Depiction of the In Vitro and Genomic Basis of Resistance to Hop and High Hydrostatic Pressure of Lactiplantibacillus plantarum Isolated from Spoiled Beer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strain and Growth Conditions and Molecular Identification
2.2. Hop Resistance of KKP 3573 Strain
2.3. HHP Application
2.4. Determination of the Number of Surviving Cells in the Pressurization Process
2.5. MALDI-TOF MS Analysis
2.6. Genome Sequencing
2.7. Phylogenomic Analysis
2.8. Genome Annotation
3. Results and Discussion
3.1. Beer-Spoiling Ability of Lp. plantarum KKP 3573
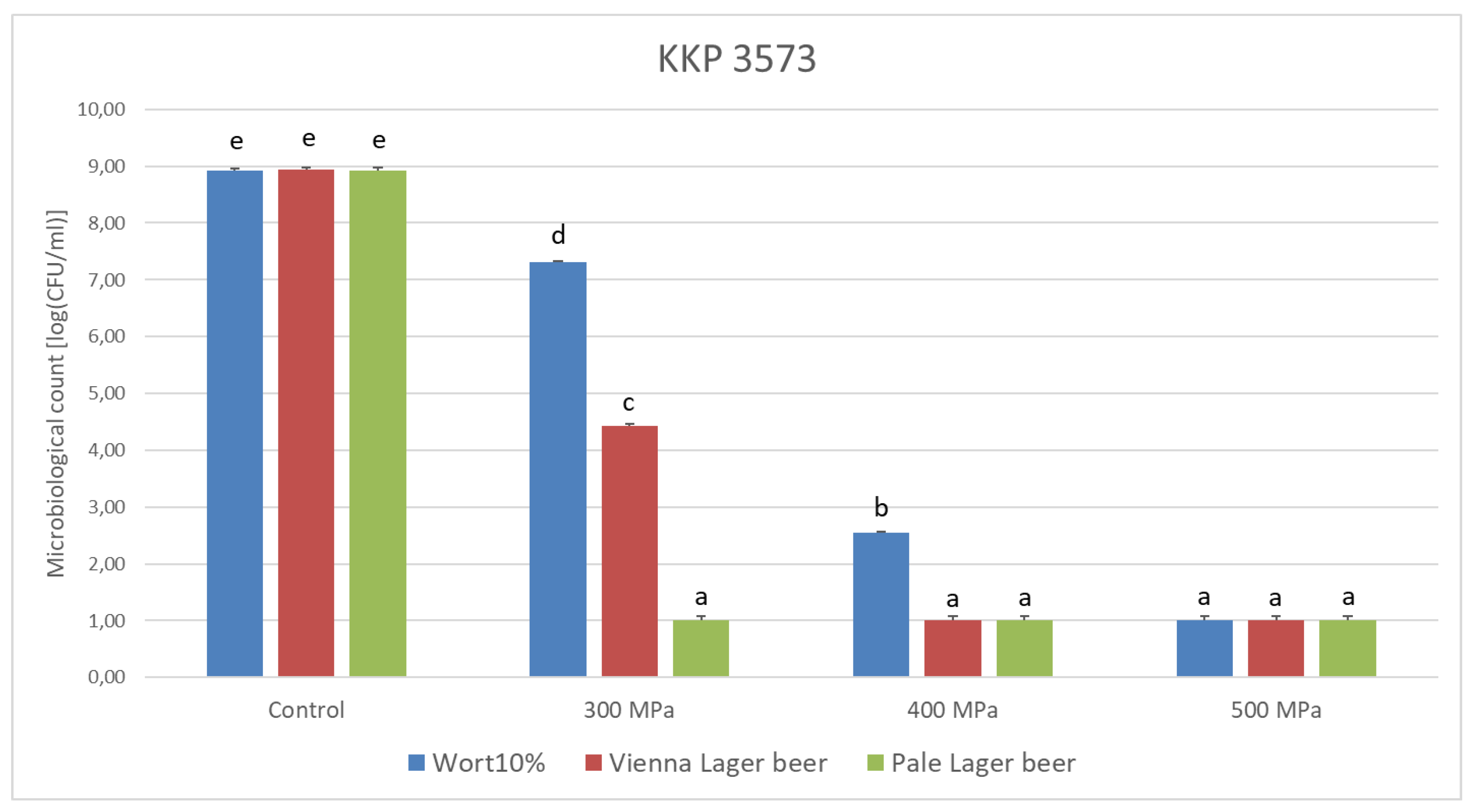
3.2. Influence of HHP on Lp. plantarum Strain Survivability
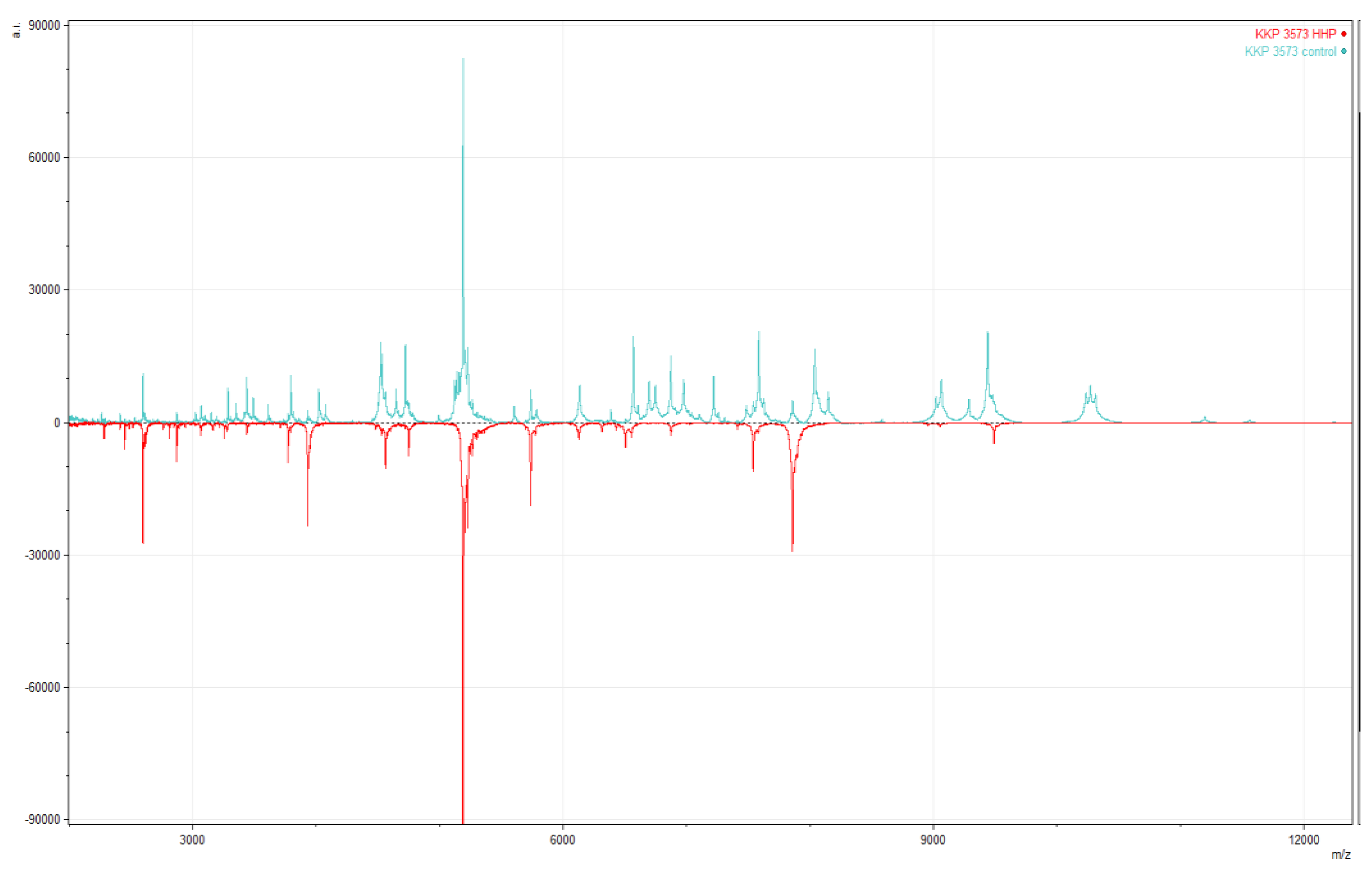
3.3. The Impact of HHP on MALDI-TOF MS Identification
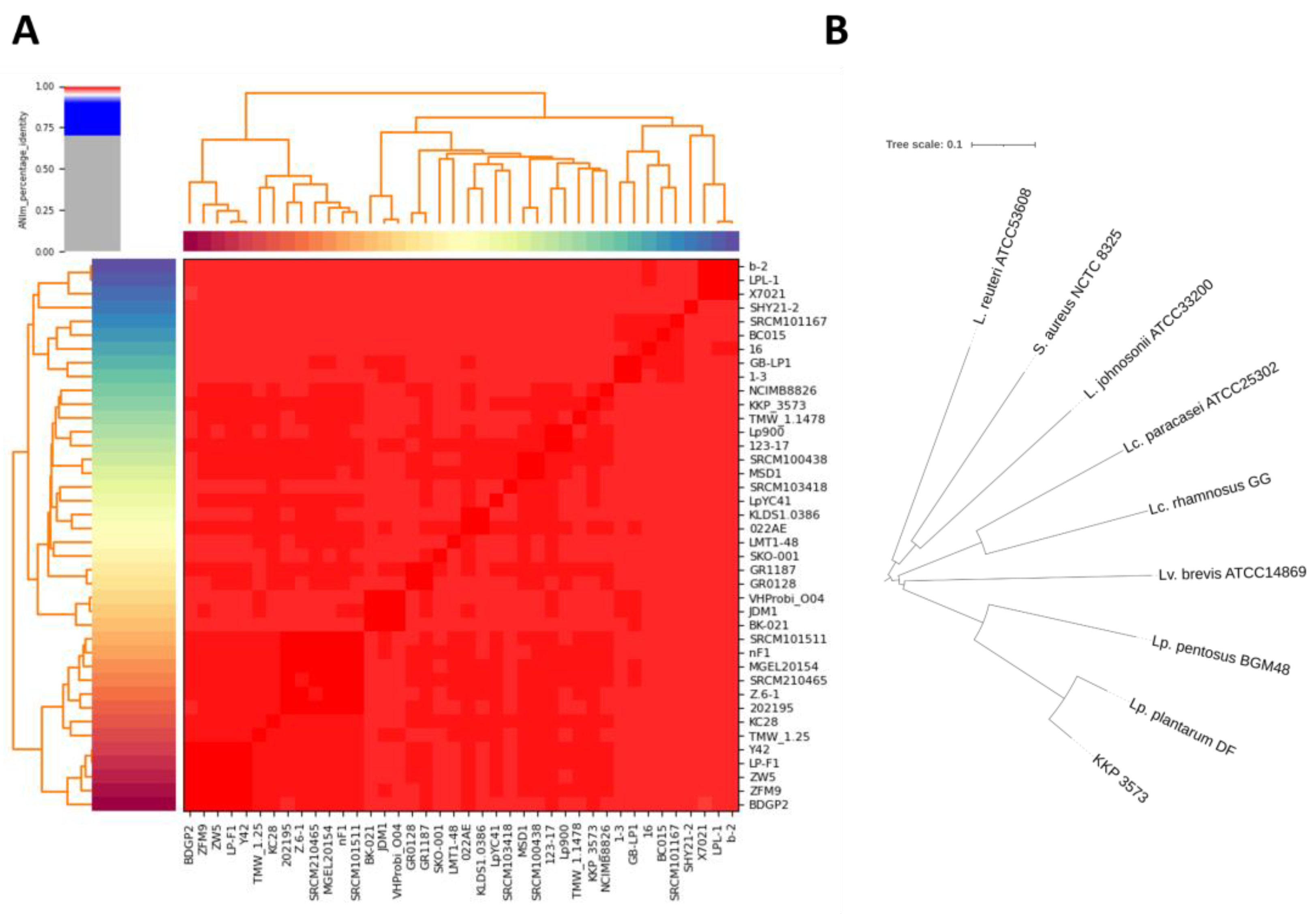
3.4. Whole-Genome Sequencing, Gene Annotation, and Phylogenomic Analysis of Strain Lp. plantarum KKP 3573
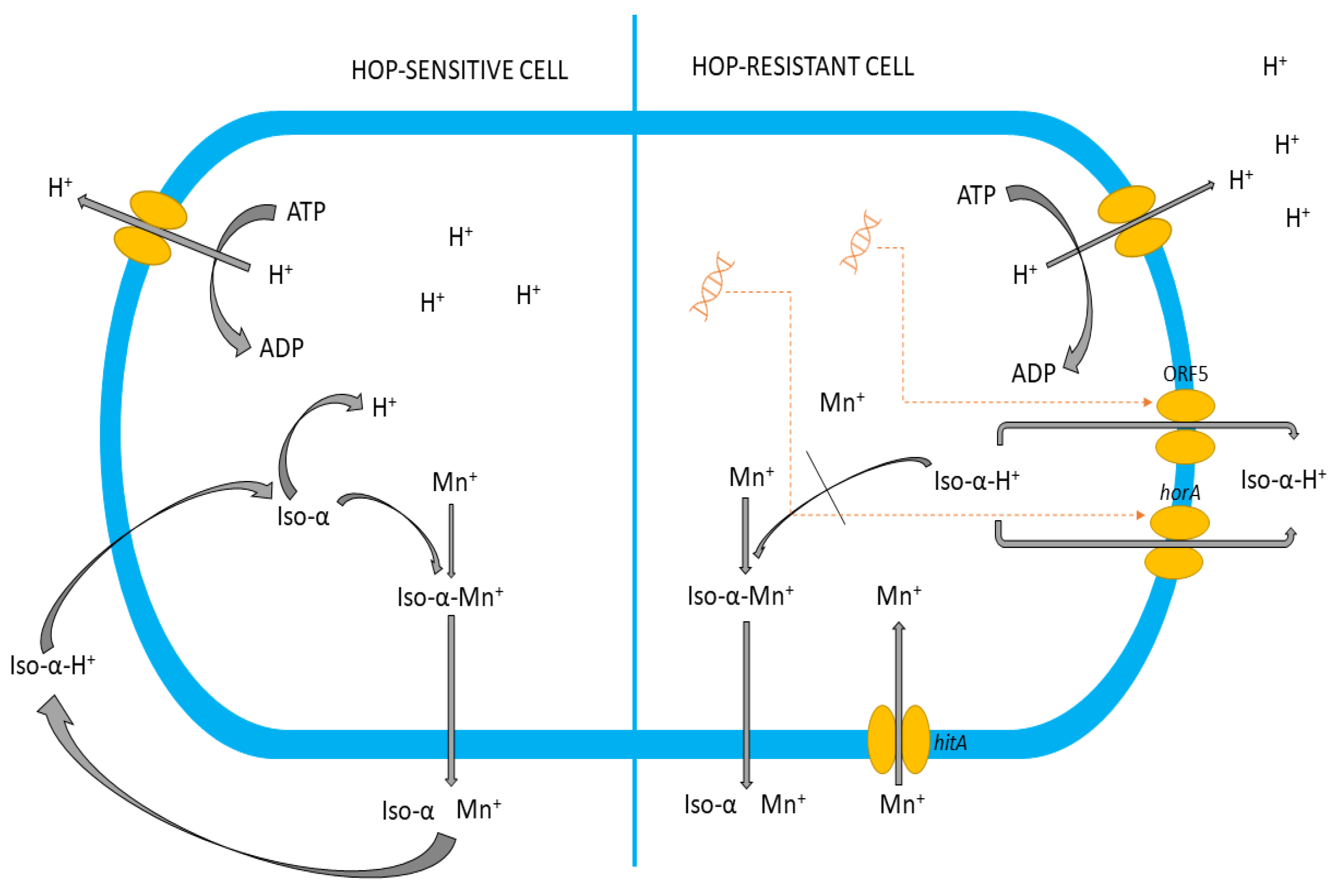
3.5. Lp. plantarum KKP 3573 Possesses Genes Involved in Tolerance to Stress and the Beer-Spoiling Phenotype
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Suzuki, K.; Iijima, K.; Sakamoto, K.; Saihi, M.; Yamashita, H. A Review of Hop Resistance in Beer Spoilage Lactic Acid Bacteria. J. Inst. Brew. 2006, 112, 173–191. [Google Scholar] [CrossRef]
- Suzuki, K. 125th Anniversary Review: Microbiological Instability of Beer Caused by Spoilage Bacteria. J. Inst. Brew. 2011, 117, 131–155. [Google Scholar] [CrossRef]
- Suzuki, K. Emergence of New Spoilage Microorganisms in the Brewing Industry and Development of Microbiological Quality Control Methods to Cope with This Phenomenon—A Review. J. Am. Soc. Brew. Chem. 2020, 78, 245–259. [Google Scholar] [CrossRef]
- Deng, Y.; Liu, J.; Li, H.; Li, L.; Tu, J.; Fang, H.; Chen, J.; Qian, F. An Improved Plate Culture Procedure for the Rapid Detection of Beer-Spoilage Lactic Acid Bacteria. J. Inst. Brew. 2014, 120, 127–132. [Google Scholar] [CrossRef]
- Bucka-Kolendo, J.; Sokołowska, B. Lactic Acid Bacteria Stress Response to Preservation Processes in the Beverage and Juice Industry. Acta Biochim. Pol. 2017, 64, 459–464. [Google Scholar] [CrossRef]
- Ulmer, H.M.; Herberhold, H.; Fahsel, S.; Gänzle, M.G.; Winter, R.; Vogel, R.F. Effects of Pressure-Induced Membrane Phase Transitions on Inactivation of HorA, an ATP-Dependent Multidrug Resistance Transporter, in Lactobacillus plantarum. Appl. Environ. Microbiol. 2002, 68, 1088–1095. [Google Scholar] [CrossRef] [PubMed]
- Buzrul, S. High Hydrostatic Pressure Treatment of Beer and Wine: A Review. Innov. Food Sci. Emerg. Technol. 2012, 13, 1–12. [Google Scholar] [CrossRef]
- Buzrul, S.; Alpas, H.; Bozoglu, F. Effect of High Hydrostatic Pressure on Quality Parameters of Lager Beer. J. Sci. Food Agric. 2005, 85, 1672–1676. [Google Scholar] [CrossRef]
- Yordanov, D.G.; Angelova, G.V. High Pressure Processing for Foods Preserving. Biotechnol. Biotechnol. Equip. 2010, 24, 1940–1945. [Google Scholar] [CrossRef]
