Abstract
Intramuscular fat (IMF) content is a key determinant of pork quality. Controlling the genetic and physiological factors of IMF and the expression patterns of various genes is important for regulating the IMF content and improving meat quality in pig breeding. Growing evidence has suggested the role of genetic factors and breeds in IMF deposition; however, research on the sex factors of IMF deposition is still lacking. The present study aimed to identify potential sex-specific biomarkers strongly associated with IMF deposition in low- and high-IMF pig populations. The GSE144780 expression dataset of IMF deposition-related genes were obtained from the Gene Expression Omnibus. Initially, differentially expressed genes (DEGs) were detected in male and female low-IMF (162 DEGs, including 64 up- and 98 down-regulated genes) and high-IMF pigs (202 DEGs, including 147 up- and 55 down-regulated genes). Moreover, hub genes were screened via PPI network construction. Furthermore, hub genes were screened for potential sex-specific biomarkers using the least absolute shrinkage and selection operator machine learning algorithm, and sex-specific biomarkers in low-IMF (troponin I (TNNI1), myosin light chain 9(MYL9), and serpin family C member 1(SERPINC1)) and high-IMF pigs (CD4 molecule (CD4), CD2 molecule (CD2), and amine oxidase copper-containing 2(AOC2)) were identified, and then verified by quantitative real-time PCR (qRT-PCR) in semimembranosus muscles. Additionally, the gene set enrichment analysis and single-sample gene set enrichment analysis of hallmark gene sets were collectively performed on the identified biomarkers. Finally, the transcription factor-biomarker and lncRNA-miRNA-mRNA (biomarker) networks were predicted. The identified potential sex-specific biomarkers may provide new insights into the molecular mechanisms of IMF deposition and the beneficial foundation for improving meat quality in pig breeding.
1. Introduction
Intramuscular fat (IMF) content is an important trait in the pig industry, which is positively correlated with meat quality, and affects the tenderness, flavor, and juiciness of the meat [1,2]. The potency of fatty acid metabolism in skeletal muscle and the development of intramuscular adipocytes have a significant impact on IMF content [3]. Many studies have shown that the pattern of muscle development and IMF deposition depended on breed, genotype, sex, muscle location, diet, and slaughter weight characteristics [4,5,6,7], but the related molecular mechanisms remain elusive.
In recent years, great progress has been made in genetic mechanisms and candidate gene exploring for IMF content based on advanced molecular biology techniques and bioinformatics. Many genes have been identified as major regulators of IMF deposition, playing important roles in regulating fat metabolism and promoting IMF deposition [8]. For example, acetyl-CoA Acyltransferase 1(ACAA1) is involved in fatty acid oxidation and lipid metabolism, and is strongly linked to PPAR signaling and fatty acid metabolism pathways [9]. Acyl-CoA Synthetase Long Chain Family Member 4(ACSL4) positively regulates adipogenesis in IMF cells, and its high expression can increase monounsaturated and polyunsaturated fatty acid contents in pig intramuscular adipocytes [10]. The higher fat deposition in Alentejano pigs (obese type) than in Bisaro Portuguese pigs (lean type) may be due to the increased synthesis of new fatty acids caused by the up-regulation of key genes such as ATP Citrate Lyase (ACLY), fatty Acid Synthase (FASN) and malic Enzyme 1(ME1) [11]. However, most studies have used pigs of different breeds or different IMF contents within the same breed [12,13], with few studies on individuals of different sexes in the same breed to identify candidate genes and molecular mechanisms. Regarding sex traits, previous studies have shown significant differences in nearly all carcass characteristics between castrates and gilts; castrates produce fattier carcasses than gilts, which deposit leaner meat, and IMF levels vary considerably between sexes at the physiological and biochemical levels [14]. The results of studies by Latorre et al. and Serrano et al. showed that the IMF content and marbling of castrates were considerably higher than those of gilts, producing, to some extent, better meat quality traits [15,16]. Previous studies have also found significantly higher total fatty acid levels in castrates than in gilts for the fatty acid composition of IMF and semimembranosus muscles [17]. Numerous studies have demonstrated a significant influence of sex on pork quality, with discernible differences in IMF content between male and female pigs [18,19]. However, most studies for the differences in pork quality and IMF content between males and females did not involve a molecular and regulatory mechanism.
Consequently, our study aimed to screen potential sex-specific biomarkers in male and female pigs with different IMF levels in the same breed using integrated bioinformatic methods and machine learning algorithms. In addition, we aimed to construct molecular regulatory networks related to sex-specific biomarkers, laying a foundation for in-depth exploration of the molecular mechanisms underlying IMF deposition.
2. Materials and Methods
The research flowchart of the present analysis is shown in Figure 1.
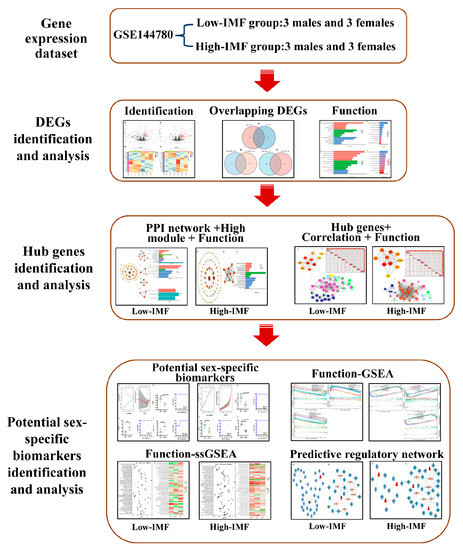
Figure 1.
Flow chart of the bioinformatics analysis in the present study.
2.1. Data Collection and Preprocessing
The GSE144780 gene expression dataset was retrieved from the public Gene Expression Omnibus database [GPL19176 Illumina HiSeq 2000 (Sus scrofa) (https://www.ncbi.nlm.nih.gov/geo/), accessed on 1 November 2022] and generated using semimembranosus muscles (SM) from Italian Large White pigs (six males and six females) [12]. The pigs were selected from 950 purebred sib-tested Italian Large White pigs, reared in the same environmental conditions on the same finishing diet and fed quasi ad libitum. Based on the extreme and divergent contents of SM IMF, twelve pigs were categorized into two groups: 6 low- and 6 high-IMF animals. In our study, the individuals with different IMF levels in the two groups were evenly distributed in the two sex subgroups, each with 3 male and 3 female pigs in the low- and high-IMF group. Details about the individuals are shown in Table 1. The original study has been approved by the Institutional Review Board of the relevant participating institutions (European rules (Council Regulation (EC) No. 1/2005 and Council Regulation (EC) No. 1099/2009)), and no additional approval was required from the ethics committee in dataset GSE144780.

Table 1.
Sample information of the GEO dataset.
2.2. Identification of Differentially Expressed Genes (DEGs)
DEGs between males and females in the low- and high-IMF groups were identified separately using the “Deseq2” package of the R software (version 4.1.2) [20]. In this study, the genes with a |log2FC (fold change)| > 1 and a p-value < 0.05 were considered as DEGs. The expression level and distribution of DEGs were visualized using the heatmap and the volcano map based on the ggplot2 package (versions 3.3.6), respectively.
2.3. DEG Functional Enrichment Analysis
Functional DEG analysis was implemented via a GO and KEGG pathway analysis using the online DAVID database (DAVID 2021, https://david.ncifcrf.gov/home.jsp, accessed on 1 November 2022). The GO term included three categories: the cellular component (CC), biological process (BP), and molecular function (MF). The functional terms with a p-value < 0.05 were considered to significantly change, and the functional terms were visualized using the R-package ggplot2.
2.4. Protein–Protein Interaction (PPI) Network Construction and Hub Gene Identification
Functional interactions between DEG-encoded proteins were analyzed by constructing a PPI network using the STRING online database (https://cn.string-db.org/, accessed on 1 November 2022) with a combined score > 0.4 and p-value < 0.05, shown by the Cytoscape software (version 3.9.1) [21]; each node represented the gene-encoding protein and edges represented the connection between nodes in the PPI network. Subsequently, the cluster analysis was performed in the whole PPI network using the Molecular Complex Detection (MCODE) algorithm by the Cytoscape plugin (Version 2.0.2) [22]. The threshold parameters were set for the degree cutoff = 2, node score cutoff = 0.2, k-score = 2, and max. depth = 100. The hub genes were identified using the Maximal Clique Centrality (MCC) method in the Cytoscape plugin cytoHubba (Version 0.1) [23], and the top 10 genes with the higher MCC scores were selected as hub genes. The interaction network of the hub genes and GO and KEGG terms were visualized by ClueGo (Version 2.5.9) and CluePedia plugin (Version 1.5.9). Thereafter, the correlations of hub genes were analyzed and visualized by the Pearson method through the ggpairs of GGally (version: 1.5.0) package based on R software.
