Implementing Core Genes and an Omnigenic Model for Behaviour Traits Prediction in Genomics
Abstract
:1. Introduction
2. Complex and Monogenic Traits
3. The Role and Limitations of Genome-Wide Association Studies
4. Practical Application of Combined Genome Variants for Complex Traits
5. Omnigenic Model Implementation
6. Biological Pathways for Behaviour Traits
7. Core Genes of Behaviour
8. Core Genes: Ethnic and Gender Perspectives
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rowe, S.J.; Tenesa, A. Human complex trait genetics: Lifting the lid of the genomics toolbox—from pathways to prediction. Curr. Genom. 2012, 13, 213–224. [Google Scholar] [CrossRef]
- Landrum, M.J.; Chitipiralla, S.; Brown, G.R.; Chen, C.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; Kaur, K.; Liu, C.; et al. ClinVar: Improvements to accessing data. Nucleic Acids Res. 2020, 48, D835–D844. [Google Scholar] [CrossRef]
- Manolio, T.A.; Collins, F.S.; Cox, N.J.; Goldstein, D.B.; Hindorff, L.A.; Hunter, D.J.; McCarthy, M.I.; Ramos, E.M.; Cardon, L.R.; Chakravarti, A.; et al. Finding the missing heritability of complex diseases. Nature 2009, 461, 747–753. [Google Scholar] [CrossRef] [PubMed]
- Visscher, P.M.; Brown, M.A.; McCarthy, M.I.; Yang, J. Five years of GWAS discovery. Am. J. Hum. Genet. 2012, 90, 7–24. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, N.; Shi, J.; Garcia-Closas, M. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat. Rev. Genet. 2016, 17, 392–406. [Google Scholar] [CrossRef]
- Khera, A.V.; Chaffin, M.; Aragam, K.G.; Haas, M.E.; Roselli, C.; Choi, S.H.; Natarajan, P.; Lander, E.S.; Lubitz, S.A.; Ellinor, P.T.; et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 2018, 50, 1219–1224. [Google Scholar] [CrossRef]
- Bray, M.S.; Hagberg, J.M.; Perusse, L.; Rankinen, T.; Roth, S.M.; Wolfarth, B.; Bouchard, C. The human gene map for performance and health-related fitness phenotypes: The 2006–2007 update. Med. Sci. Sports Exerc. 2009, 41, 35–73. [Google Scholar] [CrossRef]
- Rankinen, T.; Fuku, N.; Wolfarth, B.; Wang, G.; Sarzynski, M.A.; Alexeev, D.G.; Ahmetov, I.I.; Boulay, M.R.; Cieszczyk, P.; Eynon, N.; et al. No Evidence of a Common DNA Variant Profile Specific to World Class Endurance Athletes. PLoS ONE 2016, 11, e0147330. [Google Scholar] [CrossRef]
- Varillas-Delgado, D.; Del Coso, J.; Gutierrez-Hellin, J.; Aguilar-Navarro, M.; Munoz, A.; Maestro, A.; Morencos, E. Genetics and sports performance: The present and future in the identification of talent for sports based on DNA testing. Eur. J. Appl. Physiol. 2022, 122, 1811–1830. [Google Scholar] [CrossRef]
