Evaluation of GSTP1, GSTA4 and AChE Gene Methylation in Bovine Lymphocytes Cultured In Vitro with Miconazole Alone and in Combination with Mospilan 20SP
Abstract
:1. Introduction
2. Materials and Methods
2.1. Blood Collection and In Vitro Cultivation of Lymphocytes with Miconazole Alone and in Combination with Mospilan 20SP
2.2. DNA Isolation, Bisulphite Modification and MSP
2.3. Preparation of Fully-Methylated Bovine DNA
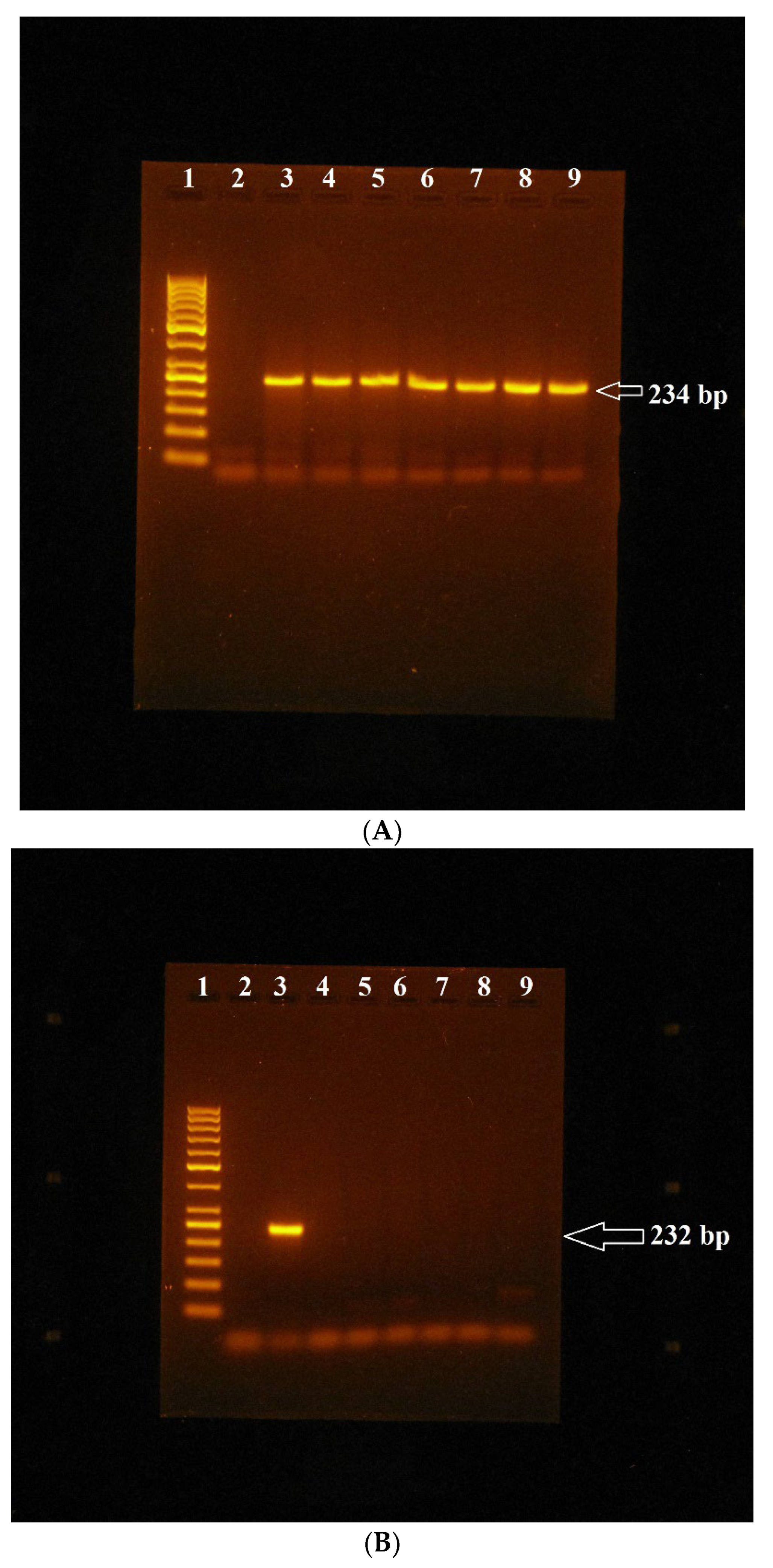
3. Results
MSP Analysis of GSTP1, GSTA4 and AChE Genes in Bovine Lymphocytes Cultivated In Vitro with Miconazole Alone and in Combination with Mospilan 20SP
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mehdipour, P.; Murphy, T.; De Carvalho, D.D. The role of DNA-demethylating agents in cancer therapy. Pharmacol. Ther. 2020, 205, 107416. [Google Scholar] [CrossRef] [PubMed]
- Neri, F.; Rapelli, S.; Krepelova, A.; Incarnato, D.; Parlato, C.; Basile, G.; Maldotti, M.; Anselmi, F.; Oliviero, S. Intragenic DNA methylation prevents spurious transcription initiation. Nature 2017, 543, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, M.V.C.; Bourc’his, D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019, 20, 590–607. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, A.M.F.; Sanglard, L.P.; Wijesena, H.R.; Ciobanu, D.C.; Horvath, S.; Spangler, M.L. DNA methylation profile in beef cattle is influenced by additive genetics and age. Sci. Rep. 2022, 12, 12016. [Google Scholar] [CrossRef]
- Rafeeiniaa, A.; Gholamreza Asadikaram, G.; Moazedd, V.; Darabi, M.K. Organochlorine pesticides may induce leukemia by methylation of CDKN2B and MGMT promoters and histone modifications. Gene 2023, 851, 146976. [Google Scholar] [CrossRef]
- Schaffner, S.L.; Kobor, M.S. DNA methylation as a mediator of genetic and environmental influences on Parkinson’s disease susceptibility: Impacts of α-Synuclein, physical activity, and pesticide exposure on the epigenome. Front. Genet. 2022, 13, 971298. [Google Scholar] [CrossRef]
- Ataei, M.; Abdollahi, M. A systematic review of mechanistic studies on the relationship between pesticide exposure and cancer induction. Toxicol. Appl. Pharm. 2022, 456, 116280. [Google Scholar] [CrossRef]
- Mahna, D.; Puri, S.; Sharma, S. DNA methylation modifications: Mediation to stipulate pesticide toxicity. Intern. J. Environ. Sci. Techn. 2021, 18, 531–544. [Google Scholar] [CrossRef]
- Piérard, G.E.; Hermanns-Lê, T.; Delvenne, P.; Piérard-Franchimont, C. Miconazole, a pharmacological barrier to skin fungal infections. Expert Opin. Pharmacother. 2012, 13, 1187–1194. [Google Scholar] [CrossRef]
- Ho, C.-Y.; Chang, A.-C.; Hsu, C.-H.; Tsai, T.-F.; Lin, Y.-C.; Chou, K.-Y.; Chen, H.-E.; Lin, J.-F.; Chen, P.-C.; Hwang, T.I.-S. Miconazole induces protective autophagy in bladder cancer cells. Environ. Toxicol. 2021, 36, 185–193. [Google Scholar] [CrossRef]
