Genomic Instability of G-Quadruplex Sequences in Escherichia coli: Roles of DinG, RecG, and RecQ Helicases
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Media
2.2. Cloning Quadruplex-Forming Repeats
2.3. Mutation Rate Measurement and Analysis of Cmr Revertants
3. Results
3.1. Confirmation of the DNA Structure Formation in G-Quadruplex-Forming DNA Repeats
3.2. Plasmids for the Analysis of the Genetic Instability of G-Quadruplex-Forming Repeats
3.3. Measurement of the Mutation Rates of Quadruplex-Forming Repeats in pBR325 and pBR235
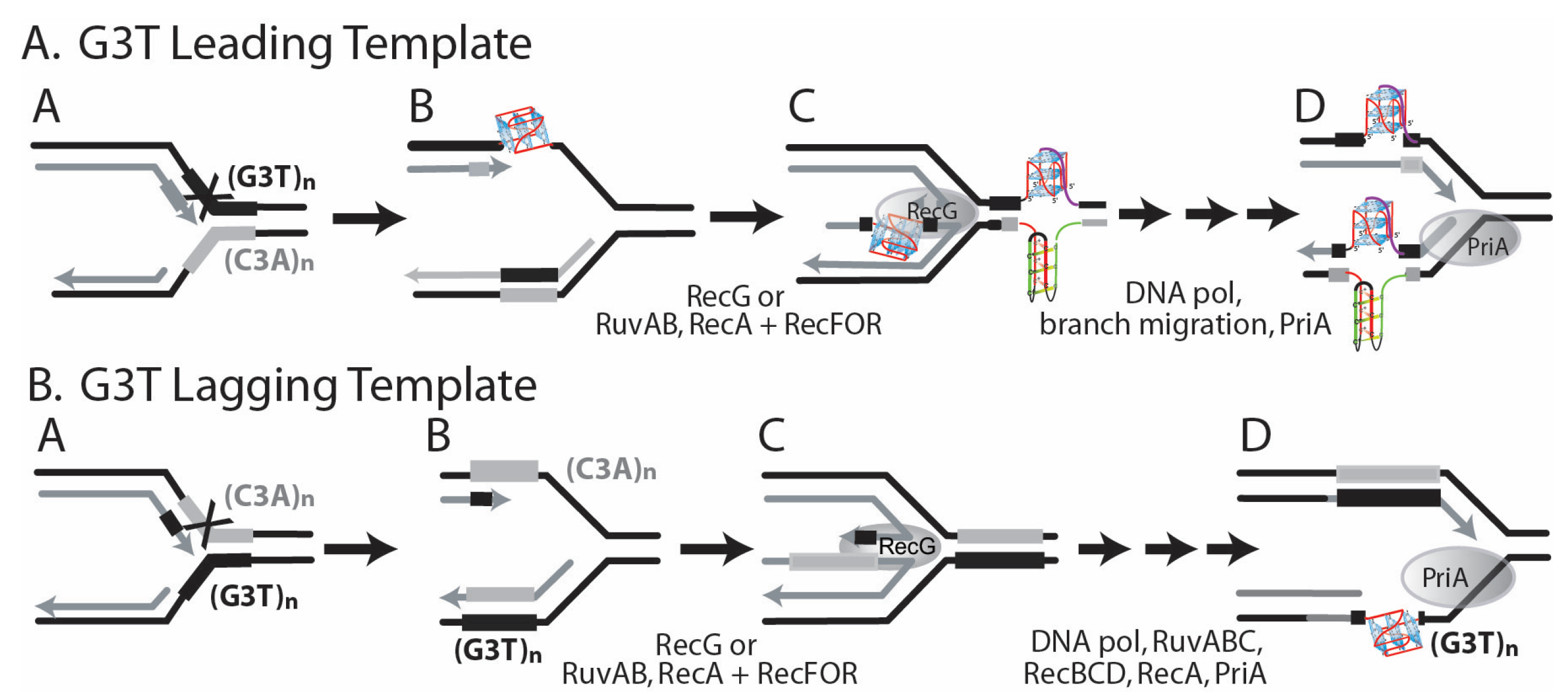
3.4. Influence of DinG on the Genetic Instability of Quadruplex-Forming Repeats
3.5. Influence of RecG and RecQ Helicases on the Instability of Quadruplex-Forming Repeats
3.6. Cmr Mutation Spectra for E. coli Containing Quadruplex-Forming Repeats
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sinden, R.R. DNA Structure and Function; Academic Press: Cambridge, MA, USA, 1994. [Google Scholar]
- Duardo, R.C.; Guerra, F.; Pepe, S.; Capranico, G. Non-B DNA structures as a booster of genome instability. Biochimie, 2023; in press. [Google Scholar] [CrossRef]
- Mirkin, S.M.; Frank-Kamenetskii, M.D. H-DNA and related structures. Annu. Rev. Biophys. Biomol. Struct. 1994, 23, 541–576. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Vasquez, K.M. Dynamic alternative DNA structures in biology and disease. Nat. Rev. Genet. 2023, 24, 211–234. [Google Scholar] [CrossRef] [PubMed]
- Sen, D.; Gilbert, W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 1988, 334, 364–366. [Google Scholar] [CrossRef]
- Chen, E.V.; Nicoludis, J.M.; Powell, B.M.; Li, K.S.; Yatsunyk, L.A. Crystal structure of a three-tetrad, parallel, K(+)-stabilized human telomeric G-quadruplex at 1.35 A resolution. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2023, 79, 144–150. [Google Scholar] [CrossRef]
- Simonsson, T. G-quadruplex DNA structures variations on a theme. Biol. Chem. 2001, 382, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Liano, D.; Monti, L.; Chowdhury, S.; Raguseo, F.; Di Antonio, M. Long-range DNA interactions: Inter-molecular G-quadruplexes and their potential biological relevance. Chem. Commun. 2022, 58, 12753–12762. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, J.; Zhang, Y.; Wang, Y.; Chen, J.; Bian, Y.; Xia, Y.; Yang, M.H.; Zheng, K.; Wang, K.B.; et al. Structure of the Major G-Quadruplex in the Human EGFR Oncogene Promoter Adopts a Unique Folding Topology with a Distinctive Snap-Back Loop. J. Am. Chem. Soc. 2023, 145, 16228–16237. [Google Scholar] [CrossRef]