- Choi, J.H.; Kang, J.W.; Mijanur Rahman, A.T.M.; Lee, S.J. Increasing Fermentable Sugar Yields by High-Pressure Treatment during Beer Mashing. J. Inst. Brew. 2016, 122, 143–146. [Google Scholar] [CrossRef]
- Perez-Lamela, C.; Reed, R.J.R.; Simal-Gandara, J. High Pressure Application to Wort and Beer. Dtsch. Leb. Rundsch. 2004, 100, 53–56. [Google Scholar]
- Yin, H.; Dong, J.; Yu, J.; Chang, Z.; Qian, Z.; Liu, M.; Huang, S.; Hu, X.; Liu, X.; Deng, Y.; et al. A Preliminary Study about the Influence of High Hydrostatic Pressure Processing on the Physicochemical and Sensorial Properties of a Cloudy Wheat Beer. J. Inst. Brew. 2016, 122, 462–467. [Google Scholar] [CrossRef]
- Iijima, K.; Suzuki, K.; Ozaki, K.; Yamashita, H. HorC Confers Beer-Spoilage Ability on Hop-Sensitive Lactobacillus brevis ABBC45cc. J. Appl. Microbiol. 2006, 100, 1282–1288. [Google Scholar] [CrossRef] [PubMed]
- Oldham, R.C.; Held, M.A.; Ryanne, M.; Oldham, C. Methods for Detection and Identification of Beer-Spoilage Microbes. Front. Microbiol. Sec. Food Microbiol. 2023, 14, 1217704. [Google Scholar] [CrossRef]
- Sakamoto, K.; Konings, W.N. Beer Spoilage Bacteria and Hop Resistance. Int. J. Food Microbiol. 2003, 89, 105–124. [Google Scholar] [CrossRef] [PubMed]
- Pittet, V.; Morrow, K.; Ziola, B. Ethanol Tolerance of Lactic Acid Bacteria, Including Relevance of the Exopolysaccharide Gene Gtf. J. Am. Soc. Brew. Chem. 2011, 69, 57–61. [Google Scholar] [CrossRef]
- Behr, J.; Gänzle, M.G.; Vogel, R.F. Characterization of a Highly Hop-Resistant Lactobacillus brevis Strain Lacking Hop Transport. Appl. Environ. Microbiol. 2006, 72, 6483–6492. [Google Scholar] [CrossRef] [PubMed]
- Fischer, S.; Ruß, W.; Meyer-Pittroff, R.; Buckow, R.; Heinz, V.; Knorr, D.; Ulmer, H.; Behr, J.; Vogel, R.F. Effects of Hydrostatic High Pressure on Micro-Biological and Technological Characteristics of Beer. Monatsschrift Brauwiss. 2006, 59, 90–99. [Google Scholar]
- Behr, J.; Geissler, A.J.; Schmid, J.; Zehe, A.; Vogel, R.F. The Identification of Novel Diagnostic Marker Genes for the Detection of Beer Spoiling Pediococcus damnosus Strains Using the BlAst Diagnostic Gene FindEr. PLoS ONE 2016, 11, e0152747. [Google Scholar] [CrossRef]
- Wouters, P.C.; Glaasker, E.; Smelt, J.P.P.M. Effects of High Pressure on Inactivation Kinetics and Events Related to Proton Efflux in Lactobacillus plantarum. Appl. Environ. Microbiol. 1998, 64, 509–514. [Google Scholar] [CrossRef]
- Zhao, Y.; Knøchel, S.; Siegumfeldt, H. Heterogeneity between and within Strains of Lactobacillus brevis Exposed to Beer Compounds. Front. Microbiol. 2017, 8, 239. [Google Scholar] [CrossRef] [PubMed]
- Behr, J.; Vogel, R.F. Mechanisms of Hop Inhibition Include the Transmembrane Redox Reaction. Appl. Environ. Microbiol. 2010, 76, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Bucka-Kolendo, J.; Sokołowska, B. Impact of High Hydrostatic Pressure on the Single Nucleotide Polymorphism of Stress-Related DnaK, HrcA, and CtsR in the Lactobacillus Strains. Qual. Assur. Saf. Crops Foods 2022, 14, 54–66. [Google Scholar] [CrossRef]