- Semenova, E.A.; Hall, E.C.R.; Ahmetov, I.I. Genes and Athletic Performance: The 2023 Update. Genes 2023, 14, 1235. [Google Scholar] [CrossRef] [PubMed]
- Webborn, N.; Williams, A.; McNamee, M.; Bouchard, C.; Pitsiladis, Y.; Ahmetov, I.; Ashley, E.; Byrne, N.; Camporesi, S.; Collins, M.; et al. Direct-to-consumer genetic testing for predicting sports performance and talent identification: Consensus statement. Br. J. Sports Med. 2015, 49, 1486–1491. [Google Scholar] [CrossRef] [PubMed]
- Wienroth, M. Socio-technical disagreements as ethical fora: Parabon NanoLab’s forensic DNA Snapshot (TM) service at the intersection of discourses around robust science, technology validation, and commerce. Biosocieties 2020, 15, 28–45. [Google Scholar] [CrossRef]
- Pospiech, E.; Teisseyre, P.; Mielniczuk, J.; Branicki, W. Predicting Physical Appearance from DNA Data-Towards Genomic Solutions. Genes 2022, 13, 121. [Google Scholar] [CrossRef] [PubMed]
- Reiss, D.; Leve, L.D.; Neiderhiser, J.M. How genes and the social environment moderate each other. Am. J. Public Health 2013, 103, S111–S121. [Google Scholar] [CrossRef] [PubMed]
- Boyle, E.A.; Li, Y.I.; Pritchard, J.K. An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell 2017, 169, 1177–1186. [Google Scholar] [CrossRef]
- Wray, N.R.; Wijmenga, C.; Sullivan, P.F.; Yang, J.; Visscher, P.M. Common Disease Is More Complex Than Implied by the Core Gene Omnigenic Model. Cell 2018, 173, 1573–1580. [Google Scholar] [CrossRef]
- Ratnakumar, A.; Weinhold, N.; Mar, J.C.; Riaz, N. Protein-Protein interactions uncover candidate ‘core genes’ within omnigenic disease networks. PLoS Genet. 2020, 16, e1008903. [Google Scholar] [CrossRef]
- Chateigner, A.; Lesage-Descauses, M.C.; Rogier, O.; Jorge, V.; Leple, J.C.; Brunaud, V.; Roux, C.P.; Soubigou-Taconnat, L.; Martin-Magniette, M.L.; Sanchez, L.; et al. Gene expression predictions and networks in natural populations supports the omnigenic theory. BMC Genom. 2020, 21, 416. [Google Scholar] [CrossRef]
- Zhang, W.; Reeves, G.R.; Tautz, D. Testing Implications of the Omnigenic Model for the Genetic Analysis of Loci Identified through Genome-wide Association. Curr. Biol. 2021, 31, 1092–1098.e6. [Google Scholar] [CrossRef]
- Iakoucheva, L.M.; Muotri, A.R.; Sebat, J. Getting to the Cores of Autism. Cell 2019, 178, 1287–1298. [Google Scholar] [CrossRef]
- Rammos, A.; Gonzalez, L.A.N.; Schizophrenia Working Group of the Psychiatric Genomics Consortium 2; Weinberger, D.R.; Mitchell, K.J.; Nicodemus, K.K. The role of polygenic risk score gene-set analysis in the context of the omnigenic model of schizophrenia. Neuropsychopharmacology 2019, 44, 1562–1569. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, M.B. Synaptic Signaling in Learning and Memory. Cold Spring Harb. Perspect. Biol. 2013, 8, a016824. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.P.; Han, W. Regulation of synaptic functions in central nervous system by endocrine hormones and the maintenance of energy homoeostasis. Biosci. Rep. 2012, 32, 423–432. [Google Scholar] [CrossRef] [PubMed]
