The Conserved Chromatin Remodeler SMARCAD1 Interacts with TFIIIC and Architectural Proteins in Human and Mouse
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Plasmids
2.3. Lysate Preparations, Cell Fractionations, Immunopreciptiations, and Western Blots
2.4. Antibodies
2.5. GST-Pulldown Assay
2.6. Chromatin Immunoprecipitation
2.7. ChIP-Seq Analysis
2.8. Gene Expression Analysis
3. Results
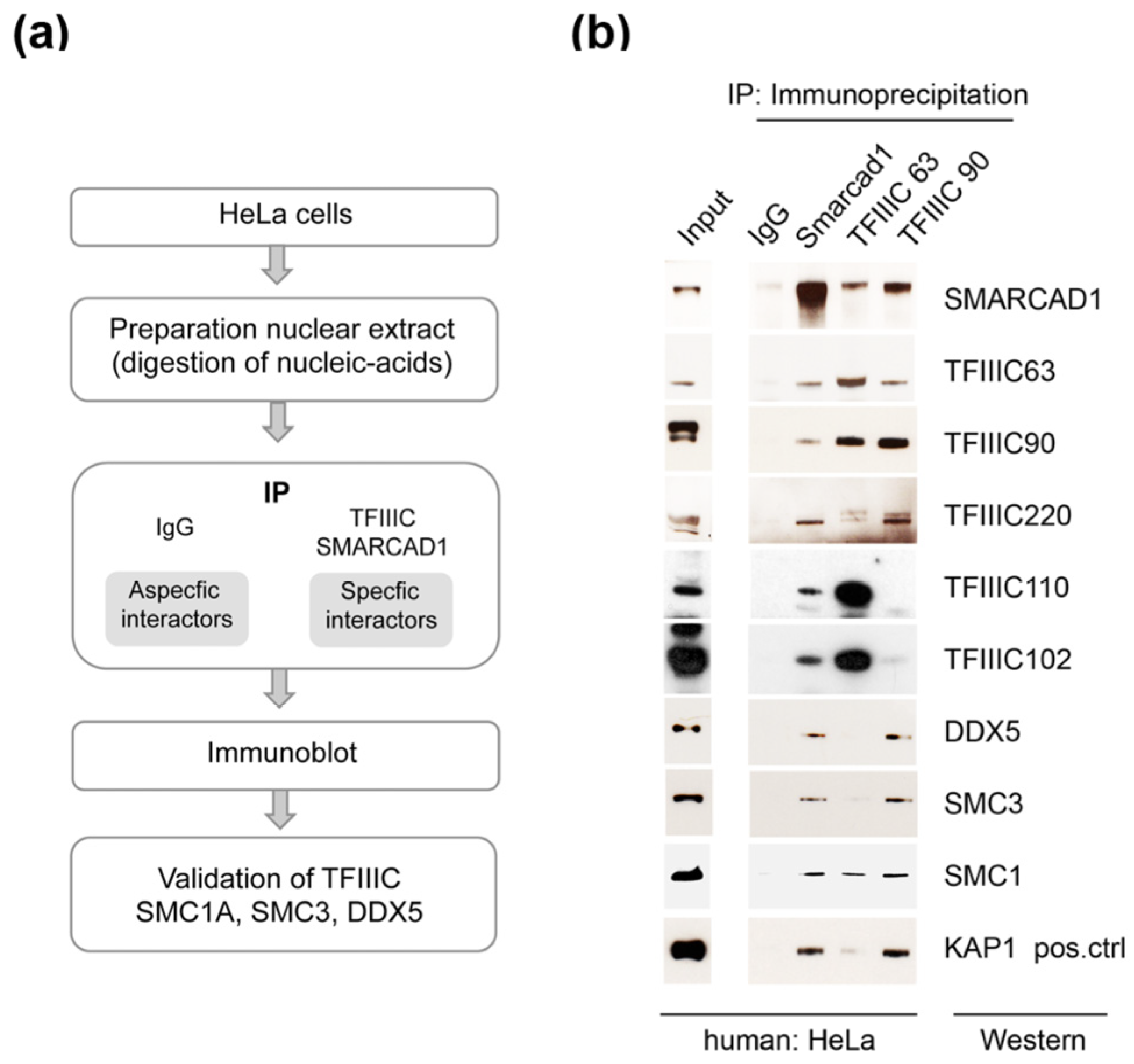
3.1. Human Remodeler SMARCAD1 Interacts with TFIIIC and Chromatin Architectural Proteins
3.2. Human TFIIIC102 Interacts Directly with SMARCAD1
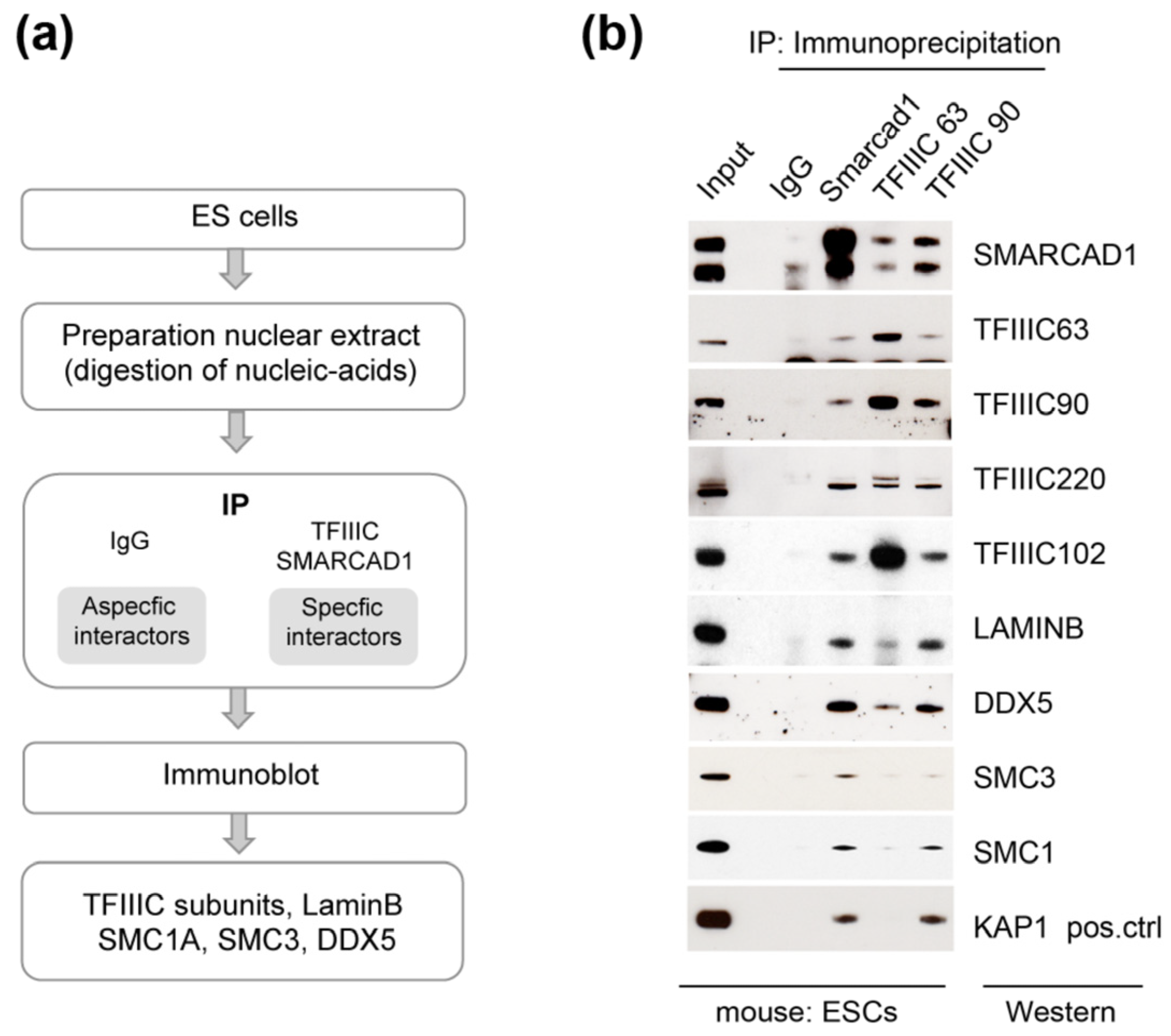
3.3. SMARCAD1 Interaction with TFIIIC Is Conserved in Pluripotent Mouse Cells
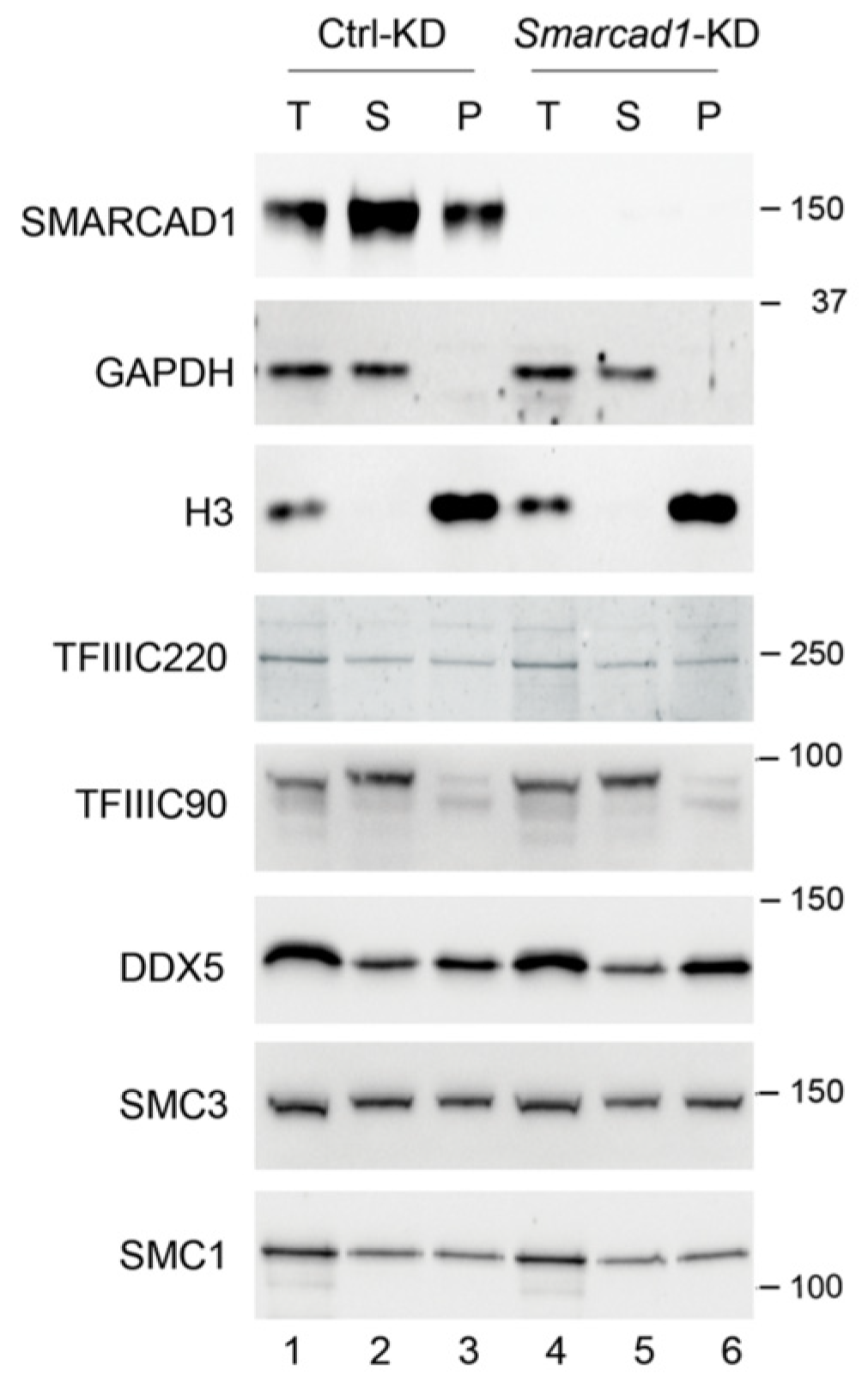
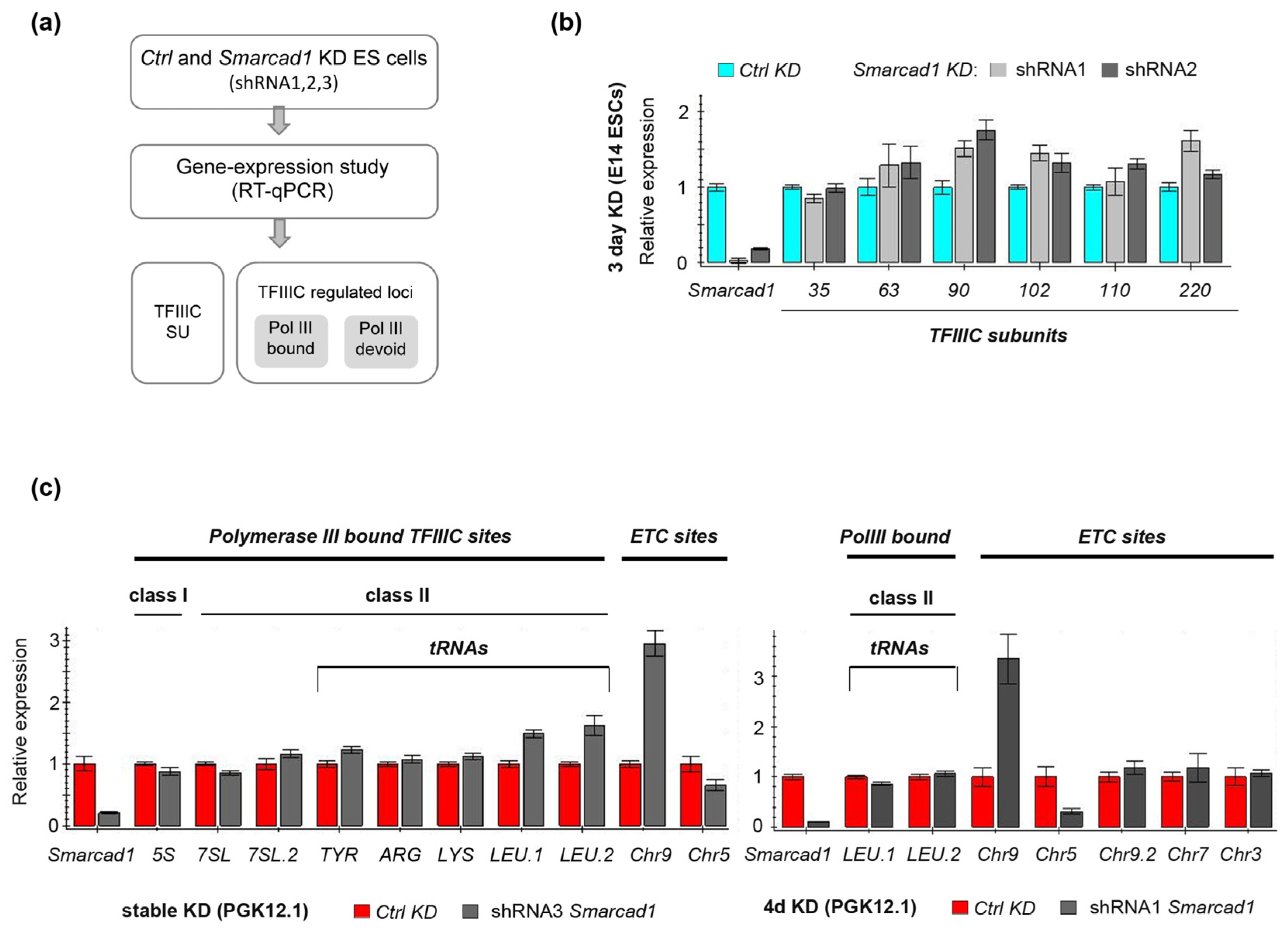
3.4. Impact of SMARCAD1 on the Expression of TFIIIC and TFIIIC Targets
3.5. SMARCAD1 and TFIIIC Binding Sites Are Distinct in the ESC Genome
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rowley, M.J.; Corces, V.G. Organizational principles of 3D genome architecture. Nat. Rev. Genet. 2018, 19, 789–800. [Google Scholar] [CrossRef]
- Zheng, H.; Xie, W. The role of 3D genome organization in development and cell differentiation. Nat. Rev. Mol. Cell Biol. 2019, 20, 535–550. [Google Scholar] [CrossRef] [PubMed]
- Clapier, C.R.; Cairns, B.R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009, 78, 273–304. [Google Scholar] [CrossRef] [PubMed]
- Ho, L.; Crabtree, G.R. Chromatin remodelling during development. Nature 2010, 463, 474–484. [Google Scholar] [CrossRef] [PubMed]
- Hargreaves, D.C.; Crabtree, G.R. ATP-dependent chromatin remodeling: Genetics, genomics and mechanisms. Cell Res. 2011, 21, 396–420. [Google Scholar] [CrossRef]
- Magaña-Acosta, M.; Valadez-Graham, V. Chromatin Remodelers in the 3D Nuclear Compartment. Front. Genet. 2020, 11, 600615. [Google Scholar] [CrossRef]
- Alpsoy, A.; Sood, S.; Dykhuizen, E.C. At the Crossroad of Gene Regulation and Genome Organization: Potential Roles for ATP-Dependent Chromatin Remodelers in the Regulation of CTCF-Mediated 3D Architecture. Biology 2021, 10, 272. [Google Scholar] [CrossRef]
- Cubeñas-Potts, C.; Corces, V.G. Architectural Proteins, Transcription, and the Three-dimensional Organization of the Genome. FEBS Lett. 2015, 589, 2923–2930. [Google Scholar] [CrossRef]
- Phillips-Cremins, J.E.; Corces, V.G. Chromatin insulators: Linking genome organization to cellular function. Mol. Cell 2013, 50, 461–474. [Google Scholar] [CrossRef]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012, 485, 376–380. [Google Scholar] [CrossRef]
- Nora, E.P.; Goloborodko, A.; Valton, A.-L.; Gibcus, J.H.; Uebersohn, A.; Abdennur, N.; Dekker, J.; Mirny, L.A.; Bruneau, B.G. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell 2017, 169, 930–944.e22. [Google Scholar] [CrossRef] [PubMed]
- van Bortle, K.; Corces, V.G. tDNA insulators and the emerging role of TFIIIC in genome organization. Transcription 2012, 3, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Sizer, R.E.; Chahid, N.; Butterfield, S.P.; Donze, D.; Bryant, N.J.; White, R.J. TFIIIC-based chromatin insulators through eukaryotic evolution. Gene 2022, 835, 146533. [Google Scholar] [CrossRef] [PubMed]
