Hub Gene Mining and Co-Expression Network Construction of Low-Temperature Response in Maize of Seedling by WGCNA
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Materials and Treatments
2.2. Clustering Analysis
2.3. Functional Enrichment of Module Genes
2.4. Construction of Gene Co-Expression Networks
3. Results and Analysis
3.1. RNA Sequencing
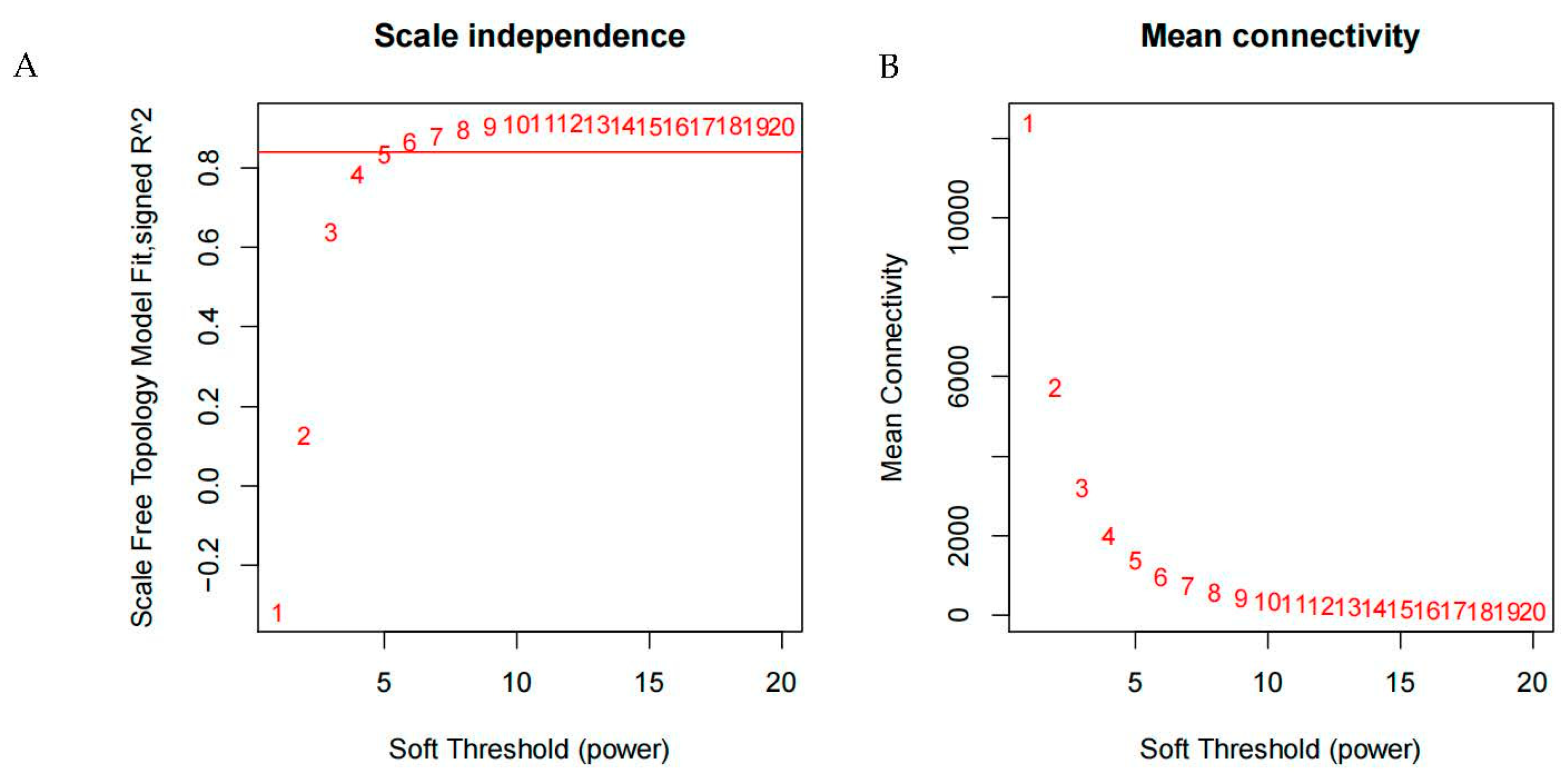
3.2. Determination of Soft Threshold in Gene Co-Expression Networks
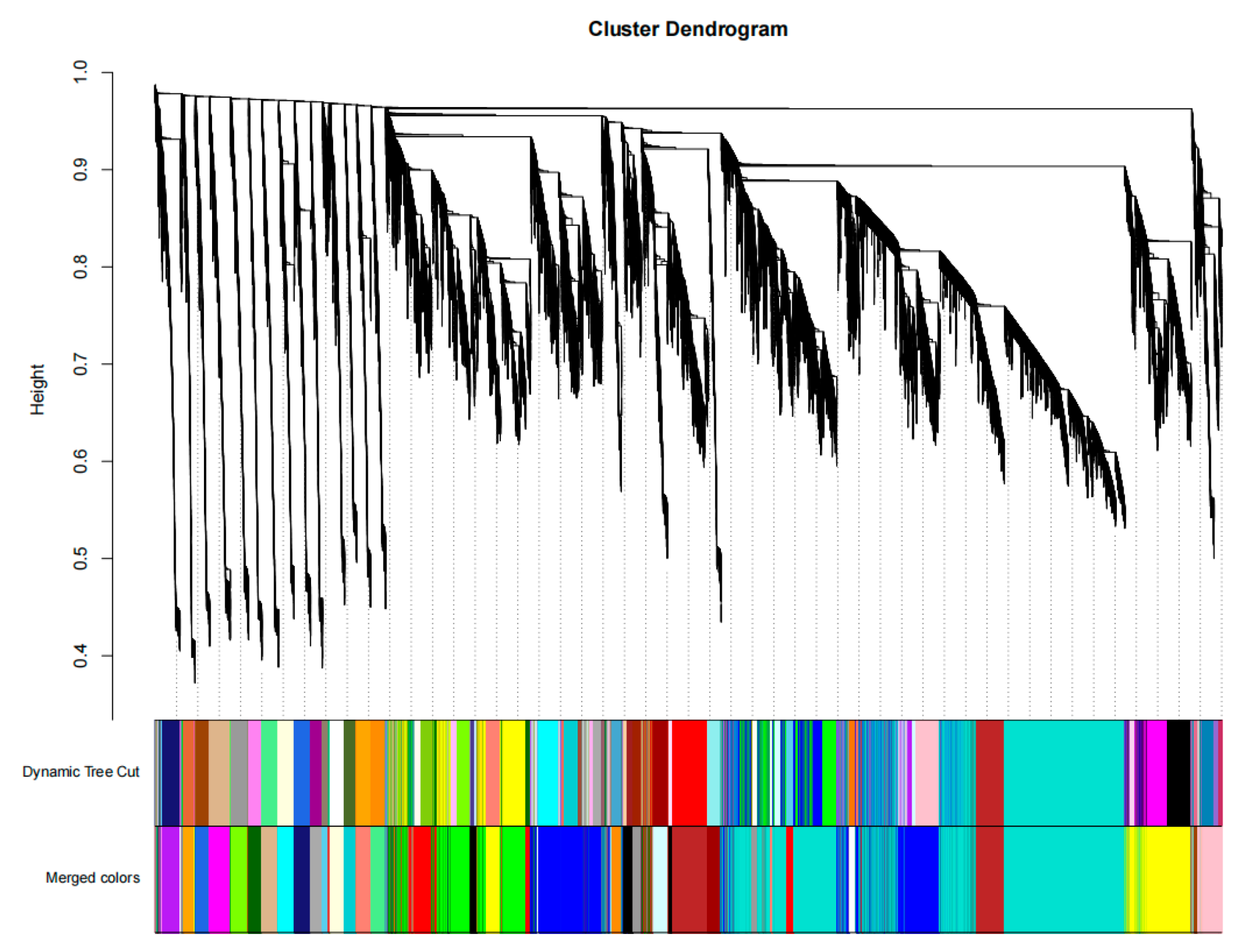
3.3. Gene Clustering and Module Segmentation in Gene Co-Expression Networks
3.4. Identification of Specific Modules under Low-Temperature Stress
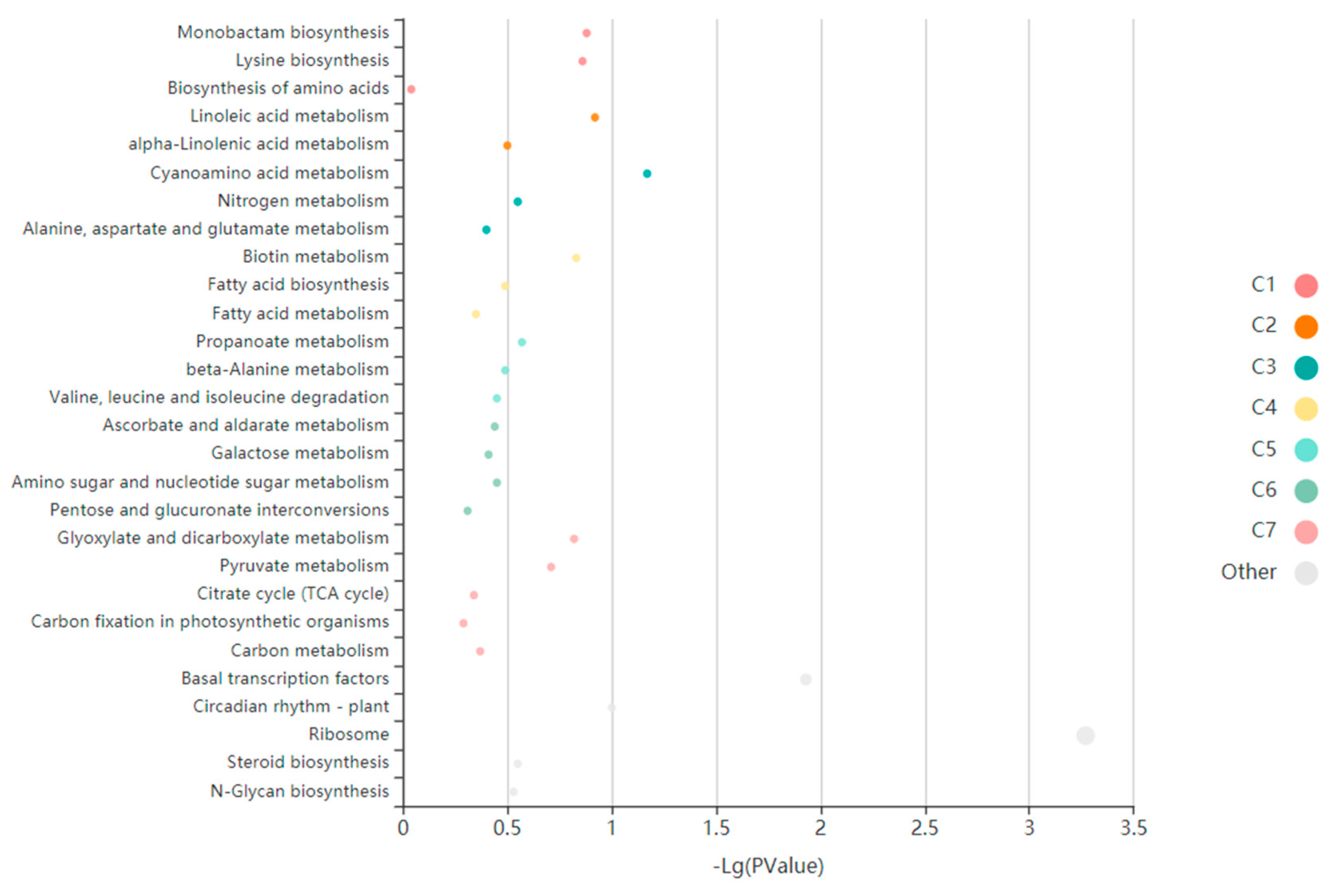
3.5. Enrichment Analysis of Specific Modules under Low-Temperature Stress
3.6. Construction of Gene Co-Expression Networks
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chinnusamy, V.; Schumaker, K.S.; Zhu, J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef]
- Liu, S.; Guo, N.; Ma, H.X.; Sun, H.; Zheng, X.; Shi, J. First report of root rot caused by bipolaris zeicola on maize in hebei province. Plant Dis. 2021, 105, 2247. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.G.; Li, S.J.; Cai, Q.; Wang, Z.H.; Cao, J.S.; Yu, T.; Xie, T.L. Exogenous diethyl aminoethyl hexanoate ameliorates low temperature stress by improving nitrogen metabolism in maize seedlings. PLoS ONE 2020, 15, e0232294. [Google Scholar] [CrossRef] [PubMed]
- Enders, T.A.; Dennis, S.S.; Oakland, J.; Callen, S.T.; Gehan, M.A.; Miller, N.D.; Spalding, E.P.; Springer, N.M.; Hirsch, C.D. Classifying cold-stress responses of inbred maize seedlings using RGB imaging. Plant Direct 2019, 3, e00104. [Google Scholar] [CrossRef] [PubMed]
- Fryer, M.J.; Andrews, J.R.; Oxborough, K.; Blowers, D.A.; Baker, N.R. Relationship between CO2 assimilation, photosynthetic electron transport, and active O2 metabolism in leaves of maize in the field during periods of low temperature. Plant Physiol. 1998, 116, 571–580. [Google Scholar] [CrossRef]
- Ying, J.; Lee, E.; Tollenaar, M. Response of maize leaf photosynthesis to low temperature during the grain-filling period. Field Crop Res. 2000, 68, 87–96. [Google Scholar] [CrossRef]