- Robinson, P.K. Enzymes: Principles and biotechnological applications. Essays Biochem. 2015, 59, 1–41. [Google Scholar] [CrossRef]
- Starka, L.; Duskova, M. What is a hormone? Physiol. Res. 2020, 69, S183–S185. [Google Scholar] [CrossRef]
- Soares, M.C.; Bshary, R.; Fusani, L.; Goymann, W.; Hau, M.; Hirschenhauser, K.; Oliveira, R.F. Hormonal mechanisms of cooperative behaviour. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010, 365, 2737–2750. [Google Scholar] [CrossRef] [PubMed]
- Gasmi, A.; Nasreen, A.; Menzel, A.; Gasmi Benahmed, A.; Pivina, L.; Noor, S.; Peana, M.; Chirumbolo, S.; Bjorklund, G. Neurotransmitters Regulation and Food Intake: The Role of Dietary Sources in Neurotransmission. Molecules 2022, 28, 210. [Google Scholar] [CrossRef]
- Teleanu, R.I.; Niculescu, A.G.; Roza, E.; Vladacenco, O.; Grumezescu, A.M.; Teleanu, D.M. Neurotransmitters-Key Factors in Neurological and Neurodegenerative Disorders of the Central Nervous System. Int. J. Mol. Sci. 2022, 23, 5954. [Google Scholar] [CrossRef]
- Edenberg, H.J.; McClintick, J.N. Alcohol Dehydrogenases, Aldehyde Dehydrogenases, and Alcohol Use Disorders: A Critical Review. Alcohol. Clin. Exp. Res. 2018, 42, 2281–2297. [Google Scholar] [CrossRef]
- Bierut, L.J.; Goate, A.M.; Breslau, N.; Johnson, E.O.; Bertelsen, S.; Fox, L.; Agrawal, A.; Bucholz, K.K.; Grucza, R.; Hesselbrock, V.; et al. ADH1B is associated with alcohol dependence and alcohol consumption in populations of European and African ancestry. Mol. Psychiatry 2012, 17, 445–450. [Google Scholar] [CrossRef]
- Hoang, Y.T.T.; Nguyen, Y.T.; Vu, L.T.; Bui, H.T.T.; Nguyen, Q.V.; Vu, N.P.; Nguyen, T.D.; Nguyen, H.H. Association of ADH1B rs1229984, ADH1C rs698, and ALDH2 rs671 with Alcohol abuse and Alcoholic Cirrhosis in People Living in Northeast Vietnam. Asian Pac. J. Cancer Prev. 2023, 24, 2073–2082. [Google Scholar] [CrossRef] [PubMed]
- Polimanti, R.; Gelernter, J. ADH1B: From alcoholism, natural selection, and cancer to the human phenome. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2018, 177, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alfoldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Di Iorio, C.R.; Carey, C.E.; Michalski, L.J.; Corral-Frias, N.S.; Conley, E.D.; Hariri, A.R.; Bogdan, R. Hypothalamic-pituitary-adrenal axis genetic variation and early stress moderates amygdala function. Psychoneuroendocrinology 2017, 80, 170–178. [Google Scholar] [CrossRef]