2.5. Screening of Potential Sex-Specific Biomarkers
The least absolute shrinkage and selection operator (LASSO) machine learning algorithm [24] was used to screen for potential sex-specific biomarkers among the top 10 hub genes identified in the low- and high-IMF groups. In addition, LASSO was built with the “glmnet” R package and performed variable screening and complexity adjustment while fitting a generalized linear model. To evaluate the predictive performance of potential sex-specific biomarkers, their expression levels were compared between the males and females using an unpaired t-test. p < 0.05 was considered to be significant. Then, the receiver operating characteristic (ROC) curves of dataset GSE144780 (source set) were established, and the area under the curve (AUC) values of potential biomarkers was calculated to assess the efficacy of the distinguishing performance of potential sex-specific biomarkers using GraphPad Prism (version 8.0.1).
2.6. Analysis of Hallmark Gene Sets of Potential Sex-Specific Biomarkers
To further elucidate the functions of the potential sex-specific biomarkers, an association analysis of the hallmark gene sets was performed using a single-sample gene set enrichment analysis (ssGSEA) [25]. Firstly, we calculated the ssGSEA scores of hallmark gene sets as well as in the males and females, and subsequently analyzed the association between hallmark gene sets and potential sex-specific biomarkers; the “corrplot” package was used to obtain the Spearman rank correlation coefficient.
2.7. Construction of Potential Transcription Factor (TF) Sex-Specific Biomarker Regulatory Network
TFs of potential sex-specific biomarkers were predicted using the online database of UCSC and JASPAR 2022 (https://genome.ucsc.edu/, accessed on 1 November 2022), and the minimum score = 600 was the screening condition. The regulatory relationships between TFs and potential sex-specific biomarkers were visualized through the Cytoscape software (version 3.9.1).
2.8. Construction of ceRNA Network
The lncRNA-miRNA-mRNA interactive relationships were elucidated by constructing a lncRNA-miRNA-mRNA (potential sex-specific biomarkers) regulatory network based on the ceRNA hypothesis [26]. The potential targeted miRNAs and lncRNAs of potential sex-specific biomarkers were predicted by the online ENCORI database (https://rna.sysu.edu.cn/encori/index.php, accessed on 1 November 2022). First, the biomarkers-miRNA relationship pairs were extracted based on at least two databases: miRanda [27], TargetScan [28], RNA22 [29], and miRmap [30]. Then, the interacting lncRNAs were predicted based on above-predicted miRNAs. Finally, the co-expression network of the lncRNAs-miRNAs-mRNA (potential sex-specific biomarkers) ceRNA network was visualized through the Cytoscape software.
2.9. Animals and Tissue Collection
Thirty pigs, including fifteen males and fifteen females of the Saba pig, were used in this study, which is an indigenous pig breed in Yunnan, China. The pigs used in this study were obtained from the national-level Saba pig conservation farm (Chuxiong City, Yunnan, China). The pigs were selected from purebred Saba pigs, raised in the same environmental conditions with free access to water and feed until slaughter (~100 kg). The SM samples were collected from the carcasses at slaughter, snap frozen in liquid nitrogen, and maintained at −80 °C until total RNA extraction; some SM was stored at −20 °C for the determination of IMF content. All animal procedures were performed according to the principles of the Laboratory Animal Care and Use Guidelines issued by the Animal Research Committee of Yunnan Agricultural University; the experimental protocol was approved by the Animal Ethics Committee of Yunnan Agricultural University (approval ID: YAUACUC01).
2.10. Measurement of IMF Content
The IMF content of SM was measured by the Soxhlet extraction method [31]. Firstly, the SM sample was dried and crushed (weight W1 g), and then transferred to the extraction chamber of the soxhlet apparatus and soaked in anhydrous ether overnight. Subsequently, the anhydrous ether backflow equipment was worked for 8 h at 80 °C. Finally, the remaining sample was dried again and weighed (weight W2 g). The IMF content of SM was calculated using the following equation: IMF (%) = [(W1 − W2)/W1] × 100. Considering the weight and IMF content as factors, three male pigs and three female pigs exhibiting exceptionally high IMF content and three male pigs and three female pigs displaying remarkably low IMF content were selected for the subsequent validation analysis.
2.11. RNA Extraction and qRT-PCR
The total RNA was extracted from SM samples using an RNA sample total Extraction Kit (Tiangen, Beijing, China). Briefly, the SM sample was frozen in liquid nitrogen and ground to a fine powder. Then, chloroform was added, the aqueous phase was collected and translated to RNase-Free columns, and the RNA was precipitated by the addition of absolute ethanol. Subsequently, buffer RD and RW were stepwise added to an RNA pellet to obtain RNA. Finally, RNA was eluted from the column by 50 µL of RNase-free water. Reverse transcription was performed using PrimeScript™ RT reagent Kit with gDNA Eraser (Takara, Dalian, China) according to the manufacturer’s instructions. The first step considers: the reaction conditions of system1: 42 °C, 2 min; 4 °C, 18 min; second, the reaction conditions: 37 °C, 15 min; 85 °C, 5 s; 4 °C, 18 min. qPCR assay was performed using TB Green® Premix Ex Taq™ II (Tli RNaseH Plus) (Takara, Dalian, China) on a qPCR system (Mx3000P, Agilent Technologies, Santa Clara, CA, USA). The reaction system was prepared for qPCR reaction. It was performed using a two-step amplification, Stage1 (predenaturation): 95 °C, 30 s; Stage2: repeat 40, 95 °C, 30 s, 60 °C, 30 s; Stage3: Dissociation. The gene-specific qPCR primers are listed in Supplementary Table S1. Each experiment was performed in triplicates, and the relative expression of mRNA were calculated through the 2−ΔΔCt method; GAPDH was used as the internal control for normalization. The relative mRNA expression levels of male and female pigs were compared by unpaired t-test using SAS software (version 9.2), p < 0.05 was considered significant, and p < 0.01 was considered extremely significant.
3. Results
3.1. DEG Identification
Totals of 162 DEGs were identified between male and female pigs in the low-IMF group (p < 0.05), 64 of which were significantly up-regulated, and 98 were significantly down-regulated in males (Figure 2A; Supplementary Table S2). A total of 202 DEGs were identified in the high-IMF group, 147 of which were significantly up-regulated, and 55 were significantly down-regulated in males (Figure 2C; Supplementary Table S3). The expression levels of these DEGs in the low- and high-IMF groups were presented using a heatmap in Figure 2B,D, respectively.
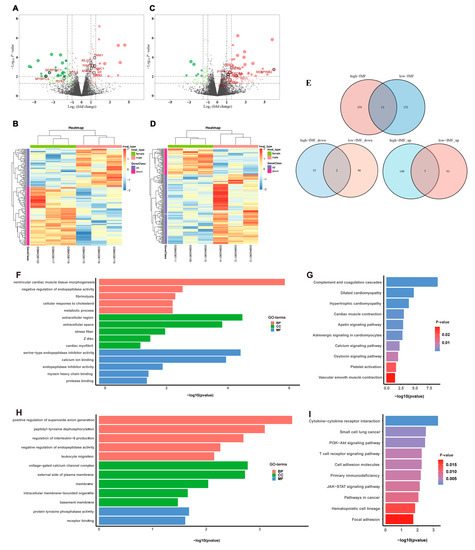
Figure 2.
Analysis of DEGs. DEGs were defined with |log2FC| > 1.5 and p-value < 0.05. (A) Volcano plot of the expression level of DEGs between male and female pigs in low-IMF group. Red dots represent a high expression of genes and green dots represent a low expression of genes. The dots with black circles were hub genes. (B) Heatmap of the expression level of DEGs between male and female pigs in low-IMF group. The abscissa indicates the sample names, and the ordinate shows the gene names. High expression of genes is shown in violet and low expression of genes is shown in red. (C) Volcano plot of the expression level of DEGs between males and females in high-IMF group. (D) Heatmap of the expression level of DEGs between males and females in high-IMF group. (E) The Venn diagram of DEGs. (F) The results shown by GO of DEGs in low-IMF group. (G) The results shown by KEGG of DEGs in low-IMF group. (H) The results shown by GO of DEGs in high-IMF group. (I) The results shown by KEGG of DEGs in high-IMF group.
The up- and down-regulated DEGs between the low- and high-IMF groups were further compared, as shown in Figure 3E. The identified overlapping, up-regulated, and down-regulated DEGs in the low- and high-IMF groups were mentioned in Table 2. Among the overlapping DEGs, GADD45G-interacting protein 1 (GADD45GIP1) and inter-α-trypsin inhibitor heavy chain 1 (ITIH1) were up-regulated in high-IMF male pigs, but down-regulated in male pigs with low IMF.
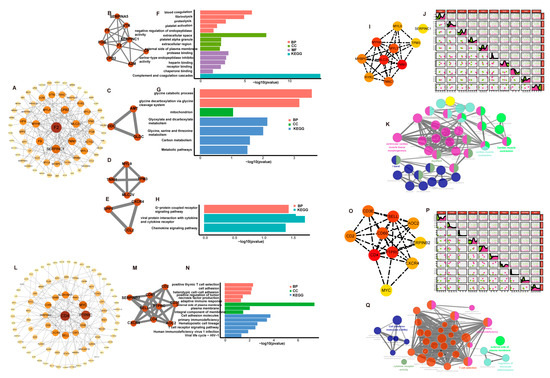
Figure 3.