- Bucka-Kolendo, J.; Juszczuk-Kubiak, E.; Sokołowska, B. Effect of High Hydrostatic Pressure on Stress-Related DnaK, HrcA, and CtsR Expression Patterns in Selected Lactobacilli Strains. Genes 2021, 12, 1720. [Google Scholar] [CrossRef]
- Kiousi, D.E.; Bucka-Kolendo, J.; Wojtczak, A.; Sokołowska, B.; Doulgeraki, A.I.; Galanis, A. Genomic Analysis and In Vitro Investigation of the Hop Resistance Phenotype of Two Novel Loigolactobacillus backii Strains, Isolated from Spoiled Beer. Microorganisms 2023, 11, 280. [Google Scholar] [CrossRef]
- Bergsveinson, J.; Baecker, N.; Pittet, V.; Ziola, B. Role of Plasmids in Lactobacillus brevis BSO 464 Hop Tolerance and Beer Spoilage. Appl. Environ. Microbiol. 2015, 81, 1234–1241. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Bamforth, C.W. The Microbiology of Malting and Brewing. Microbiol. Mol. Biol. Rev. 2013, 77, 157–172. [Google Scholar] [CrossRef]
- Suzuki, K.; Shinohara, Y.; Kurniawan, Y.N. Role of Plasmids in Beer Spoilage Lactic Acid Bacteria: A Review. J. Am. Soc. Brew. Chem. 2020, 79, 1–16. [Google Scholar] [CrossRef]
- Fujii, T.; Nakashima, K.; Hayashi, N. Random Amplified Polymorphic DNA-PCR Based Cloning of Markers to Identify the Beer-Spoilage Strains of Lactobacillus brevis, Pediococcus damnosus, Lactobacillus collinoides and Lactobacillus coryniformis. J. Appl. Microbiol. 2005, 98, 1209–1220. [Google Scholar] [CrossRef]
- Hayashi, N.; Ito, M.; Horiike, S.; Taguchi, H. Molecular Cloning of a Putative Divalent-Cation Transporter Gene as a New Genetic Marker for the Identification of Lactobacillus brevis Strains Capable of Growing in Beer. Appl. Microbiol. Biotechnol. 2001, 55, 596–603. [Google Scholar] [CrossRef] [PubMed]
- Preissler, P.; Behr, J.; Vogel, R.F. Detection of Beer-Spoilage Lactobacillus brevis Strains by Reduction of Resazurin. J. Inst. Brew. 2010, 116, 399–404. [Google Scholar] [CrossRef]
- Iijima, K.; Suzuki, K.; Asano, S.; Ogata, T.; Kitagawa, Y. HorC, a Hop-Resistance Related Protein, Presumably Functions in Homodimer Form. Biosci. Biotechnol. Biochem. 2009, 73, 1880–1882. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, A.; O’Sullivan, T.; Van Sinderen, D. Enhancing the Microbiological Stability of Malt and Beer—A Review. J. Inst. Brew. 2005, 111, 355–371. [Google Scholar] [CrossRef]
- Lado, B.H.; Yousef, A.E. Alternative Food-Preservation Technologies: Efficacy and Mechanisms. Microbes Infect. 2002, 4, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Romanek, J.; Opiela, J. Zastosowanie Wysokiego Ciśnienia Hydrostatycznego (HHP) w Przemyśle Spożywczym, Farmaceutycznym Oraz Medycynie. Wiadomości Zootech. 2015, 4, 34–40. [Google Scholar]
- Winter, R.; Jeworrek, C. Effect of Pressure on Membranes. Soft Matter 2009, 5, 3157–3173. [Google Scholar] [CrossRef]
- Wemekamp-Kamphuis, H.H.; Karatzas, A.K.; Wouters, J.A.; Abee, T. Enhanced Levels of Cold Shock Proteins in Listeria monocytogenes LO28 upon Exposure to Low Temperature and High Hydrostatic Pressure. Appl. Environ. Microbiol. 2002, 68, 456–463. [Google Scholar] [CrossRef]
