Molecular Genetic Analysis of Russian Patients with Coagulation Factor FVII Deficiency
Abstract
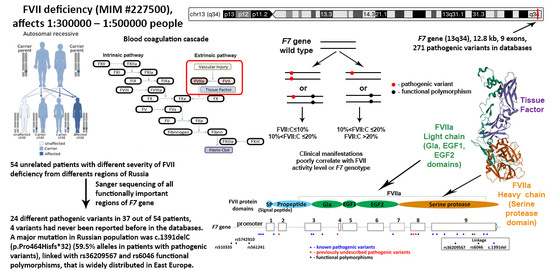
:1. Introduction
2. Materials and Methods
3. Results
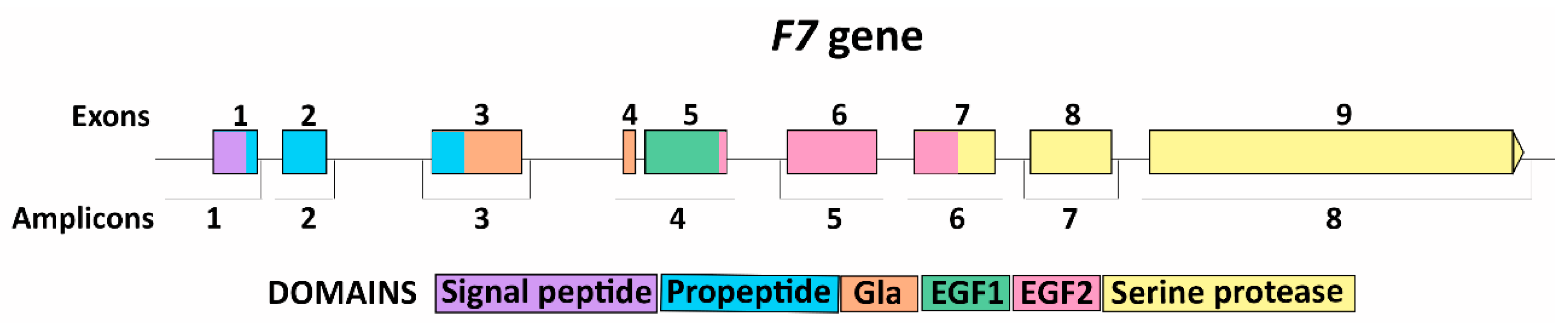
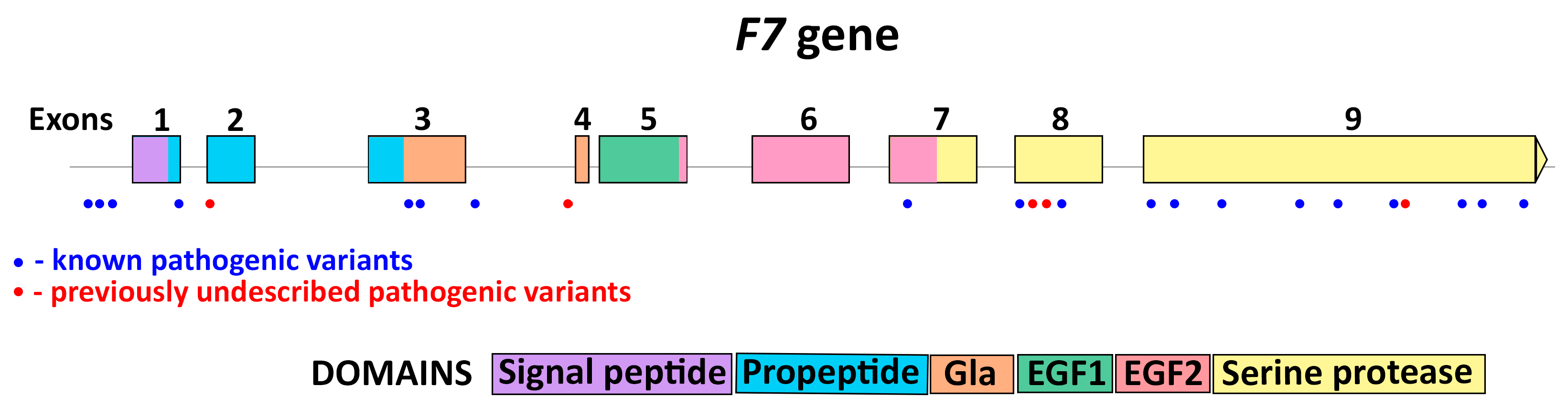
3.1. F7 Mutation Spectrum
3.2. Novel Mutations
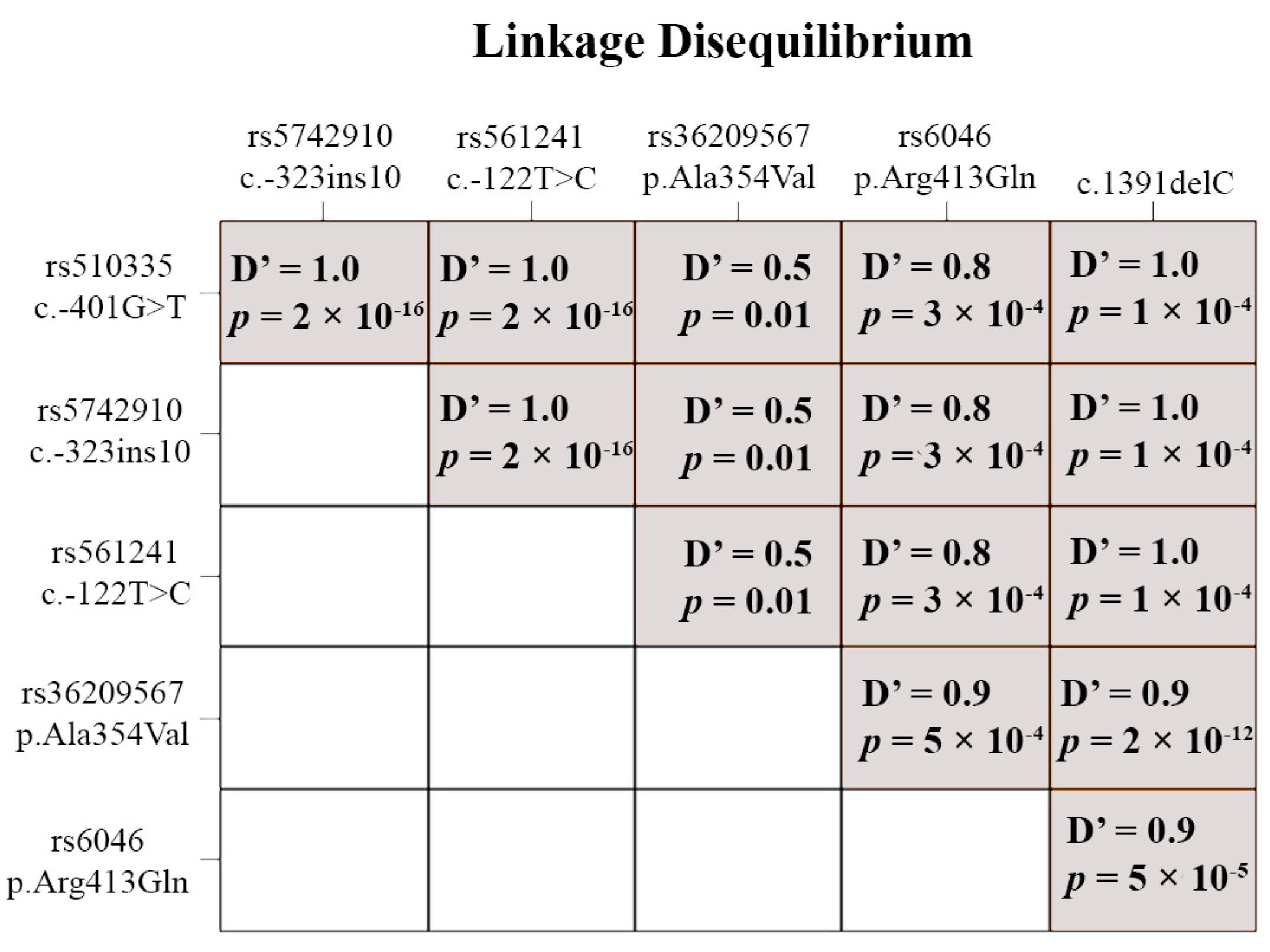
3.3. Functional Polymorphisms
3.4. Genotype and Phenotype Interaction
4. Discussion
4.1. Mutation Spectrum of F7 Gene
4.2. Gene Variant c.1061 C>T (p.Ala354Val)
4.3. Functional Polymorphisms
4.4. Genotype and Phenotype Interaction
4.5. Patients without Found Pathogenic Variants
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bernardi, F.; Mariani, G. Biochemical, molecular and clinical aspects of coagulation factor VII and its role in hemostasis and thrombosis. Haematologica 2021, 106, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Ken-Dror, G.; Drenos, F.; Humphries, S.E.; Talmud, P.J.; Hingorani, A.D.; Kivimäki, M.; Kumari, M.; Bauer, K.A.; Morrissey, J.H.; Ireland, H.A. Haplotype and genotype effects of the F7 gene on circulating factor VII, coagulation activation markers and incident coronary heart disease in UK men. J. Thromb. Haemost. 2010, 8, 2394–2403. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Yuan, Q.; Du, Z.; Liu, C.; Xu, H.; Yang, W.; Chen, L.; Zhao, J.; Xie, R.; Hu, J.; et al. Contribution of factor VII polymorphisms to coagulopathy in patients with isolated traumatic brain injury. Clin. Neurol. Neurosurg. 2021, 208, 106836. [Google Scholar] [CrossRef] [PubMed]