- Li, Z.; Hu, G.H.; Liu, X.F.; Zhou, Y.; Li, Y.; Zhang, X.; Yuan, X.H.; Zhang, Q.; Yang, D.G.; Wang, T.Y.; et al. Transcriptome sequencing identified genes and gene ontologies associated with early freezing tolerance in maize. Front. Plant Sci. 2016, 7, 1477. [Google Scholar] [CrossRef]
- Zhang, B.; Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, 17. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Kuang, J.F.; Wu, C.J.; Guo, Y.F.; Walther, D.; Shan, W.; Chen, J.Y.; Chen, L.; Lu, W.J. Deciphering transcriptional regulators of banana fruit ripening by regulatory network analysis. Plant Biotechnol. J. 2021, 19, 477–489. [Google Scholar] [CrossRef]
- Sun, S.; Xiong, X.P.; Zhu, Q.; Li, Y.J.; Sun, J. Transcriptome sequencing and metabolome analysis reveal genes involved in pigmentation of green-colored cotton fibers. Int. J. Mol. Sci. 2019, 20, 4838. [Google Scholar] [CrossRef]
- Zou, X.Y.; Liu, A.Y.; Zhang, Z.; Ge, Q.; Fan, S.M.; Gong, W.K.; Li, J.W.; Gong, J.W.; Shi, Y.Z.; Tian, B.M.; et al. Co-expression network analysis and hub gene selection for high-quality fiber in upland cotton (Gossypium hirsutum) using RNA sequencing analysis. Genes. 2019, 10, 119. [Google Scholar] [CrossRef]
- Greenham, K.; Guadagno, C.R.; Gehan, M.A.; Mockler, T.C.; Weinig, C.; Ewers, B.E.; McClung, C.R. Temporal network analysis identifies early physiological and transcriptomic indicators of mild drought in Brassica rapa. eLife 2017, 6, e29655. [Google Scholar] [CrossRef]
- Tan, M.P.; Cheng, D.; Yang, Y.N.; Zhang, G.Q.; Qin, M.J.; Chen, J.; Chen, Y.H.; Jiang, M.Y. Co-expression network analysis of the transcriptomes of rice roots exposed to various cadmium stresses reveals universal cadmium-responsive genes. BMC Plant Biol. 2017, 17, 194. [Google Scholar] [CrossRef]
- Ma, J.; Cao, Y.Y.; Wang, L.F.; Li, J.J.; Wang, H.; Fan, Y.P.; Li, H.Y. Identification of gene co-expression modules of maize plant height and ear height by WGCNA. Acta Agron. Sin. 2020, 46, 385–394, (In Chinese with English abstract). [Google Scholar]
- Yang, Y.X.; Sang, Z.Q.; Xu, C.; Dai, W.S.; Zou, C. Identification of maize flowering gene co-expression modules by WGCNA. Acta Agron. Sin. 2019, 45, 161–174, (In Chinese with English abstract). [Google Scholar] [CrossRef]