- Robey, M.; Benito, A.; Hutson, R.H.; Pascual, C.; Park, S.F.; Mackey, B.M. Variation in Resistance to High Hydrostatic Pressure and RpoS Heterogeneity in Natural Isolates of Escherichia coli O157:H7. Appl. Environ. Microbiol. 2001, 67, 4901–4907. [Google Scholar] [CrossRef]
- Bucka-Kolendo, J.; Sokołowska, B.; Winiarczyk, S. Influence of High Hydrostatic Pressure on the Identification of Lactobacillus by MALDI-TOF MS-Preliminary Study. Microorganisms 2020, 8, 813. [Google Scholar] [CrossRef]
- PN ISO 15214: 2002; Microbiology Of Food And Animal Feeding Stuffs-Horizontal Method For The Enumeration Of Mesophilic Lactic Acid Bacteria-Colony-Count Technique At 30 Degrees C. Polish Committee for Standardization: Warszawa, Poland, 2002.
- Akimowicz, M.; Bucka-Kolendo, J. MALDI-TOF MS-Application in Food Microbiology. Acta Biochim. Pol. 2020, 67, 327–332. [Google Scholar] [CrossRef]
- Babraham Bioinformatics—FastQC A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 19 March 2022).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. Original Articles SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Boetzer, M.; Henkel, C.V.; Jansen, H.J.; Butler, D.; Pirovano, W. Scaffolding Pre-Assembled Contigs Using SSPACE. Bioinform. Appl. Note 2011, 27, 578–579. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-Depth Characterization and Visualization of Bacterial Genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, L.; Glover, R.H.; Humphris, S.; Elphinstone, J.G.; Toth, I.K. Genomics and Taxonomy in Diagnostics for Food Security: Soft-Rotting Enterobacterial Plant Pathogens. Anal. Methods 2015, 8, 12–24. [Google Scholar] [CrossRef]
- Darling, A.E.; Mau, B.; Perna, N.T. ProgressiveMauve: Multiple Genome Alignment with Gene Gain, Loss and Rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v3: An Online Tool for the Display and Annotation of Phylogenetic and Other Trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Genome Analysis Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T.; Dicuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; Garciá-Fernández, A.; Larsen, M.V.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In Silico Detection and Typing of Plasmids Using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895. [Google Scholar] [CrossRef] [PubMed]
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The Reference Centre for Bacterial Insertion Sequences. Nucleic Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef] [PubMed]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A Better, Faster Version of the PHAST Phage Search Tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed]
- Biswas, A.; Staals, R.H.J.; Morales, S.E.; Fineran, P.C.; Brown, C.M. CRISPRDetect: A Flexible Algorithm to Define CRISPR Arrays. BMC Genom. 2016, 17, 356. [Google Scholar] [CrossRef]