- Vadivel, K.; Bajaj, S.P. Structural biology of factor VIIa/tissue factor initiated coagulation. Front. Biosci. 2012, 17, 2476–2494. [Google Scholar] [CrossRef]
- Millar, D.S.; Kemball-Cook, G.; McVey, J.H.; Tuddenham, E.G.; Mumford, A.D.; Attock, G.B.; Reverter, J.C.; Lanir, N.; Parapia, L.A.; Reynaud, J.; et al. Molecular analysis of the genotype-phenotype relationship in factor VII deficiency. Hum. Genet. 2000, 107, 327–342. [Google Scholar] [CrossRef]
- Herrmann, F.H.; Wulff, K.; Auerswald, G.; Schulman, S.; Astermark, J.; Batorova, A.; Kreuz, W.; Pollmann, H.; Ruiz-Saez, A.; De Bosch, N.; et al. Factor VII deficiency: Clinical manifestation of 717 subjects from Europe and Latin America with mutations in the factor 7 gene. Haemophilia 2009, 15, 267–280. [Google Scholar] [CrossRef] [PubMed]
- Quintavalle, G.; Riccardi, F.; Rivolta, G.F.; Martorana, D.; Di Perna, C.; Percesepe, A.; Tagliaferri, A.; Ad-Hoc Study Group. F7 gene variants modulate protein levels in a large cohort of patients with factor VII deficiency. Results from a genotype-phenotype study. Thromb. Haemost. 2017, 117, 1455–1464. [Google Scholar] [CrossRef] [PubMed]
- Shahbazi, S.; Mahdian, R. Factor VII Gene Defects: Review of Functional Studies and Their Clinical Implications. Iran. Biomed. J. 2019, 23, 165–174. [Google Scholar] [CrossRef]