- Schramm, L.; Hernandez, N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002, 16, 2593–2620. [Google Scholar] [CrossRef] [PubMed]
- White, R.J. Transcription by RNA polymerase III: More complex than we thought. Nat. Rev. Genet. 2011, 12, 459–463. [Google Scholar] [CrossRef]
- Orioli, A.; Pascali, C.; Pagano, A.; Teichmann, M.; Dieci, G. RNA polymerase III transcription control elements: Themes and variations. Gene 2012, 493, 185–194. [Google Scholar] [CrossRef]
- Moqtaderi, Z.; Struhl, K. Genome-Wide Occupancy Profile of the RNA Polymerase III Machinery in Saccharomyces cerevisiae Reveals Loci with Incomplete Transcription Complexes. Mol. Cell. Biol. 2004, 24, 4118–4127. [Google Scholar] [CrossRef]
- Noma, K.; Cam, H.P.; Maraia, R.J.; Grewal, S.I.S. A role for TFIIIC transcription factor complex in genome organization. Cell 2006, 125, 859–872. [Google Scholar] [CrossRef]
- van Bortle, K.; Nichols, M.H.; Li, L.; Ong, C.-T.; Takenaka, N.; Qin, Z.S.; Corces, V.G. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol. 2014, 15, R82. [Google Scholar] [CrossRef]
- Stutzman, A.V.; Liang, A.S.; Beilinson, V.; Ikegami, K. Transcription-independent TFIIIC-bound sites cluster near heterochromatin boundaries within lamina-associated domains in C. elegans. Epigenet. Chromatin 2020, 13, 1. [Google Scholar] [CrossRef]
- Carrière, L.; Graziani, S.; Alibert, O.; Ghavi-Helm, Y.; Boussouar, F.; Humbertclaude, H.; Jounier, S.; Aude, J.-C.; Keime, C.; Murvai, J.; et al. Genomic binding of Pol III transcription machinery and relationship with TFIIS transcription factor distribution in mouse embryonic stem cells. Nucleic Acids Res. 2012, 40, 270–283. [Google Scholar] [CrossRef] [PubMed]
- Moqtaderi, Z.; Wang, J.; Raha, D.; White, R.J.; Snyder, M.; Weng, Z.; Struhl, K. Genomic Binding Profiles of Functionally Distinct RNA Polymerase III Transcription Complexes in Human Cells. Nat. Struct. Mol. Biol. 2010, 17, 635–640. [Google Scholar] [CrossRef] [PubMed]
- Donze, D. Extra-transcriptional functions of RNA Polymerase III complexes: TFIIIC as a potential global chromatin bookmark. Gene 2012, 493, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Kirkland, J.G.; Raab, J.R.; Kamakaka, R.T. TFIIIC bound DNA elements in nuclear organization and insulation. Biochim. Biophys. Acta 2013, 1829, 418–424. [Google Scholar] [CrossRef]
- Pascali, C.; Teichmann, M. RNA polymerase III transcription-regulated by chromatin structure and regulator of nuclear chromatin organization. Subcell. Biochem. 2013, 61, 261–287. [Google Scholar] [CrossRef]
- Raab, J.R.; Chiu, J.; Zhu, J.; Katzman, S.; Kurukuti, S.; Wade, P.A.; Haussler, D.; Kamakaka, R.T. Human tRNA genes function as chromatin insulators. EMBO J. 2012, 31, 330–350. [Google Scholar] [CrossRef]
- Donze, D.; Adams, C.R.; Rine, J.; Kamakaka, R.T. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 1999, 13, 698–708. [Google Scholar] [CrossRef]