Hub genes identification and analysis. (A) A PPI network including 152 nodes and 165 edges in low-IMF group. (B–E) Highly correlated module with the high score in low-IMF group. (F–H) Enrichment analysis results of these four clusters; the genes of third clusters were not enriched for any GO category or KEGG pathway in low-IMF group. (I) Ten hub genes were selected in the PPI network in low-IMF group. (J) The correlation of the ten hub genes in low-IMF group. (K) Significantly functional enrichment pathway of ten hub genes in low-IMF group. (L) A PPI network including 184 nodes and 150 edges in high-IMF group. (M) Highly correlated module with the high score in high-IMF group. (N) Enrichment analysis results of cluster in high-IMF group. (O) Ten hub genes were selected in the PPI network in high-IMF group. (P) The correlation of the ten hub genes in high-IMF group. (Q) Significantly functional enrichment pathway of ten hub genes in high-IMF group.

Table 2.
Identified overlapping DEGs.
3.2. DEG Functional Analysis
In the low-IMF group, GO analysis of DEGs of male and female pigs showed that 16 BPs, 5 CCs, and 7 MFs were significantly enriched (p < 0.05) (Supplementary Table S4). The top five GO terms with the smallest p values were shown in Figure 2F. The KEGG enrichment analysis of DEGs showed 12 significantly enriched pathways (p < 0.05) (Supplementary Table S4). The top 10 KEGG terms with the smallest p values were shown in Figure 2G. The up-regulated enriched terms were mainly related to muscle processes, such as ventricular cardiac muscle tissue morphogenesis, muscle contraction, transition between fast and slow fiber, and skeletal muscle contraction (Supplementary Table S5).
In the high-IMF group, GO analysis showed that a total of 5 BPs, 5 CCs, and 2 MFs were significantly enriched (Figure 2H, Supplementary Table S6). KEGG enrichment analysis of DEGs showed 15 significantly enriched pathways (Figure 2I, Supplementary Table S6). Up-regulated enriched terms were mainly related to immune-related processes, such as T cell receptor signaling pathway, JAK-STAT signaling pathway, and PI3K-Akt signaling pathway (Supplementary Table S7).
Only one of all enriched GO terms (GO:0010951: negative regulation of endopeptidase activity) was found to overlap in a functional enrichment analysis. The results also suggested that the molecular regulatory mechanisms of male and female pigs differ substantially between the low- and high-IMF groups.
3.3. PPI Network Construction
From the low-IMF group, 162 DEGs were submitted to the STRING database, which identified a PPI network with 152 nodes and 165 edges (Figure 3A). A highly correlated module analysis showed four modules in the whole PPI network with the highest score (score = 6) including 9 nodes and 24 edges (Figure 3B–E). Enrichment analysis findings revealed that these clusters’ genes were primarily enriched by signaling pathways including a fibrinolysis, glyoxylate and dicarboxylate metabolism; glycine, serine and threonine metabolism; and glycine catabolic process (Figure 3F–H).
From the high-IMF group, 202 DEGs were submitted to the STRING database, which identified a PPI network with 184 nodes and 150 edges (Figure 3L). One module (score = 7), including 9 nodes and 24 edges, was identified in the whole PPI network (Figure 3M). A cluster enrichment analysis revealed that functions were mainly associated with immune-related response and cell molecule adhesion, such as the positive regulation of interferon-γ/interleukin-2 production, positive regulation of T cell proliferation, regulation of cytokine biosynthetic process, and positive regulation of α-β T cell activation (Figure 3N).
3.4. Identification and Analysis of Hub Genes
In the low-IMF group, the top 10 genes with the highest MCC scores were TNNI1 (score = 1092), myosin Light Chain 2 (MLC2V) (score = 1032), myosin heavy chain 6(MYH6) (score = 1008), myosin light chain 3(MYL3) (score = 888), troponin C1(TNNC1) (score = 774), myosin binding protein C3(MYBPC3) (score = 744), ryanodine receptor 2(RYR2) (score = 722), MYL9 (score = 318), tropomyosin 3(TPM3) (score = 264), and SERPINC1 (score = 230) in the whole PPI network (Figure 3I), which were identified as hub genes. Apart from MYBPC3, RYR2, MYL9, and SERPINC1, the six remaining hub genes were up-regulated in males, compared with females (Figure 2A). The expression correlations among the 10 hub genes were shown in Figure 3J. The interactive relationships between hub genes and GO and KEGG terms were shown in Figure 3K.
In the high-IMF group, the top 10 genes were CD4 (score = 308), protein tyrosine phosphatase receptor type C (PTPRC) (score = 305), selectin l (SELL) (score = 289), CD69 molecule(CD69) (score = 288), CD3E molecule(CD3E) (score = 240), CD2 (score = 121), AOC2 (score = 120), C-X-C motif chemokine receptor 4(CXCR4) (score = 38), serpin family B member 2(SERPINB2) (score = 26), and MYC proto-oncogene (MYC) (score = 15) in the whole PPI network (Figure 3O). They were identified as hub genes, which were all up-regulated in males (Figure 2C). The expression correlations among the 10 hub genes were shown in Figure 3P. The interactive relationships between hub genes and enrichment terms were shown in Figure 3Q.
3.5. Identification of Potential Sex-Specific Biomarkers
In the low-IMF group, the LASSO algorithm analysis revealed three candidate genes, TNNI1, MYL9, and SERPINC1, as potential sex-specific biomarkers (Figure 4A). The TNNI1 expression was significantly up-regulated in males, compared with females, while MYL9 and SERPINC1 showed no significant differences in expression levels (Figure 4B–D). The ROC analysis based on the GSE144780 dataset showed that the AUCs of TNNI1, MYL9, and SERPINC1 related to sex were 1, 1, and 0.8889 (AUC > 0.7), respectively, all of which showed a higher effectiveness. In particular, TNNI1 may be the sex-specific biomarker with the highest potential in low-IMF pigs (Figure 4B–D). GSEA of the three potential biomarkers detected multiple pathways, including complement and coagulation cascades, cardiac muscle contraction, and a calcium signaling pathway consistent with the DEG functional analysis results. In addition, lipid metabolism pathways and myocardial tissue damage-associated pathways, such as the cholesterol metabolism, p53 signaling pathway, and butanoate metabolism pathway, were identified, indicating that lipid deposition and myocardial tissue statuses differ between males and females in low-IMF pigs. The GSEA results are shown in Figure 5A–C, Supplementary Table S8.
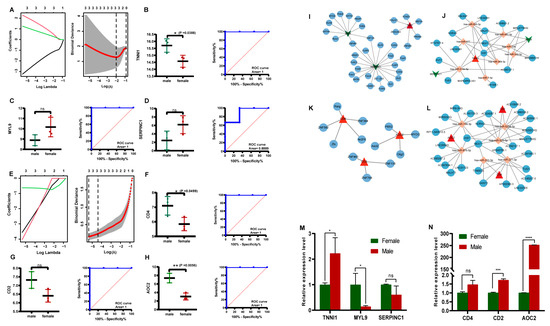
Figure 4.
Potential sex-specific biomarkers’ screening and validation. (A) LASSO regression model in low-IMF group. (B–D) Expression of potential sex-specific biomarkers in male and female pigs in low-IMF group; ROC curves of potential sex-specific biomarkers ((B) TNNI1; (C) MYL9; (D) SERPINC1). (E) LASSO regression model in high-IMF group. (F–H) Expression of potential sex-specific biomarkers in male and female pigs in high-IMF group; ROC curves of potential sex-specific biomarkers ((F) CD4; (G) CD2; (H) AOC2). (I) TFs-potential sex-specific biomarker network in low-IMF group, where the red triangles symbolize up-regulated potential biomarkers, the green arrows symbolize down-regulated potential biomarkers, and the blue nodes denote TFs. (J) lncRNA-miRNA-mRNA (potential biomarker) network in low-IMF group, where the red triangles symbolize up-regulated potential biomarkers, the green arrows symbolize down-regulated potential biomarkers, the blue nodes denote lncRNAs, and the orange nodes denote miRNAs. (K) TF-potential sex-specific biomarker network in high-IMF group. (L) lncRNA-miRNA-mRNA (potential biomarker) network in high-IMF group. Validation of mRNA expression levels of potential sex-specific biomarkers by qRT-PCR in semimembranosus muscles of Saba pigs for (M) low-IMF group, (N) high-IMF group. The symbol * means significant difference, **, *** and **** mean extremely significant difference, and ns means no significant difference.