- Wesarg, C.; Veer, I.M.; Oei, N.Y.L.; Daedelow, L.S.; Lett, T.A.; Banaschewski, T.; Barker, G.J.; Bokde, A.L.W.; Quinlan, E.B.; Desrivieres, S.; et al. The interaction of child abuse and rs1360780 of the FKBP5 gene is associated with amygdala resting-state functional connectivity in young adults. Hum. Brain Mapp. 2021, 42, 3269–3281. [Google Scholar] [CrossRef] [PubMed]
- Weeger, J.; Ising, M.; Muller-Myhsok, B.; Uhr, M.; Schmidt, U.; Steiger, A. Salivary cortisol response to psychosocial stress in the late evening depends on CRHR1 genotype. Psychoneuroendocrinology 2020, 116, 104685. [Google Scholar] [CrossRef] [PubMed]
- van der Meer, D.; Hoekstra, P.J.; Bralten, J.; van Donkelaar, M.; Heslenfeld, D.J.; Oosterlaan, J.; Faraone, S.V.; Franke, B.; Buitelaar, J.K.; Hartman, C.A. Interplay between stress response genes associated with attention-deficit hyperactivity disorder and brain volume. Genes. Brain Behav. 2016, 15, 627–636. [Google Scholar] [CrossRef]
- Chmielowiec, K.; Chmielowiec, J.; Stronska-Pluta, A.; Trybek, G.; Smiarowska, M.; Suchanecka, A.; Wozniak, G.; Jaron, A.; Grzywacz, A. Association of Polymorphism CHRNA5 and CHRNA3 Gene in People Addicted to Nicotine. Int. J. Environ. Res. Public Health 2022, 19, 10478. [Google Scholar] [CrossRef]
- Boscutti, A.; Pigoni, A.; Delvecchio, G.; Lazzaretti, M.; Mandolini, G.M.; Girardi, P.; Ferro, A.; Sala, M.; Abbiati, V.; Cappucciati, M.; et al. The Influence of 5-HTTLPR, BDNF Rs6265 and COMT Rs4680 Polymorphisms on Impulsivity in Bipolar Disorder: The Role of Gender. Genes 2022, 13, 482. [Google Scholar] [CrossRef]
- Soeiro-De-Souza, M.G.; Stanford, M.S.; Bio, D.S.; Machado-Vieira, R.; Moreno, R.A. Association of the COMT Met(1)(5)(8) allele with trait impulsivity in healthy young adults. Mol. Med. Rep. 2013, 7, 1067–1072. [Google Scholar] [CrossRef]
- Landro, N.I.; Jonassen, R.; Clark, L.; Haug, K.B.; Aker, M.; Bo, R.; Berg, J.P.; Neumeister, A.; Stiles, T.C. Serotonin transporter polymorphisms predict response inhibition in healthy volunteers. Neurosci. Lett. 2015, 584, 109–112. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Chang, H.A.; Fang, W.H.; Chang, T.C.; Huang, S.Y. Gender-specific association between serotonin transporter polymorphisms (5-HTTLPR and rs25531) and neuroticism, anxiety and depression in well-defined healthy Han Chinese. J. Affect. Disord. 2017, 207, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Kalungi, A.; Seedat, S.; Hemmings, S.M.J.; van der Merwe, L.; Joloba, M.L.; Nanteza, A.; Nakassujja, N.; Birabwa, H.; Serwanga, J.; Kaleebu, P.; et al. Association between serotonin transporter gene polymorphisms and increased suicidal risk among HIV positive patients in Uganda. BMC Genet. 2017, 18, 71. [Google Scholar] [CrossRef]