- Donze, D.; Kamakaka, R.T. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J. 2001, 20, 520–531. [Google Scholar] [CrossRef]
- Simms, T.A.; Dugas, S.L.; Gremillion, J.C.; Ibos, M.E.; Dandurand, M.N.; Toliver, T.T.; Edwards, D.J.; Donze, D. TFIIIC binding sites function as both heterochromatin barriers and chromatin insulators in Saccharomyces cerevisiae. Eukaryot. Cell 2008, 7, 2078–2086. [Google Scholar] [CrossRef]
- Scott, K.C.; White, C.V.; Willard, H.F. An RNA polymerase III-dependent heterochromatin barrier at fission yeast centromere 1. PLoS ONE 2007, 2, e1099. [Google Scholar] [CrossRef]
- Strålfors, A.; Walfridsson, J.; Bhuiyan, H.; Ekwall, K. The FUN30 chromatin remodeler, Fft3, protects centromeric and subtelomeric domains from euchromatin formation. PLoS Genet. 2011, 7, e1001334. [Google Scholar] [CrossRef] [PubMed]
- Ebersole, T.; Kim, J.-H.; Samoshkin, A.; Kouprina, N.; Pavlicek, A.; White, R.J.; Larionov, V. tRNA genes protect a reporter gene from epigenetic silencing in mouse cells. Cell Cycle 2011, 10, 2779–2791. [Google Scholar] [CrossRef]
- Yuen, K.C.; Slaughter, B.D.; Gerton, J.L. Condensin II is anchored by TFIIIC and H3K4me3 in the mammalian genome and supports the expression of active dense gene clusters. Sci. Adv. 2017, 3, e170019. [Google Scholar] [CrossRef] [PubMed]
- D’Ambrosio, C.; Schmidt, C.K.; Katou, Y.; Kelly, G.; Itoh, T.; Shirahige, K.; Uhlmann, F. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 2008, 22, 2215–2227. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, R.; de Llobet Cucalon, L.I.; Di Vona, C.; Le Dilly, F.; Vidal, E.; Lioutas, A.; Oliete, J.Q.; Jochem, L.; Cutts, E.; Dieci, G.; et al. TFIIIC Binding to Alu Elements Controls Gene Expression via Chromatin Looping and Histone Acetylation. Mol. Cell 2020, 77, 475–487.e11. [Google Scholar] [CrossRef] [PubMed]
- de Llobet Cucalon, L.; Di Vona, C.; Morselli, M.; Vezzoli, M.; Montanini, B.; Teichmann, M.; de la Luna, S.; Ferrari, R. An RNA Polymerase III General Transcription Factor Engages in Cell Type-Specific Chromatin Looping. Int. J. Mol. Sci. 2022, 23, 2260. [Google Scholar] [CrossRef]
- Hiraga, S.; Botsios, S.; Donze, D.; Donaldson, A.D. TFIIIC localizes budding yeast ETC sites to the nuclear periphery. Mol. Biol. Cell 2012, 23, 2741–2754. [Google Scholar] [CrossRef]
- Scott, K.C.; Merrett, S.L.; Willard, H.F. A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr. Biol. 2006, 16, 119–129. [Google Scholar] [CrossRef]
- Steglich, B.; Strålfors, A.; Khorosjutina, O.; Persson, J.; Smialowska, A.; Javerzat, J.-P.; Ekwall, K. The Fun30 chromatin remodeler Fft3 controls nuclear organization and chromatin structure of insulators and subtelomeres in fission yeast. PLoS Genet. 2015, 11, e1005101. [Google Scholar] [CrossRef]
- Neves-Costa, A.; Will, W.R.; Vetter, A.T.; Miller, J.R.; Varga-Weisz, P. The SNF2-family member Fun30 promotes gene silencing in heterochromatic loci. PLoS ONE 2009, 4, e8111. [Google Scholar] [CrossRef]
- Flaus, A.; Martin, D.M.A.; Barton, G.J.; Owen-Hughes, T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006, 34, 2887–2905. [Google Scholar] [CrossRef]