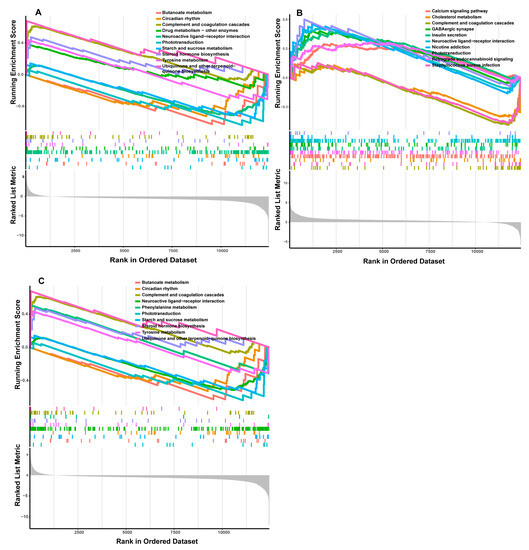
Figure 5.
GSEA functional analysis of potential sex-specific biomarkers in low-IMF group. ((A) TNNI1; (B) MYL9; (C) SERPINC1).
In the high-IMF group, three candidate genes, CD4, CD2, and AOC2, were identified as potential sex-specific biomarkers (Figure 4E). The CD4 and AOC2 expression was significantly up-regulated in males (Figure 4F–H). The AUCs of CD4, CD2, and AOC2 related to sex were all =1, all showing a higher effectiveness. In particular, AOC2 might be the sex-specific biomarker with the highest potential in high-IMF pigs (Figure 4F–H). GSEA analysis showed that potential sex-specific biomarkers in the high-IMF group were enriched in immune and inflammatory response-related pathways, such as the IL-17, and Toll-like receptor signaling pathways. Notably, males are more active than females in those pathways. In addition, lipid metabolism-related pathways, such as the regulation of lipolysis in adipocytes; PPAR signaling pathway and fat digestion and absorption; and immune/inflammation-mediated diseases, such as autoimmune thyroid disease and inflammatory bowel disease; were identified (Figure 6A–C, Supplementary Table S9). To summarize, we speculate that these sex-specific biomarkers may affect the IMF deposition by immune and inflammatory responses, as well as lipid metabolism.
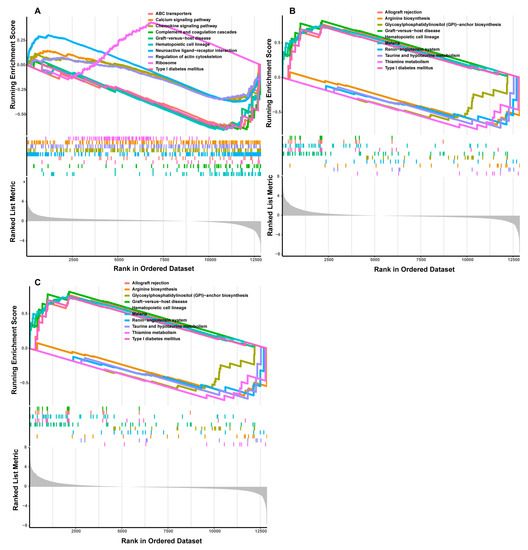
Figure 6.
GSEA functional analysis of potential sex-specific biomarkers in high-IMF group. ((A) CD4; (B) CD2; (C) AOC2).
3.6. Analysis of Hallmark Gene Sets in Difference Sex
In the low-IMF group, HALLMARK coagulation in males was lower than that in females, and HALLMARK_glycolysis was higher (Figure 7A). To the sex-specific biomarkers (Figure 7B), TNNI1 was significantly associated with estrogen_response_late (p < 0.05); MYL9 was positively correlated with complement, apoptosis, and androgen response (p < 0.05); and SERPINC1 was not observed with significant correlation with hallmark gene sets (p > 0.05).
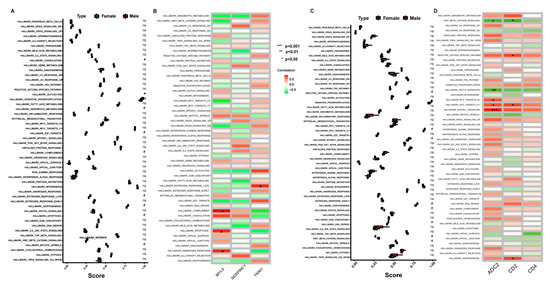
Figure 7.
Association analysis of potential sex-specific biomarkers with the hallmark gene sets. (A) ssGSEA score of hallmark gene sets and expression characteristics between male and female pigs in low-IMF group. (B) Correlation between hallmark gene sets and potential sex-specific biomarkers in low-IMF group. (C) ssGSEA score of hallmark gene sets and expression characteristics between male and female pigs in high-IMF group. (D) Correlation between hallmark gene sets and potential sex-specific biomarkers in high-IMF group.
In the high-IMF group, there were no significant differences between males and females in hallmark gene sets (Figure 7C). As shown in Figure 7D, AOC2 was negatively correlated with wnt_β_catenin_signaling (p < 0.05) and notch_signaling (p < 0.01) and positively associated with myc_targets_v2 (p < 0.05), myc_targets_v1 (p < 0.05), and mtotic1_signaling (p < 0.001). CD2 was negatively correlated with wnt_β_catenin_signaling (p < 0.05) and positively associated with oxygen species pathway (p < 0.05), myc_targets_v1 (p < 0.05), and adipogenesis (p < 0.05). However, no significant correlations were observed between CD4 and hallmark gene sets (p > 0.05)
3.7. Gene Regulatory Network Analysis of Potential Sex-Specific Biomarkers
In the low-IMF group, a total of 36 TFs were predicted based on potential sex-specific biomarkers, including six TFs predicted by TNNI1, 10 TFs predicted by MYL9, and 20 TFs predicted by SERPINC1. Subsequently, a potential sex-specific transcriptional regulatory network was constructed (Figure 4I). In the high-IMF group, a total of 12 TFs were predicted, including five TFs predicted by CD4, three TFs predicted by CD2, and four TFs predicted by AOC2. Finally, a TF-potential sex-specific biomarker network was constructed (Figure 4K).
3.8. CeRNA Network Analysis of Potential Sex-Specific Biomarkers
In the low-IMF group, a total of eight mRNA-miRNA pairs, including three mRNAs and eight miRNAs, were predicted to be related to three potential sex-specific biomarkers. The interacting lncRNAs were predicted according to these miRNAs, and six interaction pairs (including eight miRNAs and 16 lncRNAs) were predicted. Finally, the ceRNA network was constructed using Cytoscape (Figure 4J). In the high-IMF group, a total of six mRNA-miRNA relationship pairs, including three mRNAs and six miRNAs, were predicted. Then, 46 miRNA-lncRNA interaction pairs (including six miRNAs and 27 lncRNAs) were predicted, and a ceRNA network was constructed (Figure 4L).
3.9. IMF Content of SM in Saba Pigs
This set of samples consisted of 12 individuals, chosen from the population of thirty tested Saba pigs. For their extreme and divergent contents of SM IMF, the animals were divided into two groups, low- and high-IMF groups, each with six pigs (three males vs. three females), as shown in Table 3.

Table 3.
IMF content of Saba pigs. The results are expressed as mean ± SD. Different IMF level within the same gender were compared.
3.10. Validation of the Biomarkers via qRT-PCR
The expression trends for the potential sex-specific biomarkers TNNI1, MYL9, and SERPINC1 in low-IMF pigs and CD4, CD2, and AOC2 in high-IMF pigs in the SM tissues were consistent with the results of the transcriptome analysis. In addition, significant differences in mRNA expression levels of TNNI1 (p = 0.0266) and MYL9 (p = 0.0286) between the females and males in the low-IMF group were observed (Figure 4M), and CD2 (p = 0.0009) and AOC2 (p < 0.0001) in the high-IMF group were observed (Figure 4N).
4. Discussion
Pork is an important source of meat for human consumption, and IMF content is a key driver of meat quality and a polygenic characteristic in pigs. Previous studies have analyzed IMF-related hub genes and their functions [32,33,34], but the molecular mechanisms underlying IMF content still remain to be completely explored. In light of this, our study represented the pioneering application of bioinformatics methodologies to comprehensively investigate the molecular mechanisms underlying IMF deposition in male and female pigs with varying IMF levels within the same breed. The primary goals of the current work included hub gene screening, machine learning algorithm integration for potential sex-specific biomarker screening, functional analysis, and regulatory mechanism prediction of the obtained potential sex-specific biomarkers.
In the present study, 162 DEGs, including 64 up- and 98 down-regulated genes, were detected between males and females in the low-IMF group; 202 DEGs, including 147 up- and 55 down-regulated genes, were screened in the high-IMF group. Eleven overlapping DEGs were identified in low- vs. high-IMF pigs. Among the overlapping genes, GADD45GIP1 and ITIH1 were up-regulated in high-IMF male pigs, but down-regulated in low-IMF male pigs; thus, they might play opposite regulatory roles in male and female pigs with different IMF contents. GADD45GIP1, also known as adipocyte-specific Crif1, plays a vital role in regulating adipocyte oxidative phosphorylation function [35]. ITIH1 is a member of the inter-α-trypsin inhibitor family of proteins that has been implicated in multiple inflammatory diseases. The ITIH family (ITIH1, ITIH2, ITIH3, and ITIH4) is considered crucial for maintaining the uterine surface glycocalyx during placental attachment in pigs, according to a prior study [36]. Up to now, studies of GADD45GIP1 and ITIH1 on pork IMF contents of different sexes have not been reported. Our study may provide a novel insight into the role of GADD45GIP1 and ITIH1 in IMF deposition between the sexes, which requires further exploration and confirmation.