- Pourcel, C.; Touchon, M.; Villeriot, N.; Vernadet, J.P.; Couvin, D.; Toffano-Nioche, C.; Vergnaud, G. CRISPRCasdb a Successor of CRISPRdb Containing CRISPR Arrays and Cas Genes from Complete Genome Sequences, and Tools to Download and Query Lists of Repeats and Spacers. Nucleic Acids Res. 2020, 48, D535–D544. [Google Scholar] [CrossRef]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded Curation, Support for Machine Learning, and Resistome Prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef]
- Brown, C.L.; Mullet, J.; Hindi, F.; Stoll, J.E.; Gupta, S.; Choi, M.; Keenum, I.; Vikesland, P.; Pruden, A.; Zhang, L. MobileOG-Db: A Manually Curated Database of Protein Families Mediating the Life Cycle of Bacterial Mobile Genetic Elements. Appl. Environ. Microbiol. 2022, 88, e0099122. [Google Scholar] [CrossRef]
- Vernikos, G.S.; Parkhill, J. Interpolated Variable Order Motifs for Identification of Horizontally Acquired DNA: Revisiting the Salmonella Pathogenicity Islands. Bioinformatics 2006, 22, 2196–2203. [Google Scholar] [CrossRef]
- Van Heel, A.J.; De Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A User-Friendly Web Server to Thoroughly Mine RiPPs and Bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. EggNOG 5.0: A Hierarchical, Functionally and Phylogenetically Annotated Orthology Resource Based on 5090 Organisms and 2502 Viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a Reference Resource for Gene and Protein Annotation. Nucleic Acids Res. 2016, 44, D457. [Google Scholar] [CrossRef] [PubMed]
- Aertsen, A.; Van Houdt, R.; Vanoirbeek, K.; Michiels, C.W. An SOS Response Induced by High Pressure in Escherichia coli. J. Bacteriol. 2004, 186, 6133–6141. [Google Scholar] [CrossRef]
- Aertsen, A.; Vanoirbeek, K.; De Spiegeleer, P.; Sermon, J.; Hauben, K.; Farewell, A.; Nyström, T.; Michiels, C.W. Heat Shock Protein-Mediated Resistance to High Hydrostatic Pressure in Escherichia coli. Appl. Environ. Microbiol. 2004, 70, 2660–2666. [Google Scholar] [CrossRef]
- Barbieri, F.; Montanari, C.; Gardini, F.; Tabanelli, G. Biogenic Amine Production by Lactic Acid Bacteria: A Review. Foods 2019, 8, 17. [Google Scholar] [CrossRef] [PubMed]
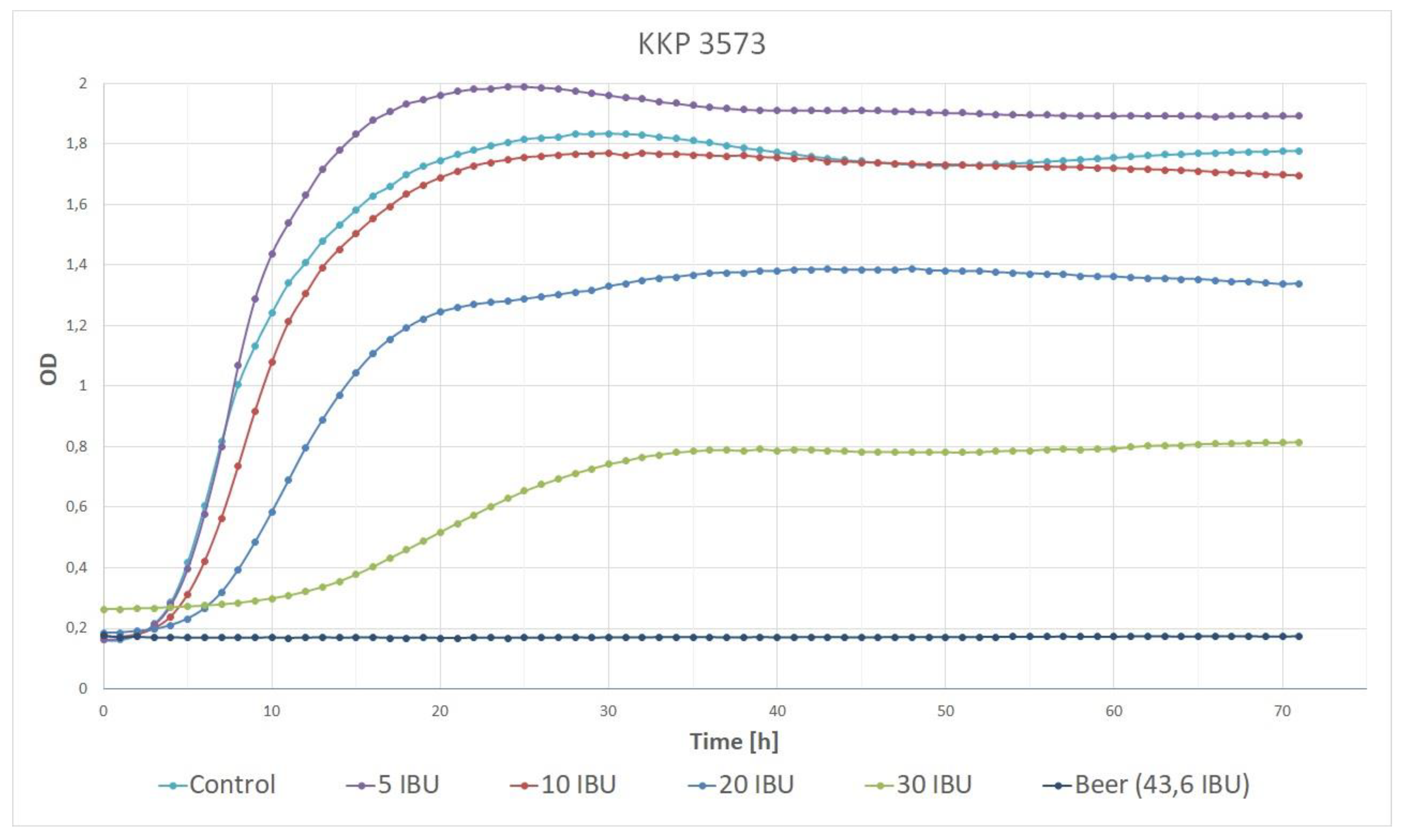

| Control | 5 IBU | 10 IBU | 20 IBU | 30 IBU | Beer 43.6 IBU | |
|---|---|---|---|---|---|---|
| MRS broth concentrate (2×) | 50% | 50% | 50% | 50% | - | - |
| MRS broth concentrate (4×) | - | - | - | - | 25% | - |
| Water | 50% | 12.5% | 25% | - | - | |
| Beer (40 IBU) | - | 37.5% | 25% | 50% | 75% | - |
| Beer (43.6 IBU) | - | - | - | - | - | 100% |
| Vienna Lager Beer | Pale Lager Beer | |
|---|---|---|
| Alcohol, % (m/m) | 4.62 ± 0.16 | 4.31 ± 0.15 |
| Alcohol, % (v/v) | 5.91 ± 0.16 | 5.45 ± 0.15 |
| Apparent Extract, % (w/w) | 2.88 ± 0.09 | <0.50 |
| Real Extract, % (w/w) | 4.99 ± 0.06 | 2.03 ± 0.03 |
| Original Wort Extract, % (m/m) | 13.85 ± 0.14 | 10.46 ± 0.11 |
| Bitterness (International Bitterness Units—IBU) | 20.0 | 20.4 |
| Medium | KKP 3573 | |
|---|---|---|
| μmax | ΔOD | |
| Control (MRS) | 0.170 ± 0.005 e | 1.675 ± 0.039 e |