- Durand-Dubief, M.; Will, W.R.; Petrini, E.; Theodorou, D.; Harris, R.R.; Crawford, M.R.; Paszkiewicz, K.; Krueger, F.; Correra, R.M.; Vetter, A.T.; et al. SWI/SNF-like chromatin remodeling factor Fun30 supports point centromere function in S. cerevisiae. PLoS Genet. 2012, 8, e1002974. [Google Scholar] [CrossRef]
- Arafat, K.; Al Kubaisy, E.; Sulaiman, S.; Karam, S.M.; Al Natour, Z.; Hassan, A.H.; Attoub, S. SMARCAD1 in Breast Cancer Progression. Cell. Physiol. Biochem. 2018, 50, 489–500. [Google Scholar] [CrossRef] [PubMed]
- Loh, A.Y.T.; Špoljar, S.; Neo, G.Y.W.; Escande-Beillard, N.; Leushacke, M.; Luijten, M.N.H.; Venkatesh, B.; Bonnard, C.; van Steensel, M.A.M.; Hamm, H.; et al. Huriez syndrome: Additional pathogenic variants supporting allelism to SMARCAD syndrome. Am. J. Med. Genet. A 2022, 188, 1752–1760. [Google Scholar] [CrossRef] [PubMed]
- Valentin, M.N.; Solomon, B.D.; Richard, G.; Ferreira, C.R.; Kirkorian, A.Y. Basan gets a new fingerprint: Mutations in the skin-specific isoform of SMARCAD1 cause ectodermal dysplasia syndromes with adermatoglyphia. Am. J. Med. Genet. A 2018, 176, 2451–2455. [Google Scholar] [CrossRef]
- Liu, F.; Xia, Z.; Zhang, M.; Ding, J.; Feng, Y.; Wu, J.; Dong, Y.; Gao, W.; Han, Z.; Liu, Y.; et al. SMARCAD1 Promotes Pancreatic Cancer Cell Growth and Metastasis through Wnt/β-catenin-Mediated EMT. Int. J. Biol. Sci. 2019, 15, 636–646. [Google Scholar] [CrossRef]
- Schoor, M.; Schuster-Gossler, K.; Roopenian, D.; Gossler, A. Skeletal dysplasias, growth retardation, reduced postnatal survival, and impaired fertility in mice lacking the SNF2/SWI2 family member ETL1. Mech. Dev. 1999, 85, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Lu, J.; Sridhar, B.; Cao, X.; Yu, P.; Zhao, T.; Chen, C.-C.; McDee, D.; Sloofman, L.; Wang, Y.; et al. SMARCAD1 Contributes to the Regulation of Naive Pluripotency by Interacting with Histone Citrullination. Cell Rep. 2017, 18, 3117–3128. [Google Scholar] [CrossRef]
- Schoor, M.; Schuster-Gossler, K.; Gossler, A. The Etl-1 gene encodes a nuclear protein differentially expressed during early mouse development. Dev. Dyn. 1993, 197, 227–237. [Google Scholar] [CrossRef]
- Hong, F.; Fang, F.; He, X.; Cao, X.; Chipperfield, H.; Xie, D.; Wong, W.H.; Ng, H.H.; Zhong, S. Dissecting early differentially expressed genes in a mixture of differentiating embryonic stem cells. PLoS Comput. Biol. 2009, 5, e1000607. [Google Scholar] [CrossRef]
- Sachs, P.; Ding, D.; Bergmaier, P.; Lamp, B.; Schlagheck, C.; Finkernagel, F.; Nist, A.; Stiewe, T.; Mermoud, J.E. SMARCAD1 ATPase activity is required to silence endogenous retroviruses in embryonic stem cells. Nat. Commun. 2019, 10, 1335. [Google Scholar] [CrossRef]
- Navarro, C.; Lyu, J.; Katsori, A.-M.; Caridha, R.; Elsässer, S.J. An embryonic stem cell-specific heterochromatin state promotes core histone exchange in the absence of DNA accessibility. Nat. Commun. 2020, 11, 5095. [Google Scholar] [CrossRef] [PubMed]
- Bren, I.; Strauss, C.; Schlesinger, S. The role of Smarcad1 in retroviral repression in mouse embryonic stem cells. bioRvix 2023. [Google Scholar] [CrossRef]