- Yu, T.; Zhang, J.; Cao, J.; Cai, Q.; Li, X.; Sun, Y.; Li, S.; Li, Y.; Hu, G.; Cao, S. Leaf transcriptomic response mediated by cold stress in two maize inbred lines with contrasting tolerance levels. Genomics 2021, 113, 782–794. [Google Scholar] [CrossRef]
- Wu, T.Z.; Hu, E.; Xu, S.B.; Chen, M.J.; Guo, P.F.; Dai, Z.H.; Feng, T.Z.; Zhou, L.; Tang, W.L.; Zhan, L.; et al. ClusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Abreu-Neto, J.B.; Turchetto-Zolet, A.C.; Oliveira, L.F.; Zanettini, M.H.; Margis-Pinheiro, M. Heavy metal-associated isoprenylated plant protein (hipp): Characterization of a family of proteins exclusive to plants. FEBS J. 2013, 280, 1604–1616. [Google Scholar] [CrossRef]
- Cui, N. CRISPR/Cas9 Technology Editing Soybean Gm HIPP26 Gene and Its Function under Cadmium Stress. Master’s Thesis, Zhejiang University, Hangzhou, China, 2021; pp. 33–38. [Google Scholar]
- Choi, J.; Lee, W.; An, G.; Kim, S.R. OsCBE1, a substrate receptor of cullin4-based E3 ubiquitin ligase, functions as a regulator of abiotic stress response and productivity in rice. Int. J. Mol. Sci. 2021, 22, 2487. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Serio, R.J.; Schofield, A.; Liu, H.; Rasmussen, S.R.; Hofius, D.; Stone, S.L. Arabidopsis RING-type E3ubiquitinligase XBAT35.2 promotes proteasome-dependent degradation of ACD11 to attenuate abiotic stress tolerance. Plant J. 2020, 104, 1712–1723. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.H.; Min, H.J.; Yu, S.G.; Byun, M.Y.; Oh, T.R.; Lee, A.; Yang, H.W.; Kim, W.T. OsATL38 mediates mono-ubiquitination of the 14-3-3 protein OsGF14d and negatively regulates the cold stress response in rice. J. Exp. Bot. 2022, 73, 307–323. [Google Scholar] [CrossRef]
- Kim, S.J.; Kim, W.T. Suppression of Arabidopsis RING E3 ubiquitin ligase AtATL78 increases tolerance to cold stress and decreases tolerance to drought stress. FEBS Lett. 2013, 587, 2584–2590. [Google Scholar] [CrossRef] [PubMed]
- Wan, D.; Li, R.; Zou, B.; Zhang, X.; Cong, J.; Wang, R.; Xia, Y.; Li, G. Calmodulin-binding protein CBP60g is a positive regulator of both disease resistance and drought tolerance in Arabidopsis. Plant Cell Rep. 2012, 31, 1269–1281. [Google Scholar] [CrossRef]