- Hassan, N.H.A. Miconazole genotoxicity in mice. J. Appl. Toxicol. 1997, 17, 313–319. [Google Scholar] [CrossRef]
- Galdíková, M.; Holečková, B.; Schwarzbacherová, V. Bovine Whole Blood Cells as a Biomarker Platform for Biological Toxicology: A Focus on Thiacloprid. In Biomarkers in Toxicology. Biomarkers in Disease: Methods, Discoveries and Applications; Patel, V.B., Preedy, V.R., Rajendram, R., Eds.; Springer: Cham, Switzerland, 2022. [Google Scholar] [CrossRef]
- Allocati, N.; Masulli, M.; Di Ilio, C.; Federici, L. Glutathione transferases: Substrates, inihibitors and pro-drugs in cancer and neurodegenerative diseases. Oncogenesis 2018, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Li, G.; Yin, J.; Li, L.; Tan, Y.; Wei, H.; Liu, B.; Deng, L.; Tang, J.; Chen, Y.; et al. GSTP1 and cancer: Expression, methylation, polymorphisms and signaling (review). Int. J. Oncol. 2020, 56, 867–878. [Google Scholar] [CrossRef] [PubMed]
- Guo, E.; Wei, H.; Liao, X.; Wu, L.; Zeng, X. Clinical significance and biological mechanism of gluthathione-S-transferase Mu gene family in colon adenocarcinoma. BMC Med. Genet. 2020, 21, 130. [Google Scholar] [CrossRef] [PubMed]
- Walczak-Nowicka, Ł.J.; Herbet, M. Acetylcholinesterase Inhibitors in the Treatment of Neurodegenerative Diseases and the Role of Acetylcholinesterase in their Pathogenesis. Int. J. Mol. Sci. 2021, 22, 9290. [Google Scholar] [CrossRef]
- Halušková, J.; Holečková, B.; Luptáková, L.; Košuth, J.; Schwarzbacherová, V.; Galdíková, M.; Koleničová, S. Study of methylation of the bovine GSTP1 gene under the influence of the pesticide Mospilan 20SP alone and in combination with the pesticide Orius 25EW. Fol. Biol. 2023; in press. [Google Scholar]
- Seyedmousavi, S.; de M G Bosco, S.; de Hoog, S.; Ebel, F.; Daniel Elad, D.; Gomes, R.R.; Jacobsen, I.D.; Jensen, H.E.; Martel, A.; Mignon, B.; et al. Fungal infections in animals: A patchwork of different situations. Med. Mycol. 2018, 56, 165–187. [Google Scholar] [CrossRef]
- Halušková, J.; Holečková, B.; Staničová, J.; Verebová, V. The preliminary study of pesticide Mospilan effect on the GSTP1 gene methylation in bovine lymphocytes. Fol. Vet. 2019, 63, 1–7. [Google Scholar] [CrossRef]
- Pallotta, M.M.; Barbato, V.; Pinton, A.; Acloque, H.; Gualtieri, R.; Talevi, R.; Jammes, H.; Capriglione, T. In vitro exposure to CPF affects bovine sperm epigenetic gene methylation pattern and the ability of sperm to support fertilization and embryo development. Environ. Mol. Mutagen. 2019, 60, 85–95. [Google Scholar] [CrossRef]
- Marçal, R.; Llorente, L.; Herrero, O.; Planelló, R.; Guilherme, S.; Pacheco, M. Intergenerational Patterns of DNA Methylation in Procambarus clarkii Following Exposure to Genotoxicants: A Conjugation in Past Simple or Past Continuous? Toxics 2021, 9, 271. [Google Scholar] [CrossRef]