- Kazakevych, J.; Denizot, J.; Liebert, A.; Portovedo, M.; Mosavie, M.; Jain, P.; Stellato, C.; Fraser, C.; Corrêa, R.O.; Célestine, M.; et al. Smarcad1 mediates microbiota-induced inflammation in mouse and coordinates gene expression in the intestinal epithelium. Genome Biol. 2020, 21, 64. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Cui, D.; Papusha, A.; Zhang, X.; Chu, C.-D.; Tang, J.; Chen, K.; Pan, X.; Ira, G. The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature 2012, 489, 576–580. [Google Scholar] [CrossRef]
- Costelloe, T.; Louge, R.; Tomimatsu, N.; Mukherjee, B.; Martini, E.; Khadaroo, B.; Dubois, K.; Wiegant, W.W.; Thierry, A.; Burma, S.; et al. The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection. Nature 2012, 489, 581–584. [Google Scholar] [CrossRef]
- Rowbotham, S.P.; Barki, L.; Neves-Costa, A.; Santos, F.; Dean, W.; Hawkes, N.; Choudhary, P.; Will, W.R.; Webster, J.; Oxley, D.; et al. Maintenance of silent chromatin through replication requires SWI/SNF-like chromatin remodeler SMARCAD1. Mol. Cell 2011, 42, 285–296. [Google Scholar] [CrossRef]
- Lo, C.S.Y.; van Toorn, M.; Gaggioli, V.; Paes Dias, M.; Zhu, Y.; Manolika, E.M.; Zhao, W.; van der Does, M.; Mukherjee, C.; G S C Souto Gonçalves, J.; et al. SMARCAD1-mediated active replication fork stability maintains genome integrity. Sci. Adv. 2021, 7, eabe7804. [Google Scholar] [CrossRef]
- Ding, D.; Bergmaier, P.; Sachs, P.; Klangwart, M.; Rückert, T.; Bartels, N.; Demmers, J.; Dekker, M.; Poot, R.A.; Mermoud, J.E. The CUE1 domain of the SNF2-like chromatin remodeler SMARCAD1 mediates its association with KRAB-associated protein 1 (KAP1) and KAP1 target genes. J. Biol. Chem. 2018, 293, 2711–2724. [Google Scholar] [CrossRef]
- Seifert-Davila, W.; Girbig, M.; Hauptmann, L.; Hoffmann, T.; Eustermann, S.; Müller, C.W. Structural insights into human TFIIIC promoter recognition. Sci. Adv. 2023, 9, eadh2019. [Google Scholar] [CrossRef]
- Dittmer, T.A.; Misteli, T. The lamin protein family. Genome Biol. 2011, 12, 222. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Brick, K.; Evrard, Y.; Xiao, T.; Camerini-Otero, R.D.; Felsenfeld, G. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 2010, 24, 2543–2555. [Google Scholar] [CrossRef] [PubMed]
- Kind, J.; van Steensel, B. Genome-nuclear lamina interactions and gene regulation. Curr. Opin. Cell Biol. 2010, 22, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Büchel, G.; Carstensen, A.; Mak, K.-Y.; Roeschert, I.; Leen, E.; Sumara, O.; Hofstetter, J.; Herold, S.; Kalb, J.; Baluapuri, A.; et al. Association with Aurora-A Controls N-MYC-Dependent Promoter Escape and Pause Release of RNA Polymerase II during the Cell Cycle. Cell Rep. 2017, 21, 3483–3497. [Google Scholar] [CrossRef]
- Giraud, G.; Terrone, S.; Bourgeois, C.F. Functions of DEAD box RNA helicases DDX5 and DDX17 in chromatin organization and transcriptional regulation. BMB Rep. 2018, 51, 613–622. [Google Scholar] [CrossRef]
- Gibson, T.J.; Seiler, M.; Veitia, R.A. The transience of transient overexpression. Nat. Methods 2013, 10, 715–721. [Google Scholar] [CrossRef] [PubMed]