- Lv, T.; Li, X.; Fan, T.; Luo, H.; Xie, C.; Zhou, Y.; Tian, C.E. The Calmodulin-Binding Protein IQM1 Interacts with CATALASE2 to Affect Pathogen Defense. Plant Physiol. 2019, 181, 1314–1327. [Google Scholar] [CrossRef]
- Zhang, Y.X.; Xu, S.H.; Ding, P.T.; Wang, D.M.; Cheng, Y.T.; He, J.; Gao, M.H.; Xu, F.; Li, Y.; Zhu, Z.H.; et al. Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc. Natl. Acad. Sci. USA 2010, 107, 18220–18225. [Google Scholar] [CrossRef]
- Du, L.Q.; Ali, S.; Simons, A.; Hou, J.G.; Yang, T.B.; Reddy, A.S.N.; Poovaiah, B.W. Ca(2+)/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 2009, 457, 1154–1158. [Google Scholar] [CrossRef]
- Prasad, V.S.K.; Abdel-Hameed, A.E.; Xing, D.H.; Reddy, S.N. Global gene expression analysis using RNA-seq uncovered a new role for SR1/CAMTA3 transcription factor in salt stress. Sci. Rep. 2016, 6, 27021. [Google Scholar] [CrossRef]
- Zeng, H.Q.; Wu, H.C.; Wang, G.P.; Dai, S.H.; Zhu, Q.Q.; Chen, H.Y.; Yi, K.K.; Du, L.Q. Arabidopsis CAMTA3/SR1 is involved in drought stress tolerance and ABA signaling. Plant Sci. 2022, 319, 111250. [Google Scholar] [CrossRef]
- Doherty, C.J.; Van, H.A.; Myers, S.J.; Thomashow, M.F. Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 2009, 21, 972–984. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, J.; Deng, X.; Wang, P.; Geng, S.; Gao, W.; Guo, P.; Chen, Q.; Li, C.; Qu, Y. Genome-wide analysis of serine carboxypeptidase-like protein (SCPL) family and functional validation of Gh_SCPL42 unchromosome conferring cotton Verticillium der Verticillium wilt stress in Gossypium hirsutum. BMC Plant Biol. 2022, 22, 421. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, L.; Zhao, W.; Fu, L.; Han, Y.; Wang, K.; Yan, L.; Li, Y.; Zhang, X.H.; Min, D.H. Genome-wide analysis of the serine carboxypeptidase-like protein family in Triticum aestivum reveals TaSCPL184-6D is involved in abiotic stress response. BMC Genom. 2021, 22, 350. [Google Scholar] [CrossRef]