- Ouardani, C.; Elmahmoudi, H.; ElBorgi, W.; Gharbi, M.; Meriem, A.; Gouider, E. Clinical phenotype and F7 gene genotype in 40 Tunisian patients with congenital factor VII deficiency. Blood Coagul. Fibrinolysis 2022, 33, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Hunault, M.; Arbini, A.A.; Lopaciuk, S.; Carew, J.A.; Bauer, K.A. The Arg353Gln polymorphism reduces the level of coagulation factor VII. In vivo and in vitro studies. Arterioscler. Thromb. Vasc. Biol. 1997, 17, 2825–2829. [Google Scholar] [CrossRef]
- Pollak, E.S.; Hung, H.L.; Godin, W.; Overton, G.C.; High, K.A. Functional characterization of the human factor VII 5′-flanking region. J. Biol. Chem. 1996, 271, 1738–1747. [Google Scholar] [CrossRef] [PubMed]
- Maniatis, T.; Fritsch, E.F.; Sambrook, J. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1982; ISBN 978-0-87969-136-3. [Google Scholar]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A Method and Server for Predicting Damaging Missense Mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, J.M.; Cooper, D.N.; Schuelke, M.; Seelow, D. MutationTaster2: Mutation Prediction for the Deep-Sequencing Age. Nat. Methods 2014, 11, 361–362. [Google Scholar] [CrossRef]
- Shihab, H.A.; Gough, J.; Cooper, D.N.; Stenson, P.D.; Barker, G.L.A.; Edwards, K.J.; Day, I.N.M.; Gaunt, T.R. Predicting the Functional, Molecular and Phenotypic Consequences of Amino Acid Substitutions using Hidden Markov Models. Hum. Mutat. 2013, 34, 57–65. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023; Available online: https://www.R-project.org/ (accessed on 22 August 2023).