- Male, G.; von Appen, A.; Glatt, S.; Taylor, N.M.I.; Cristovao, M.; Groetsch, H.; Beck, M.; Müller, C.W. Architecture of TFIIIC and its role in RNA polymerase III pre-initiation complex assembly. Nat. Commun. 2015, 6, 7387. [Google Scholar] [CrossRef]
- Hsieh, Y.J.; Wang, Z.; Kovelman, R.; Roeder, R.G. Cloning and characterization of two evolutionarily conserved subunits (TFIIIC102 and TFIIIC63) of human TFIIIC and their involvement in functional interactions with TFIIIB and RNA polymerase III. Mol. Cell. Biol. 1999, 19, 4944–4952. [Google Scholar] [CrossRef]
- Jourdain, S.; Acker, J.; Ducrot, C.; Sentenac, A.; Lefebvre, O. The tau95 subunit of yeast TFIIIC influences upstream and downstream functions of TFIIIC.DNA complexes. J. Biol. Chem. 2003, 278, 10450–10457. [Google Scholar] [CrossRef]
- Huang, Y.; Hamada, M.; Maraia, R.J. Isolation and cloning of four subunits of a fission yeast TFIIIC complex that includes an ortholog of the human regulatory protein TFIIICbeta. J. Biol. Chem. 2000, 275, 31480–31487. [Google Scholar] [CrossRef]
- Dumay-Odelot, H.; Marck, C.; Durrieu-Gaillard, S.; Lefebvre, O.; Jourdain, S.; Prochazkova, M.; Pflieger, A.; Teichmann, M. Identification, molecular cloning, and characterization of the sixth subunit of human transcription factor TFIIIC. J. Biol. Chem. 2007, 282, 17179–17189. [Google Scholar] [CrossRef]
- Efroni, S.; Duttagupta, R.; Cheng, J.; Dehghani, H.; Hoeppner, D.J.; Dash, C.; Bazett-Jones, D.P.; Le Grice, S.; McKay, R.D.G.; Buetow, K.H.; et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell 2008, 2, 437–447. [Google Scholar] [CrossRef]
- Schlesinger, S.; Meshorer, E. Open Chromatin, Epigenetic Plasticity, and Nuclear Organization in Pluripotency. Dev. Cell 2019, 48, 135–150. [Google Scholar] [CrossRef]
- Lim, P.S.L.; Meshorer, E. Organization of the Pluripotent Genome. Cold Spring Harb. Perspect. Biol. 2021, 13, a040204. [Google Scholar] [CrossRef] [PubMed]
- Pękowska, A.; Klaus, B.; Xiang, W.; Severino, J.; Daigle, N.; Klein, F.A.; Oleś, M.; Casellas, R.; Ellenberg, J.; Steinmetz, L.M.; et al. Gain of CTCF-Anchored Chromatin Loops Marks the Exit from Naive Pluripotency. Cell Syst. 2018, 7, 482–495.e10. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Choi, E.S.; Seo, H.D.; Kang, K.; Gilmore, J.M.; Florens, L.; Washburn, M.P.; Choe, J.; Workman, J.L.; Lee, D. Chromatin remodeller Fun30Fft3 induces nucleosome disassembly to facilitate RNA polymerase II elongation. Nat. Commun. 2017, 8, 14527. [Google Scholar] [CrossRef] [PubMed]
- Doiguchi, M.; Nakagawa, T.; Imamura, Y.; Yoneda, M.; Higashi, M.; Kubota, K.; Yamashita, S.; Asahara, H.; Iida, M.; Fujii, S.; et al. SMARCAD1 is an ATP-dependent stimulator of nucleosomal H2A acetylation via CBP, resulting in transcriptional regulation. Sci. Rep. 2016, 6, 20179. [Google Scholar] [CrossRef]
- Winter, A.G.; Sourvinos, G.; Allison, S.J.; Tosh, K.; Scott, P.H.; Spandidos, D.A.; White, R.J. RNA polymerase III transcription factor TFIIIC2 is overexpressed in ovarian tumors. Proc. Natl. Acad. Sci. USA 2000, 97, 12619–12624. [Google Scholar] [CrossRef]