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef] [PubMed]
- Phan, A.T.; Kuryavyi, V.; Luu, K.N.; Patel, D.J. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007, 35, 6517–6525. [Google Scholar] [CrossRef]
- Patel, D.J.; Phan, A.T.; Kuryavyi, V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: Diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007, 35, 7429–7455. [Google Scholar] [CrossRef] [PubMed]
- Dey, U.; Sarkar, S.; Teronpi, V.; Yella, V.R.; Kumar, A. G-quadruplex motifs are functionally conserved in cis-regulatory regions of pathogenic bacteria: An in-silico evaluation. Biochimie 2021, 184, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Deze, O.; Laffleur, B.; Cogne, M. Roles of G4-DNA and G4-RNA in Class Switch Recombination and Additional Regulations in B-Lymphocytes. Molecules 2023, 28, 1159. [Google Scholar] [CrossRef] [PubMed]
- Bahls, B.; Aljnadi, I.M.; Emidio, R.; Mendes, E.; Paulo, A. G-Quadruplexes in c-MYC Promoter as Targets for Cancer Therapy. Biomedicines 2023, 11, 969. [Google Scholar] [CrossRef]
- Panova, V.V.; Dolinnaya, N.G.; Novoselov, K.A.; Savitskaya, V.Y.; Chernykh, I.S.; Kubareva, E.A.; Alexeevski, A.V.; Zvereva, M.I. Conserved G-Quadruplex-Forming Sequences in Mammalian TERT Promoters and Their Effect on Mutation Frequency. Life 2023, 13, 1478. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.E.; Cao, K.; Ryvkin, P.; Wang, L.S.; Johnson, F.B. Altered gene expression in the Werner and Bloom syndromes is associated with sequences having G-quadruplex forming potential. Nucleic Acids Res. 2010, 38, 1114–1122. [Google Scholar] [CrossRef]
- Gunaratnam, M.; Neidle, S. An evaluation cascade for G-quadruplex telomere targeting agents in human cancer cells. Methods Mol. Biol. 2010, 613, 303–313. [Google Scholar] [CrossRef]
- Escaja, N.; Mir, B.; Garavis, M.; Gonzalez, C. Non-G Base Tetrads. Molecules 2022, 27, 5287. [Google Scholar] [CrossRef]
- Abou Assi, H.; Garavis, M.; Gonzalez, C.; Damha, M.J. i-Motif DNA: Structural features and significance to cell biology. Nucleic Acids Res. 2018, 46, 8038–8056. [Google Scholar] [CrossRef]
- Luo, X.; Zhang, J.; Gao, Y.; Pan, W.; Yang, Y.; Li, X.; Chen, L.; Wang, C.; Wang, Y. Emerging roles of i-motif in gene expression and disease treatment. Front. Pharmacol. 2023, 14, 1136251. [Google Scholar] [CrossRef]
- Atkinson, J.; McGlynn, P. Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res. 2009, 37, 3475–3492. [Google Scholar] [CrossRef]
- Kim, S.H.; Pytlos, M.J.; Sinden, R.R. Replication restart: A pathway for (CTG).(CAG) repeat deletion in Escherichia coli. Mutat. Res. 2006, 595, 5–22. [Google Scholar] [CrossRef]