- Warnes, G.; Gorjanc, W.C.F.G.; Leisch, F.; Man, M. Genetics: Population Genetics. R package version 1.3.8.1.3. 2021. Available online: https://CRAN.R-project.org/package=genetics (accessed on 22 August 2023).
- Carew, J.A.; Pollak, E.S.; High, K.A.; Bauer, K.A. Severe factor VII deficiency due to a mutation disrupting an Sp1 binding site in the factor VII promoter. Blood 1998, 92, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Kavlie, A.; Hiltunen, L.; Rasi, V.; Prydz, H. Two novel mutations in the human coagulation factor VII promoter. Thromb. Haemost. 2003, 90, 194–205. [Google Scholar] [CrossRef] [PubMed]
- Wulff, K.; Herrmann, F.H. Twenty two novel mutations of the factor VII gene in factor VII deficiency. Hum. Mutat. 2000, 15, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Giansily-Blaizot, M.; Aguilar-Martinez, P.; Biron-Andreani, C.; Jeanjean, P.; Igual, H.; Schved, J.F.; Study Group of Factor Seven Deficiency. Analysis of the genotypes and phenotypes of 37 unrelated patients with inherited factor VII deficiency. Eur. J. Hum. Genet. 2001, 9, 105–112. [Google Scholar] [CrossRef]
- Peyvandi, F.; Jenkins, P.V.; Mannucci, P.M.; Billio, A.; Zeinali, S.; Perkins, S.J.; Perry, D.J. Molecular characterisation and three-dimensional structural analysis of mutations in 21 unrelated families with inherited factor VII deficiency. Thromb. Haemost. 2000, 84, 250–257. [Google Scholar] [CrossRef] [PubMed]
- Alshinawi, C.; Scerri, C.; Galdies, R.; Aquilina, A.; Felice, A.E. Two new missense mutations (P134T and A244V) in the coagulation factor VII gene. Hum. Mutat. 1998, 11 (Suppl. S1), S189–S191. [Google Scholar] [CrossRef]
- Bernardi, F.; Castaman, G.; Pinotti, M.; Ferraresi, P.; Di Iasio, M.G.; Lunghi, B.; Rodeghiero, F.; Marchetti, G. Mutation pattern in clinically asymptomatic coagulation factor VII deficiency. Hum. Mutat. 1996, 8, 108–115. [Google Scholar] [CrossRef]
- Tamary, H.; Fromovich, Y.; Shalmon, L.; Reich, Z.; Dym, O.; Lanir, N.; Brenner, B.; Paz, M.; Luder, A.S.; Blau, O.; et al. Ala244Val is a common, probably ancient mutation causing factor VII deficiency in Moroccan and Iranian Jews. Thromb. Haemost. 1996, 76, 283–291. [Google Scholar] [CrossRef]
- Marchetti, G.; Patracchini, P.; Gemmati, D.; DeRosa, V.; Pinotti, M.; Rodorigo, G.; Casonato, A.; Girolami, A.; Bernardi, F. Detection of two missense mutations and characterization of a repeat polymorphism in the factor VII gene (F7). Hum. Genet. 1992, 89, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Arbini, A.A.; Bodkin, D.; Lopaciuk, S.; Bauer, K.A. Molecular analysis of Polish patients with factor VII deficiency. Blood 1994, 84, 2214–2220. [Google Scholar] [CrossRef] [PubMed]
