (Fabaceae: Phaseoleae) reveals a cryptic lineage. Plant. Divers. 2020, 42, 376–385. [Google Scholar] [CrossRef]



| Marker Name | Foward Primer (5′–3′) | Reverse Primer (3′–5′) | Amplicon Size |
|---|---|---|---|
| ITS1 | TTAGGCCGTGCGTATAGCTT | TTGCGTTCAAAGACTCGATG | 249 bp |
| trnL-F | AGAAATTCCCGGTCCAAAAC | GGCCGTTACCGAAGTATCATT | 107 bp |
| rbcL | CGGAGTTCCACCTGAAGAAG | TTGTAACGGTCAAGGCTGGT | 105 bp |
| matK | TACGCCGCTTCTGATGAATA | TCTTTAGCCAACGACCCAAT | 267 bp |
| HRM500 | GATTCGAACCGTAGACCTGCTC | CCTTAAGGTGTAGCAAGTTTCA | 115 bp |
| HRM500 (322) | ITS1 (274) | matk (241) | rbcl (73) | trnL-F (80) | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Seq # | A | C | G | T | Seq | A | C | G | T | Seq | A | C | G | T | Seq | A | C | G | T | Seq | A | C | G | T | |
| D. ac * | 312 | 106 | 49 | 45 | 112 | 214 | 63 | 52 | 56 | 43 | 233 | 75 | 28 | 39 | 91 | 67 | 13 | 18 | 17 | 19 | 76 | 23 | 15 | 12 | 26 |
| D. as | 312 | 109 | 48 | 45 | 110 | 215 | 64 | 51 | 54 | 46 | 235 | 77 | 28 | 40 | 90 | 67 | 13 | 18 | 17 | 19 | 76 | 24 | 14 | 12 | 26 |
| D. be | 313 | 108 | 48 | 45 | 112 | 216 | 66 | 52 | 53 | 45 | 234 | 75 | 30 | 39 | 90 | 67 | 13 | 19 | 17 | 18 | 71 | 24 | 22 | 10 | 15 |
| D. br | 315 | 110 | 47 | 44 | 114 | 217 | 68 | 52 | 52 | 45 | 235 | 78 | 29 | 37 | 91 | 67 | 13 | 18 | 17 | 19 | 67 | 23 | 13 | 11 | 20 |
| D. bv | 313 | 106 | 48 | 45 | 114 | 216 | 62 | 53 | 57 | 44 | 234 | 76 | 31 | 37 | 90 | 69 | 14 | 18 | 17 | 20 | 80 | 23 | 18 | 12 | 27 |
| D. du | 313 | 105 | 49 | 47 | 112 | 217 | 66 | 52 | 55 | 44 | 235 | 76 | 30 | 38 | 91 | 68 | 14 | 19 | 17 | 18 | 76 | 23 | 15 | 12 | 26 |
| D. er | 313 | 108 | 48 | 45 | 112 | 218 | 68 | 51 | 53 | 46 | 234 | 76 | 29 | 38 | 91 | 68 | 14 | 18 | 17 | 19 | 78 | 23 | 17 | 12 | 26 |
| D. ha | 313 | 108 | 48 | 45 | 112 | 217 | 70 | 50 | 52 | 45 | 235 | 77 | 29 | 38 | 91 | 68 | 14 | 18 | 17 | 19 | 78 | 23 | 17 | 12 | 26 |
| D. ib | 313 | 108 | 48 | 45 | 112 | 214 | 67 | 52 | 52 | 43 | 236 | 77 | 31 | 38 | 90 | 70 | 14 | 20 | 17 | 19 | 78 | 24 | 16 | 12 | 26 |
| D. il | 314 | 108 | 48 | 45 | 113 | 213 | 63 | 51 | 56 | 43 | 236 | 77 | 31 | 38 | 90 | 68 | 14 | 18 | 17 | 19 | 79 | 23 | 17 | 12 | 27 |
| D. mu | 310 | 109 | 48 | 44 | 109 | 215 | 64 | 51 | 56 | 44 | 239 | 78 | 31 | 38 | 92 | 69 | 15 | 19 | 17 | 18 | 79 | 24 | 16 | 12 | 27 |
| D. si | 314 | 110 | 48 | 44 | 112 | 264 | 79 | 68 | 51 | 66 | 235 | 76 | 29 | 39 | 91 | 68 | 14 | 18 | 17 | 19 | 78 | 25 | 14 | 13 | 26 |
| D. sm | 314 | 104 | 49 | 47 | 114 | 214 | 63 | 51 | 56 | 44 | 235 | 76 | 31 | 38 | 90 | 68 | 14 | 19 | 17 | 18 | 76 | 23 | 15 | 12 | 26 |
| D. te | 312 | 104 | 49 | 47 | 112 | 216 | 65 | 51 | 56 | 44 | 236 | 77 | 29 | 40 | 90 | 68 | 14 | 19 | 17 | 18 | 77 | 24 | 15 | 12 | 26 |
| D. tn | 316 | 108 | 49 | 44 | 115 | 216 | 64 | 51 | 54 | 47 | 236 | 82 | 29 | 37 | 88 | 68 | 14 | 19 | 17 | 18 | 76 | 25 | 13 | 12 | 26 |
| D. vi | 309 | 109 | 47 | 43 | 110 | 217 | 66 | 52 | 55 | 44 | 236 | 77 | 31 | 39 | 89 | 68 | 14 | 18 | 17 | 19 | 78 | 24 | 16 | 12 | 26 |
| D. vr | 315 | 110 | 48 | 46 | 111 | 215 | 67 | 48 | 52 | 48 | 233 | 77 | 32 | 35 | 89 | 67 | 13 | 18 | 17 | 19 | 79 | 23 | 16 | 14 | 26 |
| D. ac | D. as | D. be | D. br | D. bv | D. du | D. er | D. ha | D. ib | D. il | D. mu | D. si | D. sm | D. te | D. tn | D. vi | D. vr | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D. ac | 0.005 | 0.007 | 0.005 | 0.005 | 0.004 | 0.005 | 0.005 | 0.004 | 0.003 | 0.004 | 0.009 | 0.004 | 0.004 | 0.005 | 0.004 | 0.006 | |