- Terrazas-Salgado, L.; García-Gasca, A.; Betancourt-Lozano, M.; Llera-Herrera, R.; Alvarado-Cruz, I.; Yáñez-Rivera, B. Epigenetic Transgenerational Modifications Induced by Xenobiotic Exposure in Zebrafish. Front. Cell Dev. Biol. 2022, 10, 832982. [Google Scholar] [CrossRef] [PubMed]
- Rebuzzini, P.; Fabozzi, G.; Cimadomo, D.; Ubaldi, F.M.; Rienzi, L.; Zuccotti, M.; Garagna, S. Multi- and Transgenerational Effects of Environmental Toxicants on Mammalian Reproduction. Cells 2022, 11, 3163. [Google Scholar] [CrossRef] [PubMed]
- Gouin, N.; Notte, A.-M.; Kolok, S.A.; Bertin, A. Pesticide exposure affects DNA methylation patterns in natural populations of a mayfly. Sci. Total Environ. 2023, 864, 161096. [Google Scholar] [CrossRef] [PubMed]
- Holečková, B.; Koleničová, S.; Galdíková, M.; Halušková, J.; Schwarzbacherová, V. Effect of epoxiconazole on bovine acetylcholine esterase gene expression. Fol. Pharm. Cassoviensia 2022, 4, 107–113. [Google Scholar]
| Primer Name | Primer Sequence | Primer Length (bp) | PCR Product Size (bp) |
|---|---|---|---|
| Umet-GSTA4-F | GGTTGGTGTGTGTATTTTTATTAATTTGTT | 30 | 234 |
| Umet-GSTA4-R | ACATACATCTACAAACAAAACCCAAAAT | 28 | |
| Met-GSTA4-F | CGGCGTGCGTATTTTTATTAATTCGTTT | 28 | 232 |
| Met-GSTA4-R | AACGTACGTCTACGAACAAAACCCGAAAT | 29 | |
| Umet-AChE-F | TTTGTAAGTGGAATGTGGATTTAGTATTGG | 30 | 231 |
| Umet-AChE-R | CCATCAAACATCACTAAAACCCAAAAA | 27 | |
| Met-AChE-F | CGTAAGCGGAACGTGGATTTAGTATCG | 27 | 227 |
| Met-AChE-R | GTCAAACGTCGCTAAAACCCGAAAA | 25 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Halušková, J.; Holečková, B.; Schwarzbacherová, V.; Galdíková, M.; Sedláková, S.; Bučan, J. Evaluation of GSTP1, GSTA4 and AChE Gene Methylation in Bovine Lymphocytes Cultured In Vitro with Miconazole Alone and in Combination with Mospilan 20SP. Genes 2023, 14, 1791. https://doi.org/10.3390/genes14091791
Halušková J, Holečková B, Schwarzbacherová V, Galdíková M, Sedláková S, Bučan J. Evaluation of GSTP1, GSTA4 and AChE Gene Methylation in Bovine Lymphocytes Cultured In Vitro with Miconazole Alone and in Combination with Mospilan 20SP. Genes. 2023; 14(9):1791. https://doi.org/10.3390/genes14091791
Chicago/Turabian StyleHalušková, Jana, Beáta Holečková, Viera Schwarzbacherová, Martina Galdíková, Silvia Sedláková, and Jaroslav Bučan. 2023. "Evaluation of GSTP1, GSTA4 and AChE Gene Methylation in Bovine Lymphocytes Cultured In Vitro with Miconazole Alone and in Combination with Mospilan 20SP" Genes 14, no. 9: 1791. https://doi.org/10.3390/genes14091791
APA StyleHalušková, J., Holečková, B., Schwarzbacherová, V., Galdíková, M., Sedláková, S., & Bučan, J. (2023). Evaluation of GSTP1, GSTA4 and AChE Gene Methylation in Bovine Lymphocytes Cultured In Vitro with Miconazole Alone and in Combination with Mospilan 20SP. Genes, 14(9), 1791. https://doi.org/10.3390/genes14091791