- Areum, L.; Sang, L.; Won, J.; Hyun, P.; Bo, L.; Hyun, K.; Jun, A.; Hye, C. The OsCYP19-4 Gene is expressed as multiple alternatively spliced transcripts encoding isoforms with distinct cellular localizations and PPIase activities under cold stress. Int. J. Mol. Sci. 2016, 17, 1154. [Google Scholar]
- Yoon, D.H.; Lee, S.S.; Park, H.J.; Lyu, J.; Chong, W.S.; Liu, J.R.; Kim, B.G.; Ahn, J.C.; Cho, H.S. Overexpression of OsCYP19-4 increases tolerance to cold stress and enhances grain yield in rice (Oryza sativa). J. Exp. Bot. 2016, 67, 69–82. [Google Scholar] [CrossRef]
- Huang, L.C.; Liu, Y.J.; Wang, X.Q.; Jiang, C.; Zhao, Y.Q.; Lu, M.Z.; Zhang, J. Peroxisome-mediated reactive oxygen species signals modulate programmed cell death in plants. Int. J. Mol. Sci. 2022, 23, 10087. [Google Scholar] [CrossRef]
- Sandalio, L.M.; Romero-Puertas, M.C. Peroxisomes sense and respond to environmental cues by regulating ROS and RNS signalling networks. Ann. Bot. 2015, 116, 475–485. [Google Scholar] [CrossRef]
- Su, T.; Li, W.; Wang, P.; Ma, C. Dynamics of Peroxisome Homeostasis and Its Role in Stress Response and Signaling in Plants. Front. Plant Sci. 2019, 10, 705. [Google Scholar] [CrossRef]
- Heitz, T.; Widemann, E.; Lugan, R.; Miesch, L.; Ullmann, P.; Désaubry, L.; Holder, E.; Grausem, B.; Kandel, S.; Miesch, M.; et al. Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of plant hormone jasmonoyl-isoleucine for catabolic turnover. J. Biol. Chem. 2012, 287, 6296–6306. [Google Scholar] [CrossRef]
- Rabara, C.; Tripathi, P.; Reese, N.; Rushton, L.; Alexander, D.; Timko, P.; Shen, J.; Rushton, J. Tobacco drought stress responses reveal new targets for solanaceae crop improvement. BMC Genom. 2015, 16, 484. [Google Scholar]
- Chauvin, A.; Caldelari, D.; Wolfender, J.; Farmer, E.E. Four 13-lipoxygenases contribute to rapid jasmonate synthesis in wounded Arabidopsis thaliana leaves: A role for lipoxygenase 6 in responses to long-distance wound signals. New Phytol. 2013, 197, 566–575. [Google Scholar] [CrossRef] [PubMed]