- Chang, T.G.; Yen, T.T.; Wei, C.Y.; Hsiao, T.H.; Chen, I.C. Impacts of ADH1B rs1229984 and ALDH2 rs671 polymorphisms on risks of alcohol-related disorder and cancer. Cancer Med. 2023, 12, 747–759. [Google Scholar] [CrossRef] [PubMed]
- Chaity, N.I.; Apu, M.N.H. CHRNA5 rs16969968 and CHRNA3 rs578776 polymorphisms are associated with multiple nicotine dependence phenotypes in Bangladeshi smokers. Heliyon 2022, 8, e09947. [Google Scholar] [CrossRef] [PubMed]
- Rajagopal, V.M.; Watanabe, K.; Mbatchou, J.; Ayer, A.; Quon, P.; Sharma, D.; Kessler, M.D.; Praveen, K.; Gelfman, S.; Parikshak, N.; et al. Rare coding variants in CHRNB2 reduce the likelihood of smoking. Nat. Genet. 2023, 55, 1138–1148. [Google Scholar] [CrossRef] [PubMed]
- Picciotto, M.R.; Kenny, P.J. Mechanisms of Nicotine Addiction. Cold Spring Harb. Perspect. Med. 2021, 11, a039610. [Google Scholar] [CrossRef]
- Blum, K.; Han, D.; Bowirrat, A.; Downs, B.W.; Bagchi, D.; Thanos, P.K.; Baron, D.; Braverman, E.R.; Dennen, C.A.; Gupta, A.; et al. Genetic Addiction Risk and Psychological Profiling Analyses for “Preaddiction” Severity Index. J. Pers. Med. 2022, 12, 1772. [Google Scholar] [CrossRef]
- Al-Eitan, L.N.; Rababa’h, D.M.; Alghamdi, M.A. Genetic susceptibility of opioid receptor genes polymorphism to drug addiction: A candidate-gene association study. BMC Psychiatry 2021, 21, 5. [Google Scholar] [CrossRef]
- Dennis, B.B.; Bawor, M.; Thabane, L.; Sohani, Z.; Samaan, Z. Impact of ABCB1 and CYP2B6 genetic polymorphisms on methadone metabolism, dose and treatment response in patients with opioid addiction: A systematic review and meta-analysis. PLoS ONE 2014, 9, e86114. [Google Scholar] [CrossRef]
- Mihaljevic, M.; Franic, D.; Soldatovic, I.; Lukic, I.; Petrovic, S.A.; Mirjanic, T.; Stankovic, B.; Zukic, B.; Zeljic, K.; Gasic, V.; et al. The FKBP5 genotype and childhood trauma effects on FKBP5 DNA methylation in patients with psychosis, their unaffected siblings, and healthy controls. Psychoneuroendocrinology 2021, 128, 105205. [Google Scholar] [CrossRef] [PubMed]
- Brockway, D.F.; Crowley, N.A. Turning the ’Tides on Neuropsychiatric Diseases: The Role of Peptides in the Prefrontal Cortex. Front. Behav. Neurosci. 2020, 14, 588400. [Google Scholar] [CrossRef] [PubMed]