| D. as | 0.025 | 0.006 | 0.004 | 0.006 | 0.006 | 0.005 | 0.006 | 0.005 | 0.005 | 0.006 | 0.009 | 0.006 | 0.006 | 0.006 | 0.006 | 0.005 | |
| D. be | 0.049 | 0.032 | 0.007 | 0.008 | 0.007 | 0.007 | 0.008 | 0.007 | 0.007 | 0.008 | 0.010 | 0.008 | 0.008 | 0.008 | 0.008 | 0.007 | |
| D. br | 0.026 | 0.018 | 0.042 | 0.006 | 0.006 | 0.005 | 0.006 | 0.006 | 0.005 | 0.006 | 0.009 | 0.006 | 0.006 | 0.006 | 0.006 | 0.005 | |
| D. bv | 0.022 | 0.033 | 0.058 | 0.034 | 0.006 | 0.006 | 0.006 | 0.005 | 0.004 | 0.006 | 0.009 | 0.006 | 0.006 | 0.007 | 0.006 | 0.006 | |
| D. du | 0.014 | 0.029 | 0.049 | 0.030 | 0.030 | 0.005 | 0.006 | 0.005 | 0.004 | 0.005 | 0.010 | 0.003 | 0.003 | 0.006 | 0.005 | 0.006 | |
| D. er | 0.021 | 0.020 | 0.045 | 0.020 | 0.029 | 0.026 | 0.006 | 0.005 | 0.005 | 0.005 | 0.009 | 0.005 | 0.006 | 0.006 | 0.005 | 0.005 | |
| D. ha | 0.026 | 0.035 | 0.059 | 0.035 | 0.032 | 0.029 | 0.032 | 0.005 | 0.005 | 0.006 | 0.010 | 0.006 | 0.006 | 0.006 | 0.005 | 0.006 | |
| D. ib | 0.013 | 0.026 | 0.049 | 0.027 | 0.022 | 0.019 | 0.025 | 0.026 | 0.003 | 0.004 | 0.009 | 0.005 | 0.005 | 0.006 | 0.003 | 0.006 | |
| D. il | 0.011 | 0.021 | 0.046 | 0.022 | 0.014 | 0.015 | 0.020 | 0.025 | 0.009 | 0.004 | 0.009 | 0.004 | 0.004 | 0.006 | 0.004 | 0.006 | |
| D. mu | 0.014 | 0.027 | 0.052 | 0.028 | 0.028 | 0.019 | 0.023 | 0.030 | 0.018 | 0.013 | 0.010 | 0.004 | 0.004 | 0.005 | 0.003 | 0.006 | |
| D. si | 0.083 | 0.080 | 0.103 | 0.083 | 0.084 | 0.088 | 0.084 | 0.091 | 0.078 | 0.078 | 0.085 | 0.010 | 0.010 | 0.010 | 0.009 | 0.010 | |
| D. sm | 0.014 | 0.029 | 0.052 | 0.030 | 0.030 | 0.008 | 0.026 | 0.030 | 0.020 | 0.015 | 0.015 | 0.086 | 0.003 | 0.006 | 0.004 | 0.006 | |
| D. te | 0.016 | 0.032 | 0.054 | 0.033 | 0.033 | 0.006 | 0.028 | 0.033 | 0.022 | 0.018 | 0.016 | 0.091 | 0.008 | 0.005 | 0.005 | 0.006 | |
| D. tn | 0.026 | 0.029 | 0.052 | 0.030 | 0.039 | 0.030 | 0.030 | 0.036 | 0.032 | 0.027 | 0.026 | 0.092 | 0.028 | 0.026 | 0.006 | 0.006 | |
| D. vi | 0.014 | 0.027 | 0.052 | 0.028 | 0.028 | 0.019 | 0.023 | 0.026 | 0.011 | 0.013 | 0.007 | 0.084 | 0.016 | 0.019 | 0.028 | 0.006 | |
| D. vr | 0.030 | 0.020 | 0.045 | 0.022 | 0.035 | 0.035 | 0.025 | 0.036 | 0.032 | 0.027 | 0.028 | 0.087 | 0.033 | 0.035 | 0.032 | 0.028 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tripodi, P. Application of High-Resolution Melting and DNA Barcoding for Discrimination and Taxonomy Definition of Rocket Salad (Diplotaxis spp.) Species. Genes 2023, 14, 1594. https://doi.org/10.3390/genes14081594
Tripodi P. Application of High-Resolution Melting and DNA Barcoding for Discrimination and Taxonomy Definition of Rocket Salad (Diplotaxis spp.) Species. Genes. 2023; 14(8):1594. https://doi.org/10.3390/genes14081594
Chicago/Turabian StyleTripodi, Pasquale. 2023. "Application of High-Resolution Melting and DNA Barcoding for Discrimination and Taxonomy Definition of Rocket Salad (Diplotaxis spp.) Species" Genes 14, no. 8: 1594. https://doi.org/10.3390/genes14081594
APA StyleTripodi, P. (2023). Application of High-Resolution Melting and DNA Barcoding for Discrimination and Taxonomy Definition of Rocket Salad (Diplotaxis spp.) Species. Genes, 14(8), 1594. https://doi.org/10.3390/genes14081594