4.1. Low-IMF Group Analysis
Functional analysis indicated that the DEGs in the low-IMF group were primarily associated with BPs, such as ventricular cardiac muscle tissue morphogenesis, biomineral tissue development, cardiac muscle contraction, and muscle contraction. These GO terms correspond with previous studies identifying DEGs related to modulating adipogenesis in intramuscular adipocytes [37], the impact of muscle contraction on intramuscular adipose tissue [38], muscle fiber composition [39], and fat deposition in pigs [40]. KEGG enrichment analysis revealed that these genes were mainly involved in immunity and inflammation-relevant pathways, including complement and coagulation cascades and platelet activation. Complement systems and coagulation cascades might play a vital role in post-trauma and subsequent inflammatory reactions as innate immunity elements [41]. A previous study observed greater IMF deposition in Korean cattle after castration, which might lead to alterations in the complement and coagulation cascade pathways, possibly owing to obesity and the subsequent inflammation [42]. Platelet activation is regulated by the complement system, and platelet activity is mostly linked to the start of coagulation cascades [43].
Subsequently, 10 hub genes were selected from the PPI network via the topology analysis method—MCC, and were mainly involved in muscle fiber and tissue development-associated processes, especially in the development of muscle organs, such as cardiac chamber morphogenesis, ventricular cardiac muscle tissue development, and cardiac muscle tissue morphogenesis; this was consistent with the results of our previous functional analysis of DEGs. Furthermore, TNNI1, MYL9, and SERPINC1 genes were identified as potential sex-specific biomarkers in low-IMF pigs using the LASSO algorithm. Among them, TNNI1 might be the sex-specific biomarker in low-IMF pigs with the highest potential. The functional analysis of TNNI1 also indicated that it might be involved in the muscle regulatory response. TNNI1 is a slow-twitch skeletal muscle isoform expressed solely in slow-twitch skeletal fibers and is related to the muscle fiber type. A previous study has shown that TNNI1 gene polymorphisms were related to IMF content and meat color in the biceps femoris of pigs [39]. Additionally, abundant TNNI1 expression was detected in L-arginine-supplemented pigs [44], suggesting that TNNI1 might play a positive regulatory role in IMF content and meat quality improvement. Abundant TNNI1 expression was also found in cardiac muscles [45]. One study demonstrated that TNNI1 could act as a marker of pork quality [46]. In our study, TNNI1 was expressed at a higher level in males, and it had a significant positive correlation with TPM3, TNNC1, and TPM3 expression levels, which were related to muscle formation and development [47,48,49]. MYL9, which encodes myosin light chain, may regulate muscle contraction by modulating the ATPase activity of myosin heads. Previous studies have shown that MYL9 was correlated with ATP kinase activity regulation, muscle cell control, and signal transduction [50]. Several studies have found that MYL9 displayed higher transcriptome and proteome expression levels in myostatin-knockout Meishan pigs, and it was hypothesized that MYL9 might play an important role in skeletal muscle growth and development [51]. SERPINC1, a member of the serpin superfamily, contributes to the regulation of the blood coagulation cascade. A previous study showed higher SERPINC1 expression in female vs. male livers due to the different growth hormone secretion patterns between the sexes [52]. A recent study showed that SERPINC1 played a critical role in immune responses [53]. SERPINC1 expression is inherently associated with immune cell infiltration by B cells, T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells [54]. Reports have shown that the SERPINC1 gene was essentially related to fat metabolism and development during beef cattle growth [55]. Although no studies have reported a direct correlation between TNNI1, MYL9, SERPINC1 and muscle formation, and IMF deposition in male and female low-IMF pigs, we speculated that these biomarkers might be involved in advancing IMF deposition through their involvement in muscle development based on previous studies. Our results also further indicate that TNNI1, MYL9, and SERPINC1 are closely associated with muscle development and IMF content.
The GSEA functions of TNNI1, MYL9, and SERPINC1 included muscle development-related and immunity-related pathways, such as cardiac muscle contraction, calcium signaling pathway, complement and coagulation cascades, and bile secretion, which are consistent with the DEG functional analysis results described above. We also identified energy metabolism-related signaling pathways, such as the cholesterol metabolism, cAMP signaling pathway, TCA cycle, and p53 signaling pathway. CAMP, synthesized from ATP by the adenylate cyclase family, played a crucial role in both adipogenesis and lipid allocation in adipose tissue [56]. CAMP plays a vital role in the interaction between muscle contraction and calcium signaling and contributes to glycogenolysis to meet the energy requirements for muscles [57]. Previous studies have shown that cAMP pathway activation in muscles contributed to improved myofiber size and muscle strength [58]. The TCA cycle is a significant metabolic network that provides energy and supports precursors for synthetic processes and reducing factors that fuel energy production [59]. The TCA cycle is associated with IMF deposition in castrated cattle [60]. P53 was vital in regulating metabolic pathways, and its increased expression was found in obese rats and humans [61].
In addition, the qRT-PCR results showed that the expression of TNNI1 and MYL9 differed significantly between male and female in low-IMF pigs, which further indicated that TNNI1 and MYL9 might be reliable biomarkers. Especially in the TNNI1, the expression was significantly up-regulated in the male pigs in qRT-PCR and RNA-seq results.
4.2. High-IMF Group Analysis
In the high-IMF group, BPs of DEGs were mainly enriched in inflammation/ immunization-related signaling pathways, such as the regulation of interleukin-8 (IL-8) production and leukocyte migration. IL-8 facilitates macrophage infiltration in adipose tissue, which induces topical and systemic inflammation [62]. Obese individuals with a worsened glycometabolic profile and increased inflammation linked to fat tissue show higher expression of IL-8, an inflammation-related adipokine [63,64]. KEGG enrichment analysis revealed that DEGs were mainly involved in lipid metabolism-related processes, such as the MAPK signaling pathway, cytokine–cytokine receptor interaction, and JAK-STAT signaling pathway; cellular immune regulation-associated pathways, such as the T cell receptor signaling; and cell function regulation-associated pathways, such as the PI3K-Akt signaling. The MAPK pathway is one of the critical signaling systems that mediates cellular responses to the external environment and has an influential role not only in the proliferation and differentiation of adipocytes, but also in the development of muscle fibers, affecting sarcomeric traits by regulating the muscle fiber type [65,66]. A recent study also showed that MAPK signaling might be crucial for angiogenesis and adipogenesis in the male pig’s subcutaneous adipose tissue [67]. Cytokine–cytokine receptor interaction signaling pathways influence IMF deposition by regulating the upstream PPAR signaling pathway in lipid metabolism [68,69]. The JAK-STAT signaling pathway plays a vital role in IMF deposition by regulating myocyte differentiation and proliferation, thereby affecting meat quality in pigs and goats [70,71].
In addition, ten hub genes were selected from the PPI network. The functional analysis results of the above DEGs were supported by the hub genes’ considerable associations with immunity and inflammation regulation-related processes, such as the regulation of IL-8 production, T cell receptor signaling pathway, and primary immunodeficiency. Subsequently, CD4, CD2, and AOC2 genes were selected as potential sex-specific biomarkers in high-IMF pigs and further investigated. Among them, AOC2 may be the most promising sex-specific biomarker in high-IMF pigs. CD4, which encodes the CD4 membrane glycoprotein of T lymphocytes, is primarily expressed in T lymphocytes. Some investigations have shown the association between CD4+ T cells and the progression of obesity and obesity-related diseases, suggesting the role of CD4 in controlling immune and adipose tissue [72]. Activated or decreased CD4+ T cells in adipose tissue play a vital role in maintaining the pro-inflammatory state in obesity [73]. Previous studies have shown that CD4+ T cells might be activated through the expression of the main histocompatibility complex class II and co-stimulatory membrane receptors of adipocytes to satisfy the adaptive immune response that adjusted adipose tissue inflammation [74]. Additionally, the function of CD4+ T cells is regulated by adipocyte-derived factors, such as adipokines and lipids [75]. CD2, a glycoprotein expressed on the surface of most human T cells and natural killer cells, is crucial for mediating cell attachment in both T-lymphocytes and in signal transmission [76]. CD2 is crucial during the initial steps of the immune response. Several studies have documented the role of anti-CD2 monoclonal antibodies in interfering with afferent immunity, inducing CD2-antigenicity and downregulating CD3, CD4, and CD8 cell surface expression [77]. AOC2 is involved in oxidation by cytochrome P450 and meta-pathway biotransformation phases I and II, which are associated with median neuropathy and amyotrophic lateral sclerosis. The up-regulated AOC2 expression during adipocyte differentiation in vitro showed that this gene might play a vital role in adipogenesis [78,79]. Studies on gene transcriptional changes of human monoamine oxidases and adipocyte-specific markers during adipogenesis in human bone marrow mesenchymal stem cells observed that the expression pattern of AOC2 was similar to that of the adiponectin and PPARc biomarkers [80]. However, there are few studies on muscle formation and IMF deposition-related CD4, CD2, and AOC2 in male and female high-IMF pigs at present. The GSEA functions of CD4, CD2, and AOC2 included IMF deposition and immunity-related pathways, such as the JAK-STAT and T cell receptor signaling pathways; this was consistent with the results of the above DEG functional analysis.