- Durrieu-Gaillard, S.; Dumay-Odelot, H.; Boldina, G.; Tourasse, N.J.; Allard, D.; André, F.; Macari, F.; Choquet, A.; Lagarde, P.; Drutel, G.; et al. Regulation of RNA polymerase III transcription during transformation of human IMR90 fibroblasts with defined genetic elements. Cell Cycle 2018, 17, 605–615. [Google Scholar] [CrossRef]
- Graczyk, D.; Cieśla, M.; Boguta, M. Regulation of tRNA synthesis by the general transcription factors of RNA polymerase III-TFIIIB and TFIIIC, and by the MAF1 protein. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 320–329. [Google Scholar] [CrossRef]
- Dieci, G.; Fiorino, G.; Castelnuovo, M.; Teichmann, M.; Pagano, A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007, 23, 614–622. [Google Scholar] [CrossRef]
- Müller, J.; Benecke, B.J. Analysis of transcription factors binding to the human 7SL RNA gene promoter. Biochem. Cell Biol. 1999, 77, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Barski, A.; Chepelev, I.; Liko, D.; Cuddapah, S.; Fleming, A.B.; Birch, J.; Cui, K.; White, R.J.; Zhao, K. Pol II and its associated epigenetic marks are present at Pol III-transcribed noncoding RNA genes. Nat. Struct. Mol. Biol. 2010, 17, 629–634. [Google Scholar] [CrossRef]
- Oler, A.J.; Alla, R.K.; Roberts, D.N.; Wong, A.; Hollenhorst, P.C.; Chandler, K.J.; Cassiday, P.A.; Nelson, C.A.; Hagedorn, C.H.; Graves, B.J.; et al. Human RNA Polymerase III transcriptomes and relationships to Pol II promoters, enhancer-binding factors and chromatin domains. Nat. Struct. Mol. Biol. 2010, 17, 620–628. [Google Scholar] [CrossRef] [PubMed]
- Canella, D.; Praz, V.; Reina, J.H.; Cousin, P.; Hernandez, N. Defining the RNA polymerase III transcriptome: Genome-wide localization of the RNA polymerase III transcription machinery in human cells. Genome Res. 2010, 20, 710–721. [Google Scholar] [CrossRef]
- Tong, Z.-B.; Ai, H.-S.; Li, J.-B. The Mechanism of Chromatin Remodeler SMARCAD1/Fun30 in Response to DNA Damage. Front. Cell Dev. Biol. 2020, 8, 560098. [Google Scholar] [CrossRef]
- Bantele, S.C.S.; Pfander, B. Nucleosome Remodeling by Fun30SMARCAD1 in the DNA Damage Response. Front. Mol. Biosci. 2019, 6, 78. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Díaz, E.; Corces, V.G. Architectural proteins: Regulators of 3D genome organization in cell fate. Trends Cell Biol. 2014, 24, 703–711. [Google Scholar] [CrossRef]
- Byeon, B.; Wang, W.; Barski, A.; Ranallo, R.T.; Bao, K.; Schones, D.E.; Zhao, K.; Wu, C.; Wu, W.-H. The ATP-dependent Chromatin Remodeling Enzyme Fun30 Represses Transcription by Sliding Promoter-proximal Nucleosomes. J. Biol. Chem. 2013, 288, 23182–23193. [Google Scholar] [CrossRef]
- Valenzuela, L.; Dhillon, N.; Kamakaka, R.T. Transcription independent insulation at TFIIIC-dependent insulators. Genetics 2009, 183, 131–148. [Google Scholar] [CrossRef]
- Jambunathan, N.; Martinez, A.W.; Robert, E.C.; Agochukwu, N.B.; Ibos, M.E.; Dugas, S.L.; Donze, D. Multiple bromodomain genes are involved in restricting the spread of heterochromatic silencing at the Saccharomyces cerevisiae HMR-tRNA boundary. Genetics 2005, 171, 913–922. [Google Scholar] [CrossRef]