- Castro-Vale, I.; Duraes, C.; van Rossum, E.F.C.; Staufenbiel, S.M.; Severo, M.; Lemos, M.C.; Carvalho, D. The Glucocorticoid Receptor Gene (NR3C1) 9beta SNP Is Associated with Posttraumatic Stress Disorder. Healthcare 2021, 9, 173. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.H.; Chang, H.A.; Fang, W.H.; Ho, P.S.; Liu, Y.P.; Wan, F.J.; Tzeng, N.S.; Shyu, J.F.; Chang, C.C. Serotonin receptor 1A promoter polymorphism, rs6295, modulates human anxiety levels via altering parasympathetic nervous activity. Acta Psychiatr. Scand. 2018, 137, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; He, L.; Liu, J.; Guo, W.; Wang, Q.; Fang, P.; Yang, X.; Zhang, M.; Wang, C.; Gong, P. The rs6311 of serotonin receptor 2A (5-HT2A) gene is associated with alexithymia and mental health. J. Affect. Disord. 2020, 272, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Maffioletti, E.; Valsecchi, P.; Minelli, A.; Magri, C.; Bonvicini, C.; Barlati, S.; Sacchetti, E.; Vita, A.; Gennarelli, M. Association study between HTR2A rs6313 polymorphism and early response to risperidone and olanzapine in schizophrenia patients. Drug Dev. Res. 2020, 81, 754–761. [Google Scholar] [CrossRef]
- Panitz, C.; Sperl, M.F.J.; Hennig, J.; Klucken, T.; Hermann, C.; Mueller, E.M. Fearfulness, neuroticism/anxiety, and COMT Val158Met in long-term fear conditioning and extinction. Neurobiol. Learn. Mem. 2018, 155, 7–20. [Google Scholar] [CrossRef]
- Sumner, J.A.; Powers, A.; Jovanovic, T.; Koenen, K.C. Genetic influences on the neural and physiological bases of acute threat: A research domain criteria (RDoC) perspective. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2016, 171B, 44–64. [Google Scholar] [CrossRef]
- Broekman, B.F. Stress, vulnerability and resilience, a developmental approach. Eur. J. Psychotraumatol. 2011, 2, 7229. [Google Scholar] [CrossRef]
- Palumbo, S.; Mariotti, V.; Vellucci, S.; Antonelli, K.; Anderson, N.; Harenski, C.; Pietrini, P.; Kiehl, K.A.; Pellegrini, S. ANKK1 and TH gene variants in combination with paternal maltreatment increase susceptibility to both cognitive and attentive impulsivity. Front. Psychiatry 2022, 13, 868804. [Google Scholar] [CrossRef]
- Parsons, M.J.; Lester, K.J.; Barclay, N.L.; Nolan, P.M.; Eley, T.C.; Gregory, A.M. Replication of Genome-Wide Association Studies (GWAS) loci for sleep in the British G1219 cohort. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2013, 162B, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Gafarov, V.; Gromova, E.; Gagulin, I.; Panov, D.; Gafarova, A.; Krymov, E. Association of polymorphism RS934945 Gene PER2 with sleep disoders in the male population 25–44 years in Novosibirsk. Eur. Neuropsychopharmacol. 2019, 29, S923. [Google Scholar] [CrossRef]