In addition, both RNA-seq and qRT-PCR results revealed that AOC2 was significantly up-regulated in male pigs of the high-IMF group, which further indicated that AOC2 might be a reliable sex-specific biomarker.
Although sex-specific biomarkers have been identified, and some genes have been validated in previous studies, some limitations of the current study must be noted. First, the sex-specific biomarkers were obtained through a bioinformatics method; biomarker expression levels should be validated via the assessment of larger sample sizes and more pig breeds via more accurate methods. Second, the specific functions of biomarkers need to be verified using overexpression or knockdown methods in a cell or animal.
5. Conclusions
In summary, we screened potential sex-specific biomarkers in male and female pigs in the low-IMF (TNNI1, MYL9, and SERPINC1) and high-IMF groups (CD4, CD2, and AOC2). In the low-IMF group, TNNI1 was evaluated as the most valuable biomarker. AOC2 was evaluated as the most valuable biomarker in the high-IMF group. The different expression patterns of DEGs, hub genes, and potential sex-specific biomarkers were shown in the male and female pigs with different IMF level, with more genes enriched in low-IMF pigs for muscle fiber and organ-formation-associated processes, such as cardiac chamber morphogenesis, ventricular cardiac muscle tissue development, and muscle contraction, but with more genes enriched in high-IMF pigs for metabolic processes, inflammation, and the immune system, such as the MAPK signaling pathway, cytokine-cytokine receptor interaction, JAK-STAT signaling pathway, the regulation of IL-8 production, and the T cell receptor signaling pathway. The results of the present study showed that IMF deposition was closely related to sex. In addition, there were differences in the molecular and regulatory mechanisms of IMF deposition between male and female pigs with varying IMF levels. These findings provided new insights into the molecular mechanisms of pig IMF deposition and meat quality improvement.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes14091695/s1, Table S1: Primer information for potential sex-specific biomarkers and housekeeping gene GAPDH. Table S2: Total differentially expressed genes between male and female pigs in the low-IMF group. Table S3: Total differentially expressed genes between male and female pigs in the high-IMF group. Table S4: GO enrichment and KEGG pathway enrichment of differentially expressed genes between males and females in the low-IMF group. Table S5: The up- and down-regulated enrichment items of DEGs between males and females in the low-IMF group. Table S6: GO enrichment and KEGG pathway enrichment of differentially expressed genes between males and females in the high-IMF group. Table S7: The up- and down-regulated enrichment items of DEGs between males and females in the high-IMF group. Table S8: The GSEA of the three potential biomarkers in the low-IMF group. Table S9: The GSEA of the three potential biomarkers in the high-IMF group.
Author Contributions
Data curation, Y.Y. and X.W.; Formal analysis, Y.Y. and X.W.; Funding acquisition, S.L.; Investigation, S.W. and Q.C.; Methodology, M.L. and S.L.; Validation, Y.Y. and X.W.; Writing—original draft, Y.Y. and X.W.; Writing—review and editing, M.L. and S.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key Research and Development Program of China, awarded to S.L. (2022YFD1601902), Yunnan Swine Industry Technology System Program, awarded to S.L. (2020KJTX0016), Yunnan Province Important National Science & Technology Specific Projects, awarded to S.L. (202104BI090022, 202102AE090039), Yunnan Basic Research Programme Projects, awarded to X.W. (202001AU070113). These funding agencies had no role in the research design, data gathering, data analysis, or writing the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets (GSE144780) for this study can be found in the in the public NCBI GEO database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE144780).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Suzuki, K.; Irie, M.; Kadowaki, H.; Shibata, T.; Kumagai, M.; Nishida, A. Genetic parameter estimates of meat quality traits in Duroc pigs selected for average daily gain, longissimus muscle area, backfat thickness, and intramuscular fat content. J. Anim. Sci. 2005, 83, 2058–2065. [Google Scholar] [CrossRef] [PubMed]
- Aaslyng, M.; Oksama, M.; Olsen, E.; Bejerholm, C.; Baltzer, M.; Andersen, G.; Bredie, W.; Byrne, D.; Gabrielsen, G. The impact of sensory quality of pork on consumer preference. Meat Sci. 2007, 76, 61–73. [Google Scholar] [CrossRef]
- Hou, X.; Yang, Y.; Zhu, S.; Hua, C.; Zhou, R.; Mu, Y.; Tang, Z.; Li, K. Comparison of skeletal muscle miRNA and mRNA profiles among three pig breeds. Mol. Genet. Genom. 2016, 291, 559–573. [Google Scholar] [CrossRef] [PubMed]
- Tyra, M.; Ropka-Molik, K.; Terman, A.; Piórkowska, K.; Oczkowicz, M.; Bereta, A. Association between subcutaneous and intramuscular fat content in porcine ham and loin depending on age, breed and FABP3 and LEPR genes transcript abundance. Mol. Biol. Rep. 2013, 40, 2301–2308. [Google Scholar] [CrossRef]
- Lambe, N.; Wood, J.; McLean, K.; Walling, G.; Whitney, H.; Jagger, S.; Fullarton, P.; Bayntun, J.; Bünger, L. Effects of low protein diets on pigs with a lean genotype 2. Compositional traits measured with computed tomography (CT). Meat Sci. 2013, 95, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Škrlep, M.; Batorek, N.; Bonneau, M.; Prevolnik, M.; Kubale, V.; Čandek-Potokar, M. Effect of immunocastration in group-housed commercial fattening pigs on reproductive organs, malodorous compounds, carcass and meat quality. Czech J. Anim. Sci. 2012, 57, 290–299. [Google Scholar] [CrossRef]
- Ellis, M.; Webb, A.; Avery, P.; Brown, I. The influence of terminal sire genotype, sex, slaughter weight, feeding regime and slaughter-house on growth performance and carcass and meat quality in pigs and on the organoleptic properties of fresh pork. Anim. Sci. 1996, 62, 521–530. [Google Scholar] [CrossRef]
- Wang, W.; Xue, W.; Jin, B.; Zhang, X.; Ma, F.; Xu, X. Candidate gene expression affects intramuscular fat content and fatty acid composition in pigs. J. Appl. Genet. 2013, 54, 113–118. [Google Scholar] [CrossRef]
- Wang, Y.; Li, X.; Cao, Y.; Xiao, C.; Liu, Y.; Jin, H.; Cao, Y. Effect of the ACAA1 gene on preadipocyte differentiation in sheep. Front. Genet. 2021, 12, 649140. [Google Scholar] [CrossRef]
- Ren, H.; Zhang, H.; Hua, Z.; Zhu, Z.; Tao, J.; Xiao, H.; Zhang, L.; Bi, Y.; Wang, H. ACSL4 Directs Intramuscular Adipogenesis and Fatty Acid Composition in Pigs. Animals 2022, 12, 119. [Google Scholar] [CrossRef]
- Albuquerque, A.; Ovilo, C.; Nunez, Y.; Benitez, R.; Lopez-Garcia, A.; Garcia, F.; Felix, M.D.; Laranjo, M.; Charneca, R.; Martins, J.M. Comparative Transcriptomic Analysis of Subcutaneous Adipose Tissue from Local Pig Breeds. Genes 2020, 11, 422. [Google Scholar] [CrossRef]
- Zappaterra, M.; Gioiosa, S.; Chillemi, G.; Zambonelli, P.; Davoli, R. Dissecting the gene expression networks associated with variations in the major components of the fatty acid semimembranosus muscle profile in large white heavy pigs. Animals 2021, 11, 628. [Google Scholar] [CrossRef]
- Lim, K.; Lee, K.; Park, J.; Chung, W.; Jang, G.; Choi, B.; Hong, K.C.; Kim, T. Identification of differentially expressed genes in longissimus muscle of pigs with high and low intramuscular fat content using RNA sequencing. Anim. Genet. 2017, 48, 166–174. [Google Scholar] [CrossRef]