- Peyvandi, F.; Di Michele, D.; Bolton-Maggs, P.H.; Lee, C.A.; Tripodi, A.; Srivastava, A. Project on Consensus Definitions in Rare Bleeeding Disorders of the Factor VIII/Factor IX Scientific and Standardisation Committee of the International Society on Thrombosis and Haemostasis. Classification of rare bleeding disorders (RBDs) based on the association between coagulant factor activity and clinical bleeding severity. J. Thromb. Haemost. 2012, 10, 1938–1943. [Google Scholar] [CrossRef] [PubMed]
- Mariani, G.; Herrmann, F.H.; Dolce, A.; Batorova, A.; Etro, D.; Peyvandi, F.; Wulff, K.; Schved, J.F.; Auerswald, G.; Ingerslev, J.; et al. Clinical phenotypes and factor VII genotype in congenital factor VII deficiency. Thromb. Haemost. 2005, 93, 481–487. [Google Scholar] [CrossRef]
- Fromovich-Amit, Y.; Zivelin, A.; Rosenberg, N.; Tamary, H.; Landau, M.; Seligsohn, U. Characterization of mutations causing factor VII deficiency in 61 unrelated Israeli patients. J. Thromb. Haemost. 2004, 2, 1774–1781. [Google Scholar] [CrossRef] [PubMed]
- Borhany, M.; Boijout, H.; Pellequer, J.L.; Shamsi, T.; Moulis, G.; Aguilar-Martinez, P.; Schved, J.F.; Giansily-Blaizot, M. Genotype and phenotype relationships in 10 Pakistani unrelated patients with inherited factor VII deficiency. Haemophilia 2013, 19, 893–897. [Google Scholar] [CrossRef]
- Mariani, G.; Bernardi, F. Factor VII Deficiency. Semin. Thromb. Hemost. 2009, 35, 400–406. [Google Scholar] [CrossRef]
- van ‘t Hooft, F.M.; Silveira, A.; Tornvall, P.; Iliadou, A.; Ehrenborg, E.; Eriksson, P.; Hamsten, A. Two common functional polymorphisms in the promoter region of the coagulation factor VII gene determining plasma factor VII activity and mass concentration. Blood 1999, 93, 3432–3441. [Google Scholar] [CrossRef] [PubMed]
- Sabater-Lleal, M.; Chillón, M.; Howard, T.E.; Gil, E.; Almasy, L.; Blangero, J.; Fontcuberta, J.; Soria, J.M. Functional analysis of the genetic variability in the F7 gene promoter. Atherosclerosis 2007, 195, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Ferraresi, P.; Balestra, D.; Guittard, C.; Buthiau, D.; Pan-Petesh, B.; Maestri, I.; Farah, R.; Pinotti, M.; Giansily-Blaizot, M. Next-generation sequencing and recombinant expression characterized aberrant splicing mechanisms and provided correction strategies in factor VII deficiency. Haematologica 2020, 105, 829–837. [Google Scholar] [CrossRef] [PubMed]
| Amplicon | Primer | Nucleotide Sequence | Target | Fragment Length, bp |
|---|---|---|---|---|
| 1 | F7F1x | cct aag aaa cca gcc tcc ctt | Promoter; exon 1 | 686 |
| F7R1 | agc cgc cag aaa acc ctc ct | |||
| 2 | F7F2 | cta gct cac agc atg gcc tt | exon 2 | 261 |
| F7R2 | cgg tca ctt cct ctc gag ca | |||
| 3 | F7F3 | agc gcc gct ccc ctc ctc ca | exon 3 1 | 380 |