- Piazza, A.; Boule, J.B.; Lopes, J.; Mingo, K.; Largy, E.; Teulade-Fichou, M.P.; Nicolas, A. Genetic instability triggered by G-quadruplex interacting Phen-DC compounds in Saccharomyces cerevisiae. Nucleic Acids Res. 2010, 38, 4337–4348. [Google Scholar] [CrossRef]
- Parekh, V.J.; Niccum, B.A.; Shah, R.; Rivera, M.A.; Novak, M.J.; Geinguenaud, F.; Wien, F.; Arluison, V.; Sinden, R.R. Role of Hfq in Genome Evolution: Instability of G-Quadruplex Sequences in E. coli. Microorganisms 2019, 8, 28. [Google Scholar] [CrossRef] [PubMed]
- Cueny, R.R.; McMillan, S.D.; Keck, J.L. G-quadruplexes in bacteria: Insights into the regulatory roles and interacting proteins of non-canonical nucleic acid structures. Crit. Rev. Biochem. Mol. Biol. 2022, 57, 539–561. [Google Scholar] [CrossRef] [PubMed]
- Paul, T.; Voter, A.F.; Cueny, R.R.; Gavrilov, M.; Ha, T.; Keck, J.L.; Myong, S. E. coli Rep helicase and RecA recombinase unwind G4 DNA and are important for resistance to G4-stabilizing ligands. Nucleic Acids Res. 2020, 48, 6640–6653. [Google Scholar] [CrossRef]
- Estep, K.N.; Butler, T.J.; Ding, J.; Brosh, R.M. G4-Interacting DNA Helicases and Polymerases: Potential Therapeutic Targets. Curr. Med. Chem. 2019, 26, 2881–2897. [Google Scholar] [CrossRef] [PubMed]
- McGlynn, P. Helicases that underpin replication of protein-bound DNA in Escherichia coli. Biochem. Soc. Trans. 2011, 39, 606–610. [Google Scholar] [CrossRef]
- Wu, X.; Maizels, N. Substrate-specific inhibition of RecQ helicase. Nucleic Acids Res. 2001, 29, 1765–1771. [Google Scholar] [CrossRef]
- Bagchi, D.; Manosas, M.; Zhang, W.; Manthei, K.A.; Hodeib, S.; Ducos, B.; Keck, J.L.; Croquette, V. Single molecule kinetics uncover roles for E. coli RecQ DNA helicase domains and interaction with SSB. Nucleic Acids Res. 2018, 46, 8500–8515. [Google Scholar] [CrossRef]
- Khairnar, N.P.; Maurya, G.K.; Pandey, N.; Das, A.; Misra, H.S. DrRecQ regulates guanine quadruplex DNA structure dynamics and its impact on radioresistance in Deinococcus radiodurans. Mol. Microbiol. 2019, 112, 854–865. [Google Scholar] [CrossRef]
- Voter, A.F.; Qiu, Y.; Tippana, R.; Myong, S.; Keck, J.L. A guanine-flipping and sequestration mechanism for G-quadruplex unwinding by RecQ helicases. Nat. Commun. 2018, 9, 4201. [Google Scholar] [CrossRef]
- Voloshin, O.N.; Camerini-Otero, R.D. The DinG protein from Escherichia coli is a structure-specific helicase. J. Biol. Chem. 2007, 282, 18437–18447. [Google Scholar] [CrossRef]
- Voloshin, O.N.; Vanevski, F.; Khil, P.P.; Camerini-Otero, R.D. Characterization of the DNA damage-inducible helicase DinG from Escherichia coli. J. Biol. Chem. 2003, 278, 28284–28293. [Google Scholar] [CrossRef]