- Bahelka, I.; Hanusová, E.; Peskovicova, D.; Demo, P. The effect of sex and slaughter weight on intramuscular fat content and its relationship to carcass traits of pigs. Czech J. Anim. Sci. 2007, 52, 122–129. [Google Scholar] [CrossRef]
- Latorre, M.; Lázaro, R.; Gracia, M.; Nieto, M.; Mateos, G. Effect of sex and terminal sire genotype on performance, carcass characteristics, and meat quality of pigs slaughtered at 117 kg body weight. Meat Sci. 2003, 65, 1369–1377. [Google Scholar] [CrossRef]
- Serrano, M.; Valencia, D.; Nieto, M.; Lázaro, R.; Mateos, G. Influence of sex and terminal sire line on performance and carcass and meat quality of Iberian pigs reared under intensive production systems. Meat Sci. 2008, 78, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Alonso, V.; del Mar Campo, M.; Español, S.; Roncalés, P.; Beltrán, J.A. Effect of crossbreeding and gender on meat quality and fatty acid composition in pork. Meat Sci. 2009, 81, 209–217. [Google Scholar] [CrossRef]
- Channon, H.A.; Kerr, M.G.; Walker, P.J. Effect of Duroc content, sex and ageing period on meat and eating quality attributes of pork loin. Meat Sci. 2004, 66, 881–888. [Google Scholar] [CrossRef]
- Font-i-Furnols, M.; Brun, A.; Gispert, M. Intramuscular fat content in different muscles, locations, weights and genotype-sexes and its prediction in live pigs with computed tomography. Animal 2019, 13, 666–674. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Bader, G.D.; Hogue, C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef]
- Chin, C.-H.; Chen, S.-H.; Wu, H.-H.; Ho, C.-W.; Ko, M.-T.; Lin, C.-Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8, S11. [Google Scholar] [CrossRef]
- Tibshirani, R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The molecular signatures database hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- John, B.; Enright, A.J.; Aravin, A.; Tuschl, T.; Sander, C.; Marks, D.S. Human microRNA targets. PLoS Biol. 2004, 2, e363. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Loher, P.; Rigoutsos, I. Interactive exploration of RNA22 microRNA target predictions. Bioinformatics 2012, 28, 3322–3323. [Google Scholar] [CrossRef]
- Vejnar, C.E.; Zdobnov, E.M. MiRmap: Comprehensive prediction of microRNA target repression strength. Nucleic Acids Res. 2012, 40, 11673–11683. [Google Scholar] [CrossRef]
- Essengustavsson, B.; Karlsson, A.; Lundstrom, K.; Enfalt, A.C. Intramuscular fat and muscle-fiber lipid contents in halothane-gene-free pigs fed high or low-protein diets and its relations to meat quality. Meat Sci. 1994, 38, 269–277. [Google Scholar] [CrossRef]
- Muñoz, M.; García-Casco, J.M.; Caraballo, C.; Fernández-Barroso, M.Á.; Sánchez-Esquiliche, F.; Gómez, F.; Rodriguez, M.D.C.; Silió, L. Identification of candidate genes and regulatory factors underlying intramuscular fat content through longissimus dorsi transcriptome analyses in heavy Iberian pigs. Front. Genet. 2018, 9, 608. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Zhang, X.; Li, A.; Xie, L.; Miao, X. Genome-wide analysis of mRNAs and lncRNAs of intramuscular fat related to lipid metabolism in two pig breeds. Cell. Physiol. Biochem. 2018, 50, 2406–2422. [Google Scholar] [CrossRef] [PubMed]
- Zou, C.; Li, L.; Cheng, X.; Li, C.; Fu, Y.; Fang, C.; Li, C. Identification and functional analysis of long intergenic non-coding RNAs underlying intramuscular fat content in pigs. Front. Genet. 2018, 9, 102. [Google Scholar] [CrossRef]
- Choi, M.J.; Jung, S.-B.; Lee, S.E.; Kang, S.G.; Lee, J.H.; Ryu, M.J.; Chung, H.K.; Chang, J.Y.; Kim, Y.K.; Hong, H.J. An adipocyte-specific defect in oxidative phosphorylation increases systemic energy expenditure and protects against diet-induced obesity in mouse models. Diabetologia 2020, 63, 837–852. [Google Scholar] [CrossRef] [PubMed]
- Geisert, R.; Ashworth, M.; Malayer, J. Expression of inter-alpha-trypsin inhibitor heavy chains in endometrium of cyclic and pregnant gilts. Reproduction 2003, 126, 621–627. [Google Scholar] [CrossRef]
- Wu, W.; Zhang, D.; Yin, Y.; Ji, M.; Xu, K.; Huang, X.; Peng, Y.; Zhang, J. Comprehensive transcriptomic view of the role of the LGALS12 gene in porcine subcutaneous and intramuscular adipocytes. BMC Genom. 2019, 20, 509. [Google Scholar] [CrossRef]
- Biltz, N.K.; Collins, K.H.; Shen, K.C.; Schwartz, K.; Harris, C.A.; Meyer, G.A. Infiltration of intramuscular adipose tissue impairs skeletal muscle contraction. J. Physiol. 2020, 598, 2669–2683. [Google Scholar] [CrossRef]
- Yang, H.; Xu, Z.; Lei, M.; Li, F.; Deng, C.; Xiong, Y.; Zuo, B. Association of 3 polymorphisms in porcine troponin I genes (TNNI1 and TNNI2) with meat quality traits. J. Appl. Genet. 2010, 51, 51–57. [Google Scholar] [CrossRef]
- Piórkowska, K.; Żukowski, K.; Ropka-Molik, K.; Tyra, M.; Gurgul, A. A comprehensive transcriptome analysis of skeletal muscles in two Polish pig breeds differing in fat and meat quality traits. Genet. Mol. Biol. 2018, 41, 125–136. [Google Scholar] [CrossRef]
- Amara, U.; Rittirsch, D.; Flierl, M.; Bruckner, U.; Klos, A.; Gebhard, F.; Lambris, J.D.; Huber-Lang, M. Interaction between the coagulation and complement system. Adv. Exp. Med. Biol. 2008, 632, 68–76. [Google Scholar] [CrossRef]
- Na, S.W.; Park, S.J.; Hong, S.J.; Baik, M. Transcriptome changes associated with fat deposition in the longissimus thoracis of Korean cattle following castration. J. Anim. Physiol. Anim. Nutr. 2020, 104, 1637–1646. [Google Scholar] [CrossRef] [PubMed]
- Rawish, E.; Sauter, M.; Sauter, R.; Nording, H.; Langer, H.F. Complement, inflammation and thrombosis. Brit. J. Pharmacol. 2021, 178, 2892–2904. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zheng, C.; Hu, Y.; Wang, L.; Yang, X.; Jiang, Z. Dietary L-arginine supplementation affects the skeletal longissimus muscle proteome in finishing pigs. PLoS ONE 2015, 10, e0117294. [Google Scholar] [CrossRef]
- Yang, H.; Xu, Z.; Lei, M.; Li, F.; Deng, C.; Xiong, Y.; Zuo, B. Real-time reverse transcription–PCR expression profiling of porcine troponin I family in three different types of muscles during development. Mol. Biol. Rep. 2011, 38, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Pierzchala, M.; Hoekman, A.; Urbanski, P.; Kruijt, L.; Kristensen, L.; Young, J.; Oksbjerg, N.; Goluch, D.; Te Pas, M. Validation of biomarkers for loin meat quality (M. longissimus) of pigs. J. Anim. Breed. Genet. 2014, 131, 258–270. [Google Scholar] [CrossRef] [PubMed]
- Ropka-Molik, K.; Żukowski, K.; Eckert, R.; Gurgul, A.; Piórkowska, K.; Oczkowicz, M. Comprehensive analysis of the whole transcriptomes from two different pig breeds using RNA-Seq method. Anim. Genet. 2014, 45, 674–684. [Google Scholar] [CrossRef]
- Chen, G.; Su, Y.; Cai, Y.; He, L.; Yang, G. Comparative transcriptomic analysis reveals beneficial effect of dietary mulberry leaves on the muscle quality of finishing pigs. Vet. Med. Sci. 2019, 5, 526–535. [Google Scholar] [CrossRef]
- Jing, L.; Hou, Y.; Wu, H.; Miao, Y.; Li, X.; Cao, J.; Michael Brameld, J.; Parr, T.; Zhao, S. Transcriptome analysis of mRNA and miRNA in skeletal muscle indicates an important network for differential Residual Feed Intake in pigs. Sci. Rep. 2015, 5, 11953. [Google Scholar] [CrossRef]
- Tan, X.; Chen, M. MYLK and MYL9 expression in non-small cell lung cancer identified by bioinformatics analysis of public expression data. Tumor Biol. 2014, 35, 12189–12200. [Google Scholar] [CrossRef]