| 5 IBU | 0.170 ± 0.005 aE | 1.675 ± 0.039 aE |
| 10 IBU | 0.235 ± 0.005 aF | 1.828 ± 0.013 aF |
| 20 IBU | 0.163 ± 0.007 aD | 1.602 ± 0.034 aD |
| 30 IBU | 0.099 ± 0.005 aC | 1.204 ± 0.018 aC |
| Beer 43.6 IBU | 0.031 ± 0.001 aB | 0.534 ± 0.026 aB 1 |
| Genome Characteristics | Value |
|---|---|
| Length | 3,295,227 bp |
| GC content | 44.39% |
| Total genes | 3.102 |
| CDSs | 3.052 |
| rRNAs | 4 |
| tRNAs | 42 |
| ncRNAs | 4 |
| Pseudogenes | 29 |
| No. of CRISPR arrays | 0 |
| IS elements | 39 |
| Phages: | |
| Intact | 3 |
| Incomplete | 3 |
| Questionable | 0 |
| Antibiotic resistance genes: | |
| Perfect hits | 0 |
| Strict hits | 2 |
| Loose hits | 0 |
| Virulence genes | 0 |
| Clusters of Orthologous Groups | Lp. plantarum KKP 3573 | Lp. plantarum Pangenome |
|---|---|---|
| C—Energy production and conversion | 3.651505 | 2.624688 |
| D—Cell cycle control and mitosis | 1.313261 | 1.633416 |
| E—Amino acid metabolism and transport | 7.078796 | 4.538653 |
| F—Nucleotide metabolism and transport | 4.163997 | 1.826683 |
| G—Carbohydrate metabolism and transport | 9.000641 | 6.80798 |
| H—Coenzyme metabolism | 3.042921 | 2.119701 |
| I—Lipid metabolism | 2.114029 | 1.147132 |
| J—Translation | 5.477258 | 1.739401 |
| K—Transcription | 9.577194 | 6.689526 |
| L—Replication and repair | 4.836643 | 17.96758 |
| M—Cell wall/membrane/envelope biogenesis | 5.605381 | 7.325436 |
| N—Cell motility | 0.512492 | 0.361596 |
| O—Posttranslational modification, protein turnover, chaperone functions | 1.825753 | 1.209476 |
| P—Inorganic ion transport and metabolism | 5.028828 | 4.033666 |
| Q—Secondary Structure | 0.832799 | 0.891521 |
| T—Signal transduction | 2.434337 | 1.677057 |
| U—Intracellular trafficking and secretion | 2.466368 | 1.683292 |
| V—Defense mechanisms | 1.953876 | 2.718204 |
| S—Function unknown | 18.57783 | 18.45387 |
| No annotation | 10.50609 | 14.55112 |
| Total (%) | 100 | 100 |
| Locus Tag | Gene Function | Gene | E-Value |
|---|---|---|---|
| Acid tolerance | |||
| MHOBIDOO_02172 | Sodium proton antiporter | yvgP | 0.0 |
| MHOBIDOO_01677 | ATP synthase subunit α | atpA | 0.0 |
| MHOBIDOO_01673 | ATP synthase subunit a | atpB | 4.82 × 10−165 |
| MHOBIDOO_01680 | ATP synthase epsilon chain | atpC | 5.95 × 10−74 |
| MHOBIDOO_01679 | ATP synthase subunit β | atpD | 0.0 |
| MHOBIDOO_01674 | ATP synthase subunit c | atpE | 1.81 × 10-37 |
| MHOBIDOO_01675 | ATP synthase subunit b | atpF | 5.41 × 10−77 |
| MHOBIDOO_01678 | ATP synthase γ chain | atpG | 9.14 × 10−213 |
| MHOBIDOO_01676 | ATP synthase subunit delta | atpH | 2.03 × 10−118 |
| Hop resistance | |||
| MHOBIDOO_02432 | H(+)-stimulated, divalent metal cation uptake system | mntH | 0.0 |
| MHOBIDOO_00120 | H(+)-stimulated, divalent metal cation uptake system | mntH | 5.71 × 10−301 |
| MHOBIDOO_01928 | H(+)-stimulated, divalent metal cation uptake system | mntH | 1.8 × 10−290 |
| MHOBIDOO_00859 | Unsaturated fatty acid biosynthesis | fabZ | 8.55 × 10−99 |