- Mackay, R.P.; Xu, Q.; Weinberger, P.M. R-Loop Physiology and Pathology: A Brief Review. DNA Cell Biol. 2020, 39, 1914–1925. [Google Scholar] [CrossRef] [PubMed]
- Boubakri, H.; de Septenville, A.L.; Viguera, E.; Michel, B. The helicases DinG, Rep and UvrD cooperate to promote replication across transcription units in vivo. EMBO J. 2010, 29, 145–157. [Google Scholar] [CrossRef]
- McGlynn, P.; Lloyd, R.G. RecG helicase activity at three- and four-strand DNA structures. Nucleic Acids Res. 1999, 27, 3049–3056. [Google Scholar] [CrossRef]
- Trinh, T.Q.; Sinden, R.R. Preferential DNA secondary structure mutagenesis in the lagging strand of replication in E. coli. Nature 1991, 352, 544–547. [Google Scholar] [CrossRef]
- Hashem, V.I.; Rosche, W.A.; Sinden, R.R. Genetic assays for measuring rates of (CAG).(CTG) repeat instability in Escherichia coli. Mutat. Res. 2002, 502, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Cai, L.; Pytlos, M.J.; Edwards, S.F.; Sinden, R.R. Generation of long tracts of disease-associated DNA repeats. Biotechniques 2005, 38, 247–253. [Google Scholar] [CrossRef]
- Luria, S.E.; Delbruck, M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics 1943, 28, 491–511. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Ma, W.T.; Sandri, G.H. On fluctuation analysis: A new, simple and efficient method for computing the expected number of mutants. Genetica 1992, 85, 173–179. [Google Scholar] [CrossRef]
- Cappannini, A.; Mosca, K.; Mukherjee, S.; Moafinejad, S.N.; Sinden, R.R.; Arluison, V.; Bujnicki, J.; Wien, F. NACDDB: Nucleic Acid Circular Dichroism Database. Nucleic Acids Res. 2022, 51, D226–D231. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.; Pourpak, A.; Beetz-Rogers, K.; Gokhale, V.; Sun, D.; Hurley, L.H. Formation of pseudosymmetrical G-quadruplex and i-motif structures in the proximal promoter region of the RET oncogene. J. Am. Chem. Soc. 2007, 129, 10220–10228. [Google Scholar] [CrossRef] [PubMed]
- Trinh, T.Q.; Sinden, R.R. The influence of primary and secondary DNA structure in deletion and duplication between direct repeats in Escherichia coli. Genetics 1993, 134, 409–422. [Google Scholar] [CrossRef]
- Rosche, W.A.; Trinh, T.Q.; Sinden, R.R. Differential DNA secondary structure-mediated deletion mutation in the leading and lagging strands. J. Bacteriol. 1995, 177, 4385–4391. [Google Scholar] [CrossRef]
- Sinden, R.R.; Zheng, G.X.; Brankamp, R.G.; Allen, K.N. On the deletion of inverted repeated DNA in Escherichia coli: Effects of length, thermal stability, and cruciform formation in vivo. Genetics 1991, 129, 991–1005. [Google Scholar] [CrossRef]