- Parico, G.C.G.; Perez, I.; Fribourgh, J.L.; Hernandez, B.N.; Lee, H.W.; Partch, C.L. The human CRY1 tail controls circadian timing by regulating its association with CLOCK:BMAL1. Proc. Natl. Acad. Sci. USA 2020, 117, 27971–27979. [Google Scholar] [CrossRef]
- Soler, C.T.; Kanders, S.H.; Olofsdotter, S.; Vadlin, S.; Aslund, C.; Nilsson, K.W. Exploration of the Moderating Effects of Physical Activity and Early Life Stress on the Relation between Brain-Derived Neurotrophic Factor (BDNF) rs6265 Variants and Depressive Symptoms among Adolescents. Genes 2022, 13, 1236. [Google Scholar] [CrossRef]
- White, K.C.; McDonald, A.K.; Compton, D.M. 5-HTR2A Polymorphisms rs6311 and rs6313 and Major Depressive Disorder: A Meta-Analysis. J. Behav. Brain Sci. 2022, 12, 499–513. [Google Scholar] [CrossRef]
- Liu, X.; Li, Y.I.; Pritchard, J.K. Trans Effects on Gene Expression Can Drive Omnigenic Inheritance. Cell 2019, 177, 1022–1034.e6. [Google Scholar] [CrossRef] [PubMed]
- Kolla, N.J.; Bortolato, M. The role of monoamine oxidase A in the neurobiology of aggressive, antisocial, and violent behavior: A tale of mice and men. Prog. Neurobiol. 2020, 194, 101875. [Google Scholar] [CrossRef]
- Schmitz, L.L.; Gard, A.M.; Ware, E.B. Examining sex differences in pleiotropic effects for depression and smoking using polygenic and gene-region aggregation techniques. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2019, 180, 448–468. [Google Scholar] [CrossRef]
- Mendonca, M.S.; Mangiavacchi, P.M.; Mendes, A.V.; Loureiro, S.R.; Martin-Santos, R.; Gloria, L.S.; Marques, W.; De Marco, S.P.G.; Kanashiro, M.M.; Hallak, J.E.C.; et al. DNA methylation in regulatory elements of the FKBP5 and NR3C1 gene in mother-child binomials with depression. J. Affect. Disord. 2023, 331, 287–299. [Google Scholar] [CrossRef]
- Bühler, K.-M.; Rincón-Pérez, I.; Calleja-Conde, J.; Albert, J.; Hinojosa, J.A.; Giné, E.; Echeverry-Alzate, V.; López-Moreno, J.A.; Huertas, E. The genetics of self-reported trait impulsivity: Contribution of catecholaminergic gene variants in European ancestry individuals. Personal. Individ. Differ. 2023, 200, 111906. [Google Scholar] [CrossRef]
- Quinn, J.P.; Savage, A.L.; Bubb, V.J. Non-coding genetic variation shaping mental health. Curr. Opin. Psychol. 2019, 27, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Alsheikh, A.J.; Wollenhaupt, S.; King, E.A.; Reeb, J.; Ghosh, S.; Stolzenburg, L.R.; Tamim, S.; Lazar, J.; Davis, J.W.; Jacob, H.J. The landscape of GWAS validation; systematic review identifying 309 validated non-coding variants across 130 human diseases. BMC Med. Genom. 2022, 15, 74. [Google Scholar] [CrossRef]
- Zhang, F.; Lupski, J.R. Non-coding genetic variants in human disease. Hum. Mol. Genet. 2015, 24, R102–R110. [Google Scholar] [CrossRef]