| MHOBIDOO_00734 | Iron-dependent repressor | mntR | 2.94 × 10−155 |
| MHOBIDOO_00860 | Unsaturated fatty acid biosynthesis | fabH | 2.3 × 10−229 |
| MHOBIDOO_00862 | Unsaturated fatty acid biosynthesis | fabD | 2.34 × 10−213 |
| MHOBIDOO_00863 | Unsaturated fatty acid biosynthesis | fabG | 1.04 × 10−161 |
| MHOBIDOO_00864 | Unsaturated fatty acid biosynthesis | fabF | 1.54 × 10−289 |
| MHOBIDOO_00866 | Unsaturated fatty acid biosynthesis | fabZ2 | 1.71 × 10−91 |
| Bile salt tolerance: | |||
| MHOBIDOO_03052 | Linear amide C-N hydrolases, choloylglycine hydrolase family | pva2 | 4.38 × 10−243 |
| MHOBIDOO_00290 | Linear amide C-N hydrolase, choloylglycine hydrolase family protein | pva1 | 2.28 × 10−250 |
| MHOBIDOO_00482 | Linear amide C-N hydrolase, choloylglycine hydrolase family protein | cbh | 2.83 × 10−237 |
| MHOBIDOO_01519 | Linear amide C-N hydrolase, choloylglycine hydrolase family protein | yxeI | 2.83 × 10−238 |
| MHOBIDOO_03052 | Linear amide C-N hydrolases, choloylglycine hydrolase family | pva2 | 4.38 × 10−243 |
| Extreme temperature tolerance: | |||
| MHOBIDOO_02053 | ‘Cold shock’ DNA-binding domain | cspP | 6.22 × 10−43 |
| MHOBIDOO_03112 | Cold shock protein | cspA | 2.54 × 10−42 |
| MHOBIDOO_00316 | Cold shock protein domain | cspL | 4.37 × 10−43 |
| MHOBIDOO_00735 | Cold shock protein | cspC | 1.78 × 10−42 |
| MHOBIDOO_01147 | Heat shock 40 kDa protein | dnaJ | 2.54 × 10−266 |
| MHOBIDOO_01148 | Heat shock 70 kDa protein | dnaK | 0.0 |
| MHOBIDOO_01457 | Belongs to the small heat shock protein (HSP20) family | hsp2 | 2.61 × 10−96 |
| MHOBIDOO_03043 | Belongs to the small heat shock protein (HSP20) family | hsp3 | 4.58 ×10−103 |
| MHOBIDOO_00239 | Belongs to the small heat shock protein (HSP20) family | hsp1 | 2.31 × 10−95 |
| MHOBIDOO_01942 | Recovery of the cell from heat-induced damage, in cooperation with DnaK, DnaJ, and GrpE | clpC | 0.0 |
| MHOBIDOO_02252 | Molecular chaperone | GroEL | 0.0 |
| MHOBIDOO_02253 | Cochaperonin | GroES | 1.7 × 10−59 |
| MHOBIDOO_01942 | Part of a stress-induced multichaperone system, it is involved in the recovery of the cell from heat-induced damage, in cooperation with DnaK, DnaJ, and GrpE | clpC | 0.0 |
| MHOBIDOO_02146 | Belongs to the ClpA ClpB family | clpE | 0.0 |
| MHOBIDOO_02199 | Cleaves peptides in various proteins in a process that requires ATP hydrolysis. Has chymotrypsin-like activity. Plays a major role in the degradation of misfolded proteins | clpP | 5.11 × 10−133 |
| MHOBIDOO_00438 | C-terminal, D2 small domain, of ClpB protein | clpL | 0.0 |
| MHOBIDOO_01059 | Part of a stress-induced multichaperone system, it is involved in the recovery of the cell from heat-induced damage, in cooperation with DnaK, DnaJ, and GrpE | clpB | 0.0 |
| MHOBIDOO_01230 | ATP-dependent specificity component of the Clp protease. It directs the protease to specific substrates. Can perform chaperone functions in the absence of ClpP | clpX | 7.8 × 10−300 |
| Osmotic shock tolerance | |||
| MHOBIDOO_01149 | Response to hyperosmotic and heat shock | grpE | 4.94 × 10−115 |