- Edwards, S.F.; Hashem, V.I.; Klysik, E.A.; Sinden, R.R. Genetic instabilities of (CCTG).(CAGG) and (ATTCT).(AGAAT) disease-associated repeats reveal multiple pathways for repeat deletion. Mol. Carcinog. 2009, 48, 336–349. [Google Scholar] [CrossRef]
- Wang, Y.; Patel, D.J. Solution structure of the Tetrahymena telomeric repeat d(T2G4)4 G-tetraplex. Structure 1994, 2, 1141–1156. [Google Scholar] [CrossRef]
- Kumar, N.; Sahoo, B.; Varun, K.A.; Maiti, S.; Maiti, S. Effect of loop length variation on quadruplex-Watson Crick duplex competition. Nucleic Acids Res. 2008, 36, 4433–4442. [Google Scholar] [CrossRef]
- Nambiar, M.; Goldsmith, G.; Moorthy, B.T.; Lieber, M.R.; Joshi, M.V.; Choudhary, B.; Hosur, R.V.; Raghavan, S.C. Formation of a G-quadruplex at the BCL2 major breakpoint region of the t(14;18) translocation in follicular lymphoma. Nucleic Acids Res. 2011, 39, 936–948. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Hurley, L.H. The importance of negative superhelicity in inducing the formation of G-quadruplex and i-motif structures in the c-Myc promoter: Implications for drug targeting and control of gene expression. J. Med. Chem. 2009, 52, 2863–2874. [Google Scholar] [CrossRef]
- Sekibo, D.A.T.; Fox, K.R. The effects of DNA supercoiling on G-quadruplex formation. Nucleic Acids Res. 2017, 45, 12069–12079. [Google Scholar] [CrossRef] [PubMed]
- Duquette, M.L.; Huber, M.D.; Maizels, N. G-rich proto-oncogenes are targeted for genomic instability in B-cell lymphomas. Cancer Res. 2007, 67, 2586–2594. [Google Scholar] [CrossRef] [PubMed]
- Duquette, M.L.; Handa, P.; Vincent, J.A.; Taylor, A.F.; Maizels, N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes. Dev. 2004, 18, 1618–1629. [Google Scholar] [CrossRef] [PubMed]
- Belotserkovskii, B.P.; Liu, R.; Tornaletti, S.; Krasilnikova, M.M.; Mirkin, S.M.; Hanawalt, P.C. Mechanisms and implications of transcription blockage by guanine-rich DNA sequences. Proc. Natl. Acad. Sci. USA 2010, 107, 12816–12821. [Google Scholar] [CrossRef]
- Hashem, V.I.; Rosche, W.A.; Sinden, R.R. Genetic recombination destabilizes (CTG)n.(CAG)n repeats in E. coli. Mutat. Res. 2004, 554, 95–109. [Google Scholar] [CrossRef]
- Hashem, V.I.; Sinden, R.R. Duplications between direct repeats stabilized by DNA secondary structure occur preferentially in the leading strand during DNA replication. Mutat. Res. 2005, 570, 215–226. [Google Scholar] [CrossRef]
- Glickman, B.W.; Ripley, L.S. Structural intermediates of deletion mutagenesis: A role for palindromic DNA. Proc. Natl. Acad. Sci. USA 1984, 81, 512–516. [Google Scholar] [CrossRef]