- Tomson, K.; Vaht, M.; Laas, K.; Veidebaum, T.; Harro, J. Effect of a human serotonin 5-HT(2A) receptor gene polymorphism on impulsivity: Dependence on cholesterol levels. J. Affect. Disord. 2016, 206, 23–30. [Google Scholar] [CrossRef]
- Villafuerte, S.; Strumba, V.; Stoltenberg, S.F.; Zucker, R.A.; Burmeister, M. Impulsiveness mediates the association between GABRA2 SNPs and lifetime alcohol problems. Genes. Brain Behav. 2013, 12, 525–531. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Roige, S.; Jennings, M.V.; Thorpe, H.H.A.; Mallari, J.E.; van der Werf, L.C.; Bianchi, S.B.; Huang, Y.; Lee, C.; Mallard, T.T.; Barnes, S.A.; et al. CADM2 is implicated in impulsive personality and numerous other traits by genome- and phenome-wide association studies in humans and mice. Transl. Psychiatry 2023, 13, 167. [Google Scholar] [CrossRef] [PubMed]
- Tansey, K.E.; Hill, M.J.; Cochrane, L.E.; Gill, M.; Anney, R.J.; Gallagher, L. Functionality of promoter microsatellites of arginine vasopressin receptor 1A (AVPR1A): Implications for autism. Mol. Autism 2011, 2, 3. [Google Scholar] [CrossRef]
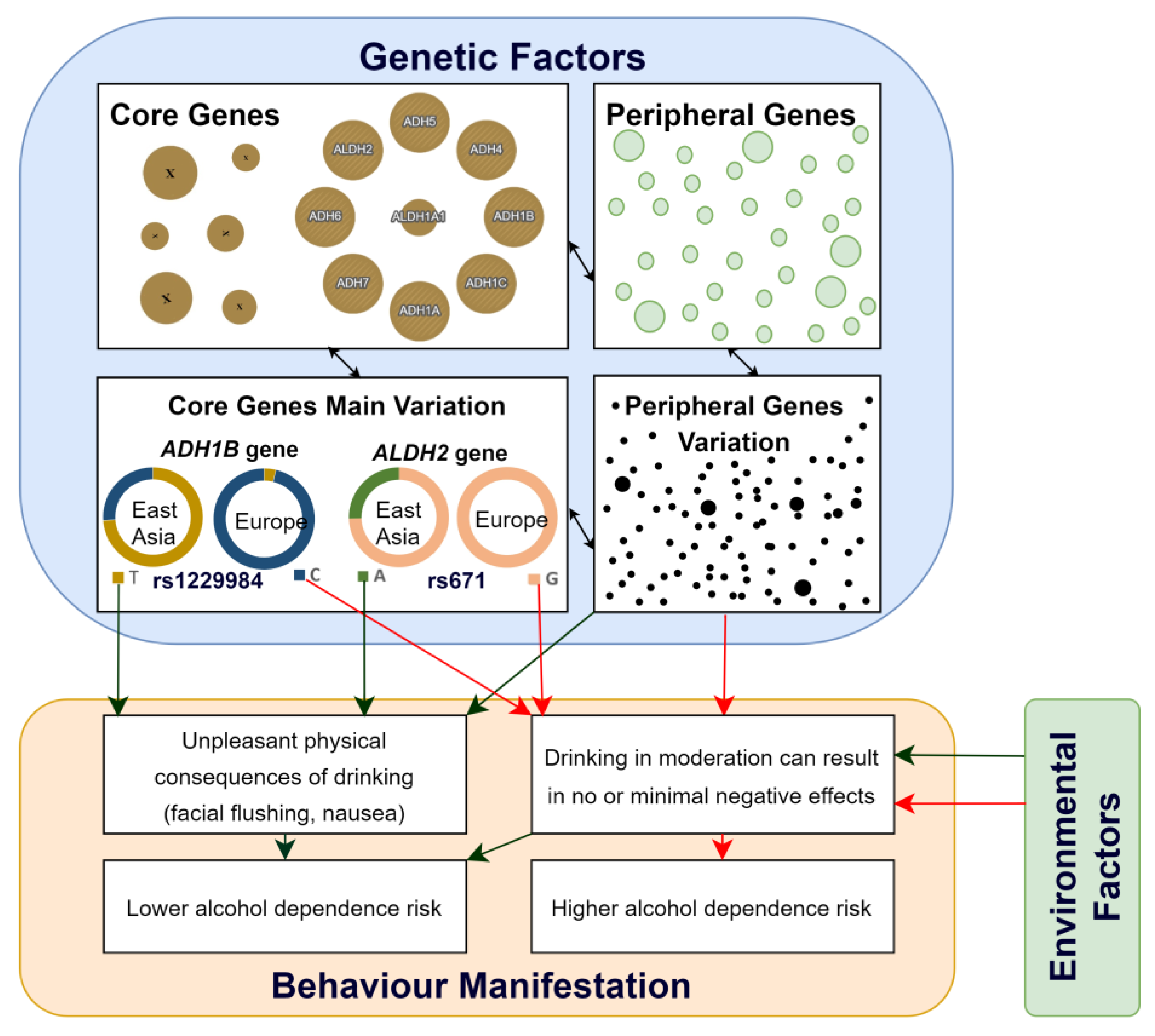
| Behaviour Trait | Core Genes | Genome Variants | References |
|---|---|---|---|
| Alcohol Use | ADH1B, ADH1C, ALDH2 | rs1229984, rs698, rs671 | [30,31,44] |
| Smoking | CHRNA5, CHRNA3, CHRNB4 | rs16969968, rs1051730 | [38,45,46,47] |
| Drug Use | DRD2, OPRM1, ABCB1 | rs1800497, rs1799971, rs1045642 | [48,49,50] |
| Stress | FKBP5, CRHR1, NR3C1 | rs1360780, rs110402, rs6195 | [51,52,53] |
| Anxiety | SLC6A4, HTR1A, HTR2A | rs6295, rs6311, rs6313 | [54,55,56] |
| Fearfulness | SLC6A4, COMT, MAOA | rs4680, rs6323, rs6354 | [57,58,59] |
| Impulsivity | DRD4, SLC6A4, COMT, HTR2A | rs1800955, rs25531, rs4680 | [40,41,60] |
| Lack of Sleep | ABCC9, PER2, PER3, CRY1 | rs11046209, rs934945, rs228697 | [61,62,63] |
| Depression | SLC6A4, BDNF, HTR2A | rs25531, rs6265, rs6313 | [42,64,65] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rancelis, T.; Domarkiene, I.; Ambrozaityte, L.; Utkus, A. Implementing Core Genes and an Omnigenic Model for Behaviour Traits Prediction in Genomics. Genes 2023, 14, 1630. https://doi.org/10.3390/genes14081630
Rancelis T, Domarkiene I, Ambrozaityte L, Utkus A. Implementing Core Genes and an Omnigenic Model for Behaviour Traits Prediction in Genomics. Genes. 2023; 14(8):1630. https://doi.org/10.3390/genes14081630
Chicago/Turabian StyleRancelis, Tautvydas, Ingrida Domarkiene, Laima Ambrozaityte, and Algirdas Utkus. 2023. "Implementing Core Genes and an Omnigenic Model for Behaviour Traits Prediction in Genomics" Genes 14, no. 8: 1630. https://doi.org/10.3390/genes14081630