| F7R3 | gcc gca gcc aaa gag acg ca | |||
| 4 | F7F45 | gtc caa gtc ccc caa ccc ca | exon 4; exon 5 | 439 |
| F7R45 | cac cta gac caa ttt cca act | |||
| 5 | F7F6 | aga aca cca ctg ctg acc ca | exon 6 | 384 |
| F7R6x | tgc cca gat ccc acc tca ca | |||
| 6 | F7F7 | tcc cat agc ctc ggc ctc aa | exon 7 | 315 |
| F7R7 | cct gcc cat ttt ccc ttc ca | |||
| 7 | F7F8x | tgc aat gcc aga ggt tcc tt | exon 8 1 | 477 |
| F7R8x | gca cga agc cca gag cca ca | |||
| 8 | F7DF9 F7R9 | tgg cca cag ccc atc ccc at | exon 9 | 775 |
| cct cct cta ccc cat taa ct |
| # | Pathogenic Variant | Exon/Intron | Domain | Defect Type | Allele Count | Reference |
|---|---|---|---|---|---|---|
| 1 | c. −55C>T | Promoter | Promoter | Regulatory | 4 | [19] |
| 2 | c. −32A>C | Promoter | Promoter | Regulatory | 1 | [20] |
| 3 | c. −30A>C | Promoter | Promoter | Regulatory | 1 | [5] |
| 4 | c.64G>A (p.Gly22Ser) | Exon 1 | Propeptide | Missense/ splicing | 3 | [21] |
| 5 | c.65G>A (p.Gly22Asp) | Exon 2 | Propeptide | Missense/ splicing | 1 | New |
| 6 | c.218G>A (p.Leu73Gln) | Exon 3 | Gla | Missense | 1 | [5] |
| 7 | c.226G>A (p.Glu76Lys) | Exon 3 | Gla | Missense | 1 | [22] |
| 8 | c.291+1G>A | Intron 3 | Gla | Splicing | 2 | [23] |
| 9 | c.347−1G>A | Intron 3 | Gla | Splicing | 1 | New |
| 10 | c.580C>A (p.Pro194Thr) | Exon 7 | EGF2 | Missense | 1 | [24] |
| 11 | c.691_693del (p.Leu231del) | Exon 8 | Serine protease | Inframe (microdeletion) | 2 | [6] |
| 12 | c.713G>C (p.Cys238Ser) | Exon 8 | Serine protease | Missense | 1 | New |
| 13 | c.728 T>C (p.Ile243Thr) | Exon 8 | Serine protease | Missense | 1 | New |
| 14 | c.760G>C (p.Cys254Arg) | Exon 8 | Serine protease | Missense | 1 | [6] |
| 15 | c.817_831del (p.Leu273_Asp277del) | Exon 9 | Serine protease | Inframe (microdeletion) | 2 | [21] |
| 16 | c.847C>T (p.Arg283Trp) | Exon 9 | Serine protease | Missense | 1 | [25] |
| 17 | c.911C>T (p.Ala304Val) | Exon 9 | Serine protease | Missense | 2 | [26] |
| 18 | c.1072A>G (p.Met358Val) | Exon 9 | Serine protease | Missense | 1 | [25] |
| 19 | c.1109G>T (p.Cys370Phe) | Exon 9 | Serine protease | Missense | 1 | [27] |
| 20 | c.1171G>A (p.Gly391Ser) | Exon 9 | Serine protease | Missense | 1 | [27] |
| 21 | c.1175A>G (p.Tyr392Cys) | Exon 9 | Serine protease | Missense | 1 | New |
| 22 | c.1285G>A (p.Ala429Thr) | Exon 9 | Serine protease | Missense | 2 | [22] |
| 23 | c.1310A>T (p.Tyr437Phe) | Exon 9 | Serine protease | Missense | 1 | [6] |
| 24 | c.1391delC (p.Pro464Hisfs*32) | Exon 9 | Serine protease | Frameshift (microdeletion) | 29 | [28] |
| rs ID | Functional Polymorphism | MAF (Global) | MAF (Russia, RUSeq Data) | MAF (Our Sample) | chi-Square | p-Value |
|---|---|---|---|---|---|---|