- Euskirchen, G.M.; Auerbach, R.K.; Davidov, E.; Gianoulis, T.A.; Zhong, G.; Rozowsky, J.; Bhardwaj, N.; Gerstein, M.B.; Snyder, M. Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS Genet. 2011, 7, e1002008. [Google Scholar] [CrossRef]
- Markert, J.; Zhou, K.; Luger, K. SMARCAD1 is an ATP-dependent histone octamer exchange factor with de novo nucleosome assembly activity. Sci. Adv. 2021, 7, eabk2380. [Google Scholar] [CrossRef] [PubMed]
- Leeb, M.; Dietmann, S.; Paramor, M.; Niwa, H.; Smith, A. Genetic exploration of the exit from self-renewal using haploid embryonic stem cells. Cell Stem Cell 2014, 14, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Vezzoli, M.; de Llobet Cucalon, L.I.; Di Vona, C.; Morselli, M.; Montanini, B.; de la Luna, S.; Teichmann, M.; Dieci, G.; Ferrari, R. TFIIIC as a Potential Epigenetic Modulator of Histone Acetylation in Human Stem Cells. Int. J. Mol. Sci. 2023, 24, 3624. [Google Scholar] [CrossRef]
- Phillips-Cremins, J.E.; Sauria, M.E.G.; Sanyal, A.; Gerasimova, T.I.; Lajoie, B.R.; Bell, J.S.K.; Ong, C.-T.; Hookway, T.A.; Guo, C.; Sun, Y.; et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 2013, 153, 1281–1295. [Google Scholar] [CrossRef]
- Crepaldi, L.; Policarpi, C.; Coatti, A.; Sherlock, W.T.; Jongbloets, B.C.; Down, T.A.; Riccio, A. Binding of TFIIIC to sine elements controls the relocation of activity-dependent neuronal genes to transcription factories. PLoS Genet. 2013, 9, e1003699. [Google Scholar] [CrossRef]
- Varshney, D.; Vavrova-Anderson, J.; Oler, A.J.; Cowling, V.H.; Cairns, B.R.; White, R.J. SINE transcription by RNA polymerase III is suppressed by histone methylation but not by DNA methylation. Nat. Commun. 2015, 6, 6569. [Google Scholar] [CrossRef] [PubMed]
- Cournac, A.; Koszul, R.; Mozziconacci, J. The 3D folding of metazoan genomes correlates with the association of similar repetitive elements. Nucleic Acids Res. 2016, 44, 245–255. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sachs, P.; Bergmaier, P.; Treutwein, K.; Mermoud, J.E. The Conserved Chromatin Remodeler SMARCAD1 Interacts with TFIIIC and Architectural Proteins in Human and Mouse. Genes 2023, 14, 1793. https://doi.org/10.3390/genes14091793
Sachs P, Bergmaier P, Treutwein K, Mermoud JE. The Conserved Chromatin Remodeler SMARCAD1 Interacts with TFIIIC and Architectural Proteins in Human and Mouse. Genes. 2023; 14(9):1793. https://doi.org/10.3390/genes14091793
Chicago/Turabian StyleSachs, Parysatis, Philipp Bergmaier, Katrin Treutwein, and Jacqueline E. Mermoud. 2023. "The Conserved Chromatin Remodeler SMARCAD1 Interacts with TFIIIC and Architectural Proteins in Human and Mouse" Genes 14, no. 9: 1793. https://doi.org/10.3390/genes14091793
APA StyleSachs, P., Bergmaier, P., Treutwein, K., & Mermoud, J. E. (2023). The Conserved Chromatin Remodeler SMARCAD1 Interacts with TFIIIC and Architectural Proteins in Human and Mouse. Genes, 14(9), 1793. https://doi.org/10.3390/genes14091793





