The Potential of Instrumental Insemination for Sustainable Honeybee Breeding
Abstract
:1. Introduction
2. Materials and Methods
2.1. Genetic Model
2.2. Selection
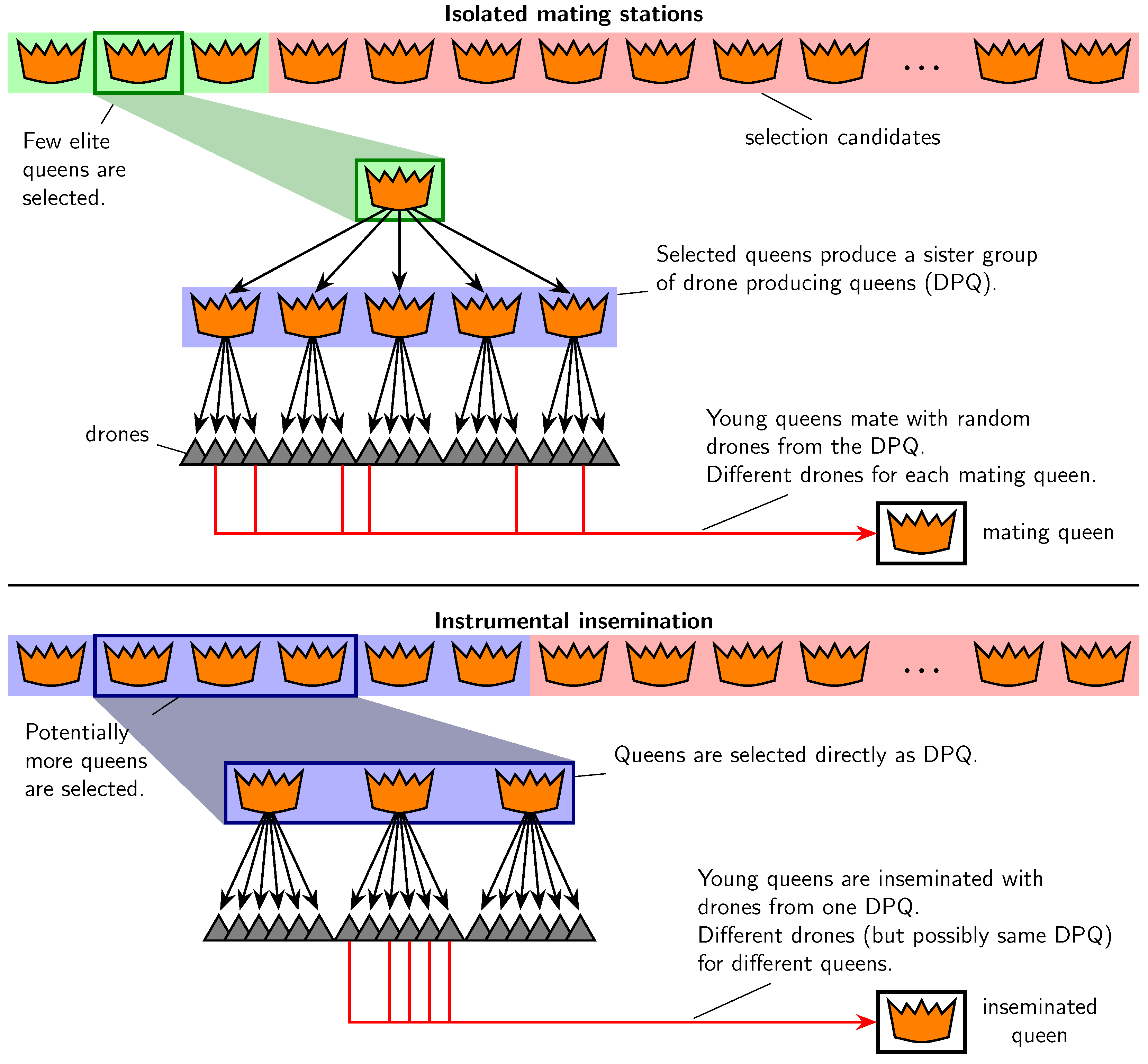