| rs510335 | c. −401G>T | 0.1622 | No data | 0.2593 | 7.44 | 0.006 |
| rs5742910 | c. −323ins10 | 0.0523 | No data | 0.2593 | 92.9 | <0.00001 |
| rs561241 | c. −122T>C | 0.1128 | 0.1239 | 0.2593 | 22.9 | <0.00001 |
| rs36209567 | c.1061 C>T (p.Ala354Val) | 0.0006 | 0.0005 | 0.3889 | 23472.6 | <0.00001 |
| rs6046 | c.1238 G>A (p.Arg413Gln) | 0.1265 | 0.1294 | 0.6111 | 229.1 | <0.00001 |
| Clinical Severity Peyvandi (Vertically)/ Mariani (Horizontally) | Severe N (%) | Moderate N (%) | Mild N (%) | Asymptomatic N (%) | No Data N (%) | Total |
|---|---|---|---|---|---|---|
| Severe (FVII:C ≤ 10%) | 3 (11.1%) | 9 (33.4%) | 10 (37%) | 4 (14.8%) | 1 (3.7%) | 27 (50%) |
| Moderate (10% < FVII:C ≤ 20%) | 0 | 2 (40%) | 2 (40%) | 1 (20%) | 0 | 5 (9.3%) |
| Mild (FVII:C > 20%) | 0 | 4 (18.2%) | 11 (50%) | 5 (22.7%) | 2 (9.1%) | 22 (40.7%) |
| Total | 3 (5.6%) | 15 (27.7%) | 23 (42.6%) | 10 (18.5%) | 3 (5.6%) |
| Pathogenic Variant Status | Total | Clinical Severity Peyvandi [29] | Clinical Severity Mariani [30] | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Severe (FVII:C ≤ 10%) N (%) | Moderate (10% < FVII:C ≤ 20%) N (%) | Mild (FVII:C > 20%) N (%) | Severe N (%) | Moderate N (%) | Mild N (%) | Asymptomatic N (%) | No Data N (%) | ||
| homozygous | 8 | 8 (100%) | 0 | 0 | 1 (12.5%) | 4 (50%) | 2 (25%) | 0 | 1 (12.5%) |
| heterozygous compound | 17 | 15 (88.2%) | 2 (11.8%) | 0 | 2 (11.8%) | 3 (17.7%) | 8 (47%) | 4 (23.5%) | 0 |
| heterozygous | 9 | 0 | 1 (11.1%) | 8 (88.9%) | 0 | 3 (33.3%) | 3 (33.3%) | 3 (33.4%) | 0 |
| heterozygous + homozygous by functional polymorphisms | 3 | 3 (100%) | 0 | 0 | 0 | 2 (66.7%) | 0 | 1 (33.3%) | 0 |
| no mutation found + homozygous by functional polymorphisms | 12 | 0 | 2 (16.7%) | 10 (83.3%) | 0 | 2 (16.7%) | 7 (58.3%) | 1 (8.3%) | 2 (16.7%) |
| no mutation found | 5 | 1 (20%) | 0 | 4 (80%) | 0 | 1 (20%) | 3 (60%) | 1 (20%) | 0 |
| Total | 54 | 27 | 5 | 22 | 3 | 15 | 23 | 10 | 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pshenichnikova, O.; Selivanova, D.; Shchemeleva, E.; Abramova, T.; Zozulya, N.; Surin, V. Molecular Genetic Analysis of Russian Patients with Coagulation Factor FVII Deficiency. Genes 2023, 14, 1767. https://doi.org/10.3390/genes14091767
Pshenichnikova O, Selivanova D, Shchemeleva E, Abramova T, Zozulya N, Surin V. Molecular Genetic Analysis of Russian Patients with Coagulation Factor FVII Deficiency. Genes. 2023; 14(9):1767. https://doi.org/10.3390/genes14091767
Chicago/Turabian StylePshenichnikova, Olesya, Daria Selivanova, Ekaterina Shchemeleva, Tatiana Abramova, Nadezhda Zozulya, and Vadim Surin. 2023. "Molecular Genetic Analysis of Russian Patients with Coagulation Factor FVII Deficiency" Genes 14, no. 9: 1767. https://doi.org/10.3390/genes14091767