- Babenko, L.M.; Shcherbatiuk, M.M.; Skaterna, T.D.; Kosakivska, I.V. Lipoxygenases and their metabolites in formation of plant stress tolerance. Ukr. Biochem. J. 2017, 89, 5–21. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.L.; Xu, J.; Li, L.C.; Guo, Z.; Si, Q.X.; Zhu, G.Z.; Wang, X.Y.; Guo, W.Z. GbCYP86A1-1 from Gossypium barbadense positively regulates defence against Verticillium dahliae by cell wall modification and activation of immune pathways. Plant Biotechnol. J. 2020, 18, 222–238. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Lai, J.B.; Wu, Q.; Zhang, S.C.; Chen, L. Jasmonate complements the function of Arabidopsis lipoxygenase3 in salinity stress response. Plant Sci. 2016, 244, 1–7. [Google Scholar] [CrossRef]
- Shaban, M.; Ahmed, M.M.; Sun, H.; Ullah, A.; Zhu, L. Genome-wide identification of lipoxygenase gene family in cotton and functional characterization in response to abiotic stresses. BMC Genom. 2018, 19, 599. [Google Scholar] [CrossRef]
- Upadhyay, R.K.; Handa, A.K.; Mattoo, A.K. Transcript Abundance Patterns of 9- and 13-Lipoxygenase Subfamily Gene Members in Response to Abiotic Stresses (Heat, Cold, Drought or Salt) in Tomato (Solanum lycopersicum L.) Highlights Member-Specific Dynamics Relevant to Each Stress. Genes 2019, 10, 683. [Google Scholar] [CrossRef]
- Gao, X.Q.; Starr, J.; Göbel, C.; Engelberth, J.; Feussner, I.; Tumlinson, J.; Kolomiets, M. Maize 9-lipoxygenase ZmLOX3 controls development, root-specific expression of defense genes, and resistance to root-knot nematodes. Mol. Plant-Microbe Interact. 2008, 21, 98–109. [Google Scholar] [CrossRef]
- Zhu, J.H.; Jeong, C.; Zhu, Y.M.; Sokolchik, I.; Miyazaki, S.; Zhu, J.K.; Hasegawa, M.; Bohnert, J.; Shi, H.Z.; Yun, D.J.; et al. Involvement of Arabidopsis HOS15 in histone deacetylation and cold tolerance. Proc. Natl. Acad. Sci. USA 2008, 105, 4945–4950. [Google Scholar] [CrossRef]
- Ananieva, E.A.; Gillaspy, G.E.; Ely, A.; Burnette, R.N.; Erickson, F.L. Interaction of the WD40 Domain of a Myoinositol Polyphosphate 5-Phosphatase with SnRK1 Links Inositol, Sugar, and Stress Signaling. Plant Physiol. 2008, 148, 1868–1882. [Google Scholar] [CrossRef]
- Sascha, B.; Hanjo, H. WD40 and CUL4-based E3 ligases: Lubricating all aspects of life. Trends Plant Sci. 2010, 16, 38–46. [Google Scholar]
- Hu, R.; Xiao, J.; Gu, T.; Yu, X.; Zhang, Y.; Chang, J.; Yang, G.; He, G. Genome-wide identification and analysis of WD40 proteins in wheat (Triticum aestivum L.). BMC Genom. 2018, 19, 803. [Google Scholar]