| MHOBIDOO_00805 | ABC transporter, ATP-binding protein | opuCA | 4.51 × 10−284 |
| MHOBIDOO_00806 | ABC transporter permease | opuCB | 7.11 × 10−135 |
| MHOBIDOO_00807 | Periplasmic glycine betaine choline-binding (lipo)protein of an ABC-type transport system (osmoprotectant binding protein) | opuCC | 3.8 × 10−224 |
| MHOBIDOO_00808 | Binding-protein-dependent transport system inner membrane component | opuCD | 1.36 × 10−136 |
| Oxidative stress survival: | |||
| MHOBIDOO_02472 | Redox-regulated molecular chaperone | hslO | 1.93 × 10−209 |
| MHOBIDOO_01980 | NADH dehydrogenase | ndh | 0.0 |
| MHOBIDOO_02214 | Pyridine nucleotide–disulfide oxidoreductase, dimerization domain | nox | 0.0 |
| MHOBIDOO_02226 | NADH oxidase | nox | 0.0 |
| MHOBIDOO_00557 | NADH oxidase | nox | 0.0 |
| MHOBIDOO_01078 | Pyridine nucleotide–disulfide oxidoreductase, dimerization domain | nox | 0.0 |
| MHOBIDOO_01095 | Pyridine nucleotide–disulfide oxidoreductase, dimerization domain | nox | 0.0 |
| MHOBIDOO_00163 | Member of the glutathione peroxidase family | gpo | 6.07 × 10−117 |
| MHOBIDOO_02589 | Thiol-specific peroxidase | tpx | 3.56 × 10−116 |
| MHOBIDOO_00571 | Peroxidase | ywbN | 2.4 × 10−230 |
| Biofilm formation: | |||
| MHOBIDOO_02087 | Capsular polysaccharide biosynthesis protein | epsB | 7.88 × 10−169 |
| MHOBIDOO_01363 | Glycosyl transferase family 2 | epsV | 1.4 × 10−181 |
| MHOBIDOO_02088 | Capsular exopolysaccharide family | ywqD | 1.43 × 10−164 |
| MHOBIDOO_02836 | Acetyltransferase (GNAT) domain | ywnH | 1.66 × 10−116 |
| MHOBIDOO_02208 | S-ribosylhomocysteine lyase | luxS | 2.21 × 10−113 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bucka-Kolendo, J.; Kiousi, D.E.; Wojtczak, A.; Doulgeraki, A.I.; Galanis, A.; Sokołowska, B. Depiction of the In Vitro and Genomic Basis of Resistance to Hop and High Hydrostatic Pressure of Lactiplantibacillus plantarum Isolated from Spoiled Beer. Genes 2023, 14, 1710. https://doi.org/10.3390/genes14091710
Bucka-Kolendo J, Kiousi DE, Wojtczak A, Doulgeraki AI, Galanis A, Sokołowska B. Depiction of the In Vitro and Genomic Basis of Resistance to Hop and High Hydrostatic Pressure of Lactiplantibacillus plantarum Isolated from Spoiled Beer. Genes. 2023; 14(9):1710. https://doi.org/10.3390/genes14091710
Chicago/Turabian StyleBucka-Kolendo, Joanna, Despoina Eugenia Kiousi, Adrian Wojtczak, Agapi I. Doulgeraki, Alex Galanis, and Barbara Sokołowska. 2023. "Depiction of the In Vitro and Genomic Basis of Resistance to Hop and High Hydrostatic Pressure of Lactiplantibacillus plantarum Isolated from Spoiled Beer" Genes 14, no. 9: 1710. https://doi.org/10.3390/genes14091710
APA StyleBucka-Kolendo, J., Kiousi, D. E., Wojtczak, A., Doulgeraki, A. I., Galanis, A., & Sokołowska, B. (2023). Depiction of the In Vitro and Genomic Basis of Resistance to Hop and High Hydrostatic Pressure of Lactiplantibacillus plantarum Isolated from Spoiled Beer. Genes, 14(9), 1710. https://doi.org/10.3390/genes14091710