- Li, X.; Xie, S.; Qian, L.; Cai, C.; Bi, H.; Cui, W. Identification of genes related to skeletal muscle growth and development by integrated analysis of transcriptome and proteome in myostatin-edited Meishan pigs. J. Proteom. 2020, 213, 103628. [Google Scholar] [CrossRef]
- Wong, J.H.; Dukes, J.; Levy, R.E.; Sos, B.; Mason, S.E.; Fong, T.S.; Weiss, E.J. Sex differences in thrombosis in mice are mediated by sex-specific growth hormone secretion patterns. J. Clin. Investig. 2008, 118, 2969–2978. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhang, D.; Li, X.; Zhang, Y.; Zhao, Y.; Xu, D.; Cheng, J.; Wang, J.; Li, W.; Lin, C. Comparative proteomics reveals genetic mechanisms of body weight in Hu sheep and Dorper sheep. J. Proteom. 2022, 267, 104699. [Google Scholar] [CrossRef]
- Li, M.; Wang, Z.; Zhu, L.; Shui, Y.; Zhang, S.; Guo, W. Down-regulation of RBP4 indicates a poor prognosis and correlates with immune cell infiltration in hepatocellular carcinoma. Biosci. Rep. 2021, 41, BSR20210328. [Google Scholar] [CrossRef] [PubMed]
- Romao, J.; He, M.; McAllister, T.; Guan, L. Effect of age on bovine subcutaneous fat proteome: Molecular mechanisms of physiological variations during beef cattle growth. J. Anim. Sci. 2014, 92, 3316–3327. [Google Scholar] [CrossRef] [PubMed]
- Petersen, R.K.; Madsen, L.; Pedersen, L.M.; Hallenborg, P.; Hagland, H.; Viste, K.; Døskeland, S.O.; Kristiansen, K. Cyclic AMP (cAMP)-mediated stimulation of adipocyte differentiation requires the synergistic action of Epac-and cAMP-dependent protein kinase-dependent processes. Mol. Cell. Biochem. 2008, 28, 3804–3816. [Google Scholar] [CrossRef]
- Ravnskjaer, K.; Madiraju, A.; Montminy, M. Role of the cAMP pathway in glucose and lipid metabolism. Metab. Control 2016, 233, 29–49. [Google Scholar] [CrossRef]
- Chen, M.; Feng, H.-Z.; Gupta, D.; Kelleher, J.; Dickerson, K.E.; Wang, J.; Hunt, D.; Jou, W.; Gavrilova, O.; Jin, J.-P. Gsα deficiency in skeletal muscle leads to reduced muscle mass, fiber-type switching, and glucose intolerance without insulin resistance or deficiency. Am. J. Physiol.-Cell Ph. 2009, 296, C930–C940. [Google Scholar] [CrossRef]
- Akram, M. Citric Acid Cycle and Role of its Intermediates in Metabolism. Cell Biochem. Biophys. 2014, 68, 475–478. [Google Scholar] [CrossRef]
- Jeong, J.; Bong, J.; Kim, G.D.; Joo, S.T.; Lee, H.J.; Baik, M. Transcriptome changes favoring intramuscular fat deposition in the longissimus muscle following castration of bulls. J. Anim. Sci. 2013, 91, 4692–4704. [Google Scholar] [CrossRef]
- Krstic, J.; Reinisch, I.; Schupp, M.; Schulz, T.J.; Prokesch, A. p53 Functions in Adipose Tissue Metabolism and Homeostasis. Int. J. Mol. Sci. 2018, 19, 2622. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, R.; Yamamoto, T.; Sakamoto, A.; Ishimaru, Y.; Narahara, S.; Sugiuchi, H.; Yamaguchi, Y. Chemokine profiles of human visceral adipocytes from cryopreserved preadipocytes: Neutrophil activation and induction of nuclear factor-kappa B repressing factor. Life Sci. 2015, 143, 225–230. [Google Scholar] [CrossRef]
- Trayhurn, P. Endocrine and signalling role of adipose tissue: New perspectives on fat. Acta Physiol. Scand. 2005, 184, 285–293. [Google Scholar] [CrossRef]
- Cimini, F.A.; Barchetta, I.; Porzia, A.; Mainiero, F.; Costantino, C.; Bertoccini, L.; Ceccarelli, V.; Morini, S.; Baroni, M.G.; Lenzi, A.; et al. Circulating IL-8 levels are increased in patients with type 2 diabetes and associated with worse inflammatory and cardiometabolic profile. Acta Diabetol. 2017, 54, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.X.; Wang, H.H.; Jiang, B.C.; Zhao, Y.Y.; Xie, Z.R.; Xiong, K.; Chen, J. Global comparison of gene expression between subcutaneous and intramuscular adipose tissue of mature Erhualian pig. Genet. Mol. Res. 2013, 12, 5085–5101. [Google Scholar] [CrossRef]
- Wu, H.; Olson, E.N. Activation of the MEF2 transcription factor in skeletal muscles from myotonic mice. J. Clin. Investig. 2002, 109, 1327–1333. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Y.; Wang, X.Y.; Li, M.L.; Wang, S.Y.; Chen, Q.; Lu, S.X. Identification of key sex-specific pathways and genes in the subcutaneous adipose tissue from pigs using WGCNA method. BMC Genom. Data 2022, 23, 35. [Google Scholar] [CrossRef]
- Liu, L.; Cui, H.X.; Fu, R.Q.; Zheng, M.Q.; Liu, R.R.; Zhao, G.P.; Wen, J. The regulation of IMF deposition in pectoralis major of fast- and slow- growing chickens at hatching. J. Anim. Sci. Biotechnol. 2017, 8, 77. [Google Scholar] [CrossRef]
- Szatmari, I.; Töröcsik, D.; Agostini, M.; Nagy, T.; Gurnell, M.; Barta, E.; Chatterjee, K.; Nagy, L. PPARγ regulates the function of human dendritic cells primarily by altering lipid metabolism. Blood 2007, 110, 3271–3280. [Google Scholar] [CrossRef]
- Qi, K.L.; Liu, Y.K.; Li, C.L.; Li, X.J.; Li, X.L.; Wang, K.J.; Qiao, R.M.; Han, X.L. Construction of circRNA-related ceRNA networks in longissimus dorsi muscle of Queshan Black and Large White pigs. Mol. Genet. Genom. 2022, 297, 101–112. [Google Scholar] [CrossRef]
- Shen, J.Y.; Hao, Z.Y.; Wang, J.Q.; Hu, J.; Liu, X.; Li, S.B.; Ke, N.; Song, Y.Z.; Lu, Y.J.; Hu, L.Y.; et al. Comparative transcriptome profile analysis of longissimus dorsi muscle tissues from two goat breeds with different meat production performance using RNA-Seq. Front. Genet. 2021, 11, 619399. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.S.; Lin, L.; Li, J.; Xiao, Z.G.; Chen, B.; Wan, L.; Li, M.X.; Wu, X.; Cho, C.H.; Shen, J. CD4(+) T cells in obesity and obesity-associated diseases. Cell. Mol. Immunol. 2018, 332, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Ioan-Facsinay, A.; Kwekkeboom, J.C.; Westhoff, S.; Giera, M.; Rombouts, Y.; van Harmelen, V.; Huizinga, T.W.J.; Deelder, A.; Kloppenburg, M.; Toes, R.E.M. Adipocyte-derived lipids modulate CD4(+) T-cell function. Eur. J. Immunol. 2013, 43, 1578–1587. [Google Scholar] [CrossRef]
- Deng, T.; Lyon, C.J.; Minze, L.J.; Lin, J.X.; Zou, J.; Liu, J.Z.; Ren, Y.L.; Yin, Z.; Hamilton, D.J.; Reardon, P.R.; et al. Class II Major Histocompatibility Complex Plays an Essential Role in Obesity-Induced Adipose Inflammation. Cell Metab. 2013, 17, 411–422. [Google Scholar] [CrossRef]
- Cohen, S.; Danzaki, K.; MacIver, N.J. Nutritional effects on T-cell immunometabolism. Eur. J. Immunol. 2017, 47, 225–235. [Google Scholar] [CrossRef]
- Yang, J.J.; Ye, Y.M.; Carroll, A.; Yang, W.; Lee, H. Structural biology of the cell adhesion protein CD2: Alternatively folded states and structure-function relation. Curr. Protein Pept. Sci. 2001, 2, 1–17. [Google Scholar] [CrossRef]
- Chavin, K.D.; Lau, H.T.; Bromberg, J.S. Prolongation of allograft and xenograft survival in mice by anti-CD2 monoclonal antibodies. Transplantation 1992, 54, 286–291. [Google Scholar] [CrossRef]
- Bour, S.; Daviaud, D.; Gres, S.; Lefort, C.; Prevot, D.; Zorzano, A.; Wabitsch, M.; Saulnier-Blache, J.S.; Valet, P.; Carpene, C. Adipogenesis-related increase of semicarbazide-sensitive amine oxidase and monoamine oxidase in human adipocytes. Biochimie 2007, 89, 916–925. [Google Scholar] [CrossRef]
- Heniquez, A.; Meissonnier, G.; Visentin, V.; Prevot, D.; Carpéné, C. High expression of semicarbazide-sensitive amine oxidase genes AOC2 and AOC3, but not the diamine oxidase gene AOC1 in human adipocytes. Inflamm. Res. 2003, 52, S74–S75. [Google Scholar] [CrossRef]
- Byun, Y.; Park, J.; Hong, S.H.; Han, M.H.; Park, S.; Jung, H.I.; Noh, M. The opposite effect of isotype-selective monoamine oxidase inhibitors on adipogenesis in human bone marrow mesenchymal stem cells. Bloorg. Med. Chem. Lett. 2013, 23, 3273–3276. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).