- Rosche, W.A.; Trinh, T.Q.; Sinden, R.R. Leading strand specific spontaneous mutation corrects a quasipalindrome by an intermolecular strand switch mechanism. J. Mol. Biol. 1997, 269, 176–187. [Google Scholar] [CrossRef]
- Rosche, W.A.; Ripley, L.S.; Sinden, R.R. Primer-template misalignments during leading strand DNA synthesis account for the most frequent spontaneous mutations in a quasipalindromic region in Escherichia coli. J. Mol. Biol. 1998, 284, 633–646. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, M.; Lacirignola, J.J.; Hurley, R.L.; Lovett, S.T. A novel mutational hotspot in a natural quasipalindrome in Escherichia coli. J. Mol. Biol. 2000, 302, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Gordenin, D.A.; Lobachev, K.S.; Degtyareva, N.P.; Malkova, A.L.; Perkins, E.; Resnick, M.A. Inverted DNA repeats: A source of eukaryotic genomic instability. Mol. Cell Biol. 1993, 13, 5315–5322. [Google Scholar] [CrossRef] [PubMed]
- Rosche, W.A.; Jaworski, A.; Kang, S.; Kramer, S.F.; Larson, J.E.; Geidroc, D.P.; Wells, R.D.; Sinden, R.R. Single-stranded DNA-binding protein enhances the stability of CTG triplet repeats in Escherichia coli. J. Bacteriol. 1996, 178, 5042–5044. [Google Scholar] [CrossRef]
- Sun, H.; Karow, J.K.; Hickson, I.D.; Maizels, N. The Bloom’s syndrome helicase unwinds G4 DNA. J. Biol. Chem. 1998, 273, 27587–27592. [Google Scholar] [CrossRef]
- London, T.B.; Barber, L.J.; Mosedale, G.; Kelly, G.P.; Balasubramanian, S.; Hickson, I.D.; Boulton, S.J.; Hiom, K. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J. Biol. Chem. 2008, 283, 36132–36139. [Google Scholar] [CrossRef]
- Krasilnikova, M.M.; Mirkin, S.M. Replication stalling at Friedreich’s ataxia (GAA)n repeats in vivo. Mol. Cell Biol. 2004, 24, 2286–2295. [Google Scholar] [CrossRef] [PubMed]
- Maizels, N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat. Struct. Mol. Biol. 2006, 13, 1055–1059. [Google Scholar] [CrossRef]
- Eddy, J.; Maizels, N. Selection for the G4 DNA motif at the 5′ end of human genes. Mol. Carcinog. 2009, 48, 319–325. [Google Scholar] [CrossRef]
- Cahoon, L.A.; Seifert, H.S. An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science 2009, 325, 764–767. [Google Scholar] [CrossRef]
- Gaffke, L.; Kubiak, K.; Cyske, Z.; Wegrzyn, G. Differential Chromosome- and Plasmid-Borne Resistance of Escherichia coli hfq Mutants to High Concentrations of Various Antibiotics. Int. J. Mol. Sci. 2021, 22, 8886. [Google Scholar] [CrossRef] [PubMed]