- Rahman, M.M.; Mian, M.K.; Ahmed, A.; Rohman, M.M. Roles of Glutathione s-transferease in maize (Zea mays L.) under cold stress. Res. Agric. Livest. Fish. 2015, 2, 9–15. [Google Scholar] [CrossRef]
- Smita, K.; Mehar, A.; Debasis, C.; Rudra, T.; Rama, D.; Prabodh, T. Expression of a rice Lambda class of glutathione S-transferase, 0sGSTL2, in Arabidopsis provides tolerance to heavy metal and other abiotic stresses. J. Hazard. Mater. 2013, 248–249, 228–237. [Google Scholar]
- Duan, X.; Yu, X.; Wang, Y.; Fu, W.; Cao, R.; Yang, L.; Ye, X. Genome-wide identification and expression analysis of glutathione S-transferase gene family to reveal their role in cold stress response in cucumber. Front. Genet. 2022, 13, 1009883. [Google Scholar] [CrossRef]




| Module Name | Hub Gene | Homologous Gene in Rice | Gene Annotation in Maize |
|---|---|---|---|
| darkorange | LOC103646333 | LOC4336080 | heavy metal-associated isoprenylated plant protein 47 |
| LOC100193294 | LOC4331934 | sulfate transporter3 | |
| LOC103647664 | LOC4343201 | argonaute protein 18 | |
| LOC103626411 | - | putative disease resistance protein RGA3 | |
| LOC103635071 | LOC107277552 | E3 ubiquitin-protein ligase EL5 | |
| LOC100284905 | LOC4331734 | uncharacterized | |
| LOC107548101 | LOC4326262 | uncharacterized | |
| LOC542717 | LOC4326149 | isoflavone reductase-like 1 | |
| 100274353 | LOC9269286 | uncharacterized | |
| 103642911 | LOC4346124 | exocyst complex component EXO70B1 | |
| greenyellow | LOC541914 | LOC4326376 | aldehyde dehydrogenase 5 |
| LOC100502288 | LOC4325649 | lipid phosphate phosphatase 2 | |
| LOC100191783 | LOC4349708 | carbohydrate transporter/sugar porter/transporter | |
| LOC103640644 | LOC4352852 | thaumatin-like protein | |
| LOC100286314 | LOC4341938 | phosphate import ATP-binding protein pstB 1 | |
| LOC100283475 | LOC4342022 | peptidyl-prolyl cis-trans isomerase CYP19-4 | |
| LOC103632710 | LOC4346970 | type IV inositol polyphosphate 5-phosphatase 9 | |
| LOC103639446 | LOC4332030 | plant calmodulin-binding protein-related | |
| LOC100191502 | LOC4347942 | serine carboxypeptidase-like 19 | |
| LOC542740 | LOC4325704 | glutathione transferase 8 | |