- Butler, T.J.; Estep, K.N.; Sommers, J.A.; Maul, R.W.; Moore, A.Z.; Bandinelli, S.; Cucca, F.; Tuke, M.A.; Wood, A.R.; Bharti, S.K.; et al. Mitochondrial genetic variation is enriched in G-quadruplex regions that stall DNA synthesis in vitro. Hum. Mol. Genet. 2020, 29, 1292–1309. [Google Scholar] [CrossRef] [PubMed]
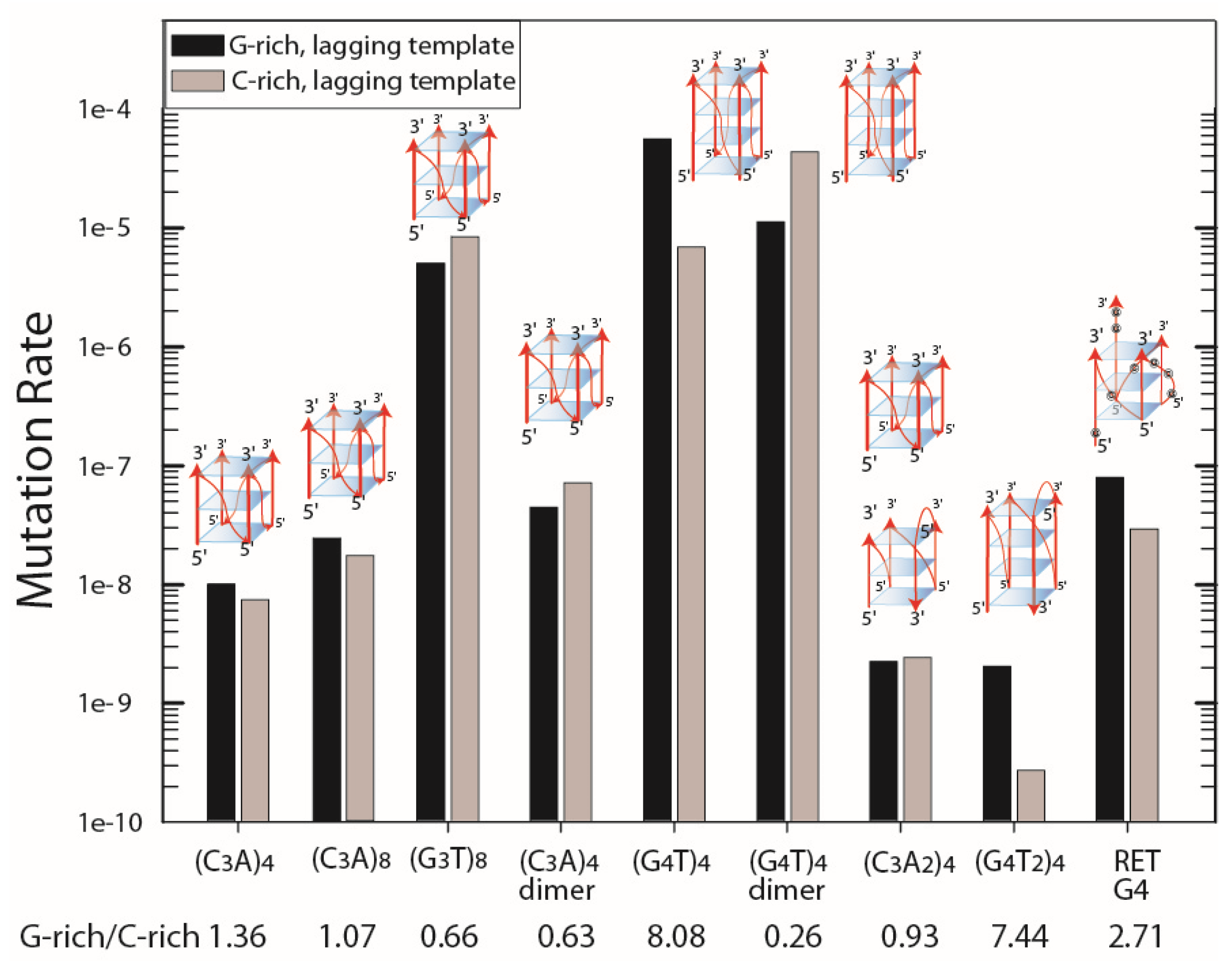

| Mutation Rate | ||||||
|---|---|---|---|---|---|---|
| Quadruplex | G-rich Strand Lagging Template | C-rich Strand Lagging Template | ||||
| wt | dinG | dinG/wt | wt | dinG | dinG/wt | |
| (G3T)4 | 9.93 × 10−9 | 3.27 × 10−8 | 3.29 | 7.32 × 10−9 | 2.47 × 10−8 | 3.37 |
| (G3T)8 | 4.99 × 10−6 | 3.18 × 10−5 | 6.37 | 8.28 × 10−6 | 5.94 × 10−5 | 7.17 |
| (G4T)4 | 5.50 × 10−5 | 5.50 × 10−5 | 1.00 | 6.81 × 10−6 | 1.04 × 10−5 | 1.53 |
| (G4T)4 D | 1.10 × 10−5 | 4.78 × 10−6 | 0.43 | 4.27 × 10−5 | 4.00 × 10−5 | 0.94 |
| RET G4 | 2.88 × 10−8 | 5.04 × 10−8 | 1.75 | 7.80 × 10−8 | 6.58 × 10−8 | 0.84 |
| Genotype | Mutation Rate | Ratio | Mutant/wt | ||
|---|---|---|---|---|---|
| G-rich Lag | C-rich Lag | G-rich/C-rich | G-rich | C-rich | |
| (G3T)8 repeat | |||||
| wild type | 1.09 × 10−7 | 5.95 × 10−9 | 18.34 | 1 | 1 |
| recG | 3.12 × 10−8 | 6.25 × 10−9 | 5.00 | 0.29 | 1.05 |
| recQ | 2.12 × 10−7 | 1.39 × 10−8 | 15.23 | 1.95 | 2.34 |
| recG recQ | 1.17 × 10−7 | 6.79 × 10−9 | 173.63 | 10.80 | 1.14 |
| (G4T)4-dimer repeat | |||||
| wild type | 2.70 × 10−5 | 1.87 × 10−6 | 14.41 | 1 | 1 |
| recG | 3.86 × 10−5 | 1.24 × 10−6 | 31.12 | 1.43 | 0.66 |
| recQ | 1.56 × 10−5 | 2.55 × 10−6 | 6.11 | 0.58 | 1.36 |
| recG recQ | 1.72 × 10−5 | 2.38 × 10−6 | 7.23 | 0.64 | 1.27 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parekh, V.J.; Węgrzyn, G.; Arluison, V.; Sinden, R.R. Genomic Instability of G-Quadruplex Sequences in Escherichia coli: Roles of DinG, RecG, and RecQ Helicases. Genes 2023, 14, 1720. https://doi.org/10.3390/genes14091720
Parekh VJ, Węgrzyn G, Arluison V, Sinden RR. Genomic Instability of G-Quadruplex Sequences in Escherichia coli: Roles of DinG, RecG, and RecQ Helicases. Genes. 2023; 14(9):1720. https://doi.org/10.3390/genes14091720
Chicago/Turabian StyleParekh, Virali J., Grzegorz Węgrzyn, Véronique Arluison, and Richard R. Sinden. 2023. "Genomic Instability of G-Quadruplex Sequences in Escherichia coli: Roles of DinG, RecG, and RecQ Helicases" Genes 14, no. 9: 1720. https://doi.org/10.3390/genes14091720
APA StyleParekh, V. J., Węgrzyn, G., Arluison, V., & Sinden, R. R. (2023). Genomic Instability of G-Quadruplex Sequences in Escherichia coli: Roles of DinG, RecG, and RecQ Helicases. Genes, 14(9), 1720. https://doi.org/10.3390/genes14091720