| lightyellow | LOC100381507 | LOC4341247 | peroxidase 52 |
| LOC100101525 | LOC4327535 | cysteine proteinase inhibitor | |
| LOC100286092 | LOC4337884 | uncharacterized | |
| LOC100272818 | LOC4339940 | aspartyl protease AED1 | |
| LOC100383295 | CYP714B2 | cytochrome P450 714B3 | |
| LOC100283004 | LOC4334796 | uncharacterized | |
| LOC100282551 | LOC4336977 | UDP-N-acetylglucosamine diphosphorylase | |
| LOC100275470 | LOC4337397 | uncharacterized | |
| LOC100191608 | LOC4330180 | HXXXD-type acyl-transferase family protein | |
| LOC100284786 | - | uncharacterized | |
| purple | LOC100037802 | LOC4334049 | lox2 linoleate 9S-lipoxygenase2 |
| LOC100285766 | LOC4327677 | IAA-amino acid hydrolase ILR1-like 4 | |
| LOC541856 | - | lox1 linoleate 9S-lipoxygenase1 | |
| LOC100272711 | - | uncharacterized | |
| LOC100037804 | - | linoleate 9S-lipoxygenase5 | |
| LOC100276188 | - | uncharacterized | |
| LOC103641407 | LOC4332648 | putative WD40-like β propeller repeat family protein | |
| LOC103641187 | LOC4344635 | (E)-β-farnesene synthase-like | |
| LOC100037810 | LOC4328603 | lox11 linoleate 13S-lipoxygenase11 | |
| LOC541838 | LOC4347319 | glutathione transferase25 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, T.; Zhang, J.; Cao, J.; Ma, X.; Li, W.; Yang, G. Hub Gene Mining and Co-Expression Network Construction of Low-Temperature Response in Maize of Seedling by WGCNA. Genes 2023, 14, 1598. https://doi.org/10.3390/genes14081598
Yu T, Zhang J, Cao J, Ma X, Li W, Yang G. Hub Gene Mining and Co-Expression Network Construction of Low-Temperature Response in Maize of Seedling by WGCNA. Genes. 2023; 14(8):1598. https://doi.org/10.3390/genes14081598
Chicago/Turabian StyleYu, Tao, Jianguo Zhang, Jingsheng Cao, Xuena Ma, Wenyue Li, and Gengbin Yang. 2023. "Hub Gene Mining and Co-Expression Network Construction of Low-Temperature Response in Maize of Seedling by WGCNA" Genes 14, no. 8: 1598. https://doi.org/10.3390/genes14081598
APA StyleYu, T., Zhang, J., Cao, J., Ma, X., Li, W., & Yang, G. (2023). Hub Gene Mining and Co-Expression Network Construction of Low-Temperature Response in Maize of Seedling by WGCNA. Genes, 14(8), 1598. https://doi.org/10.3390/genes14081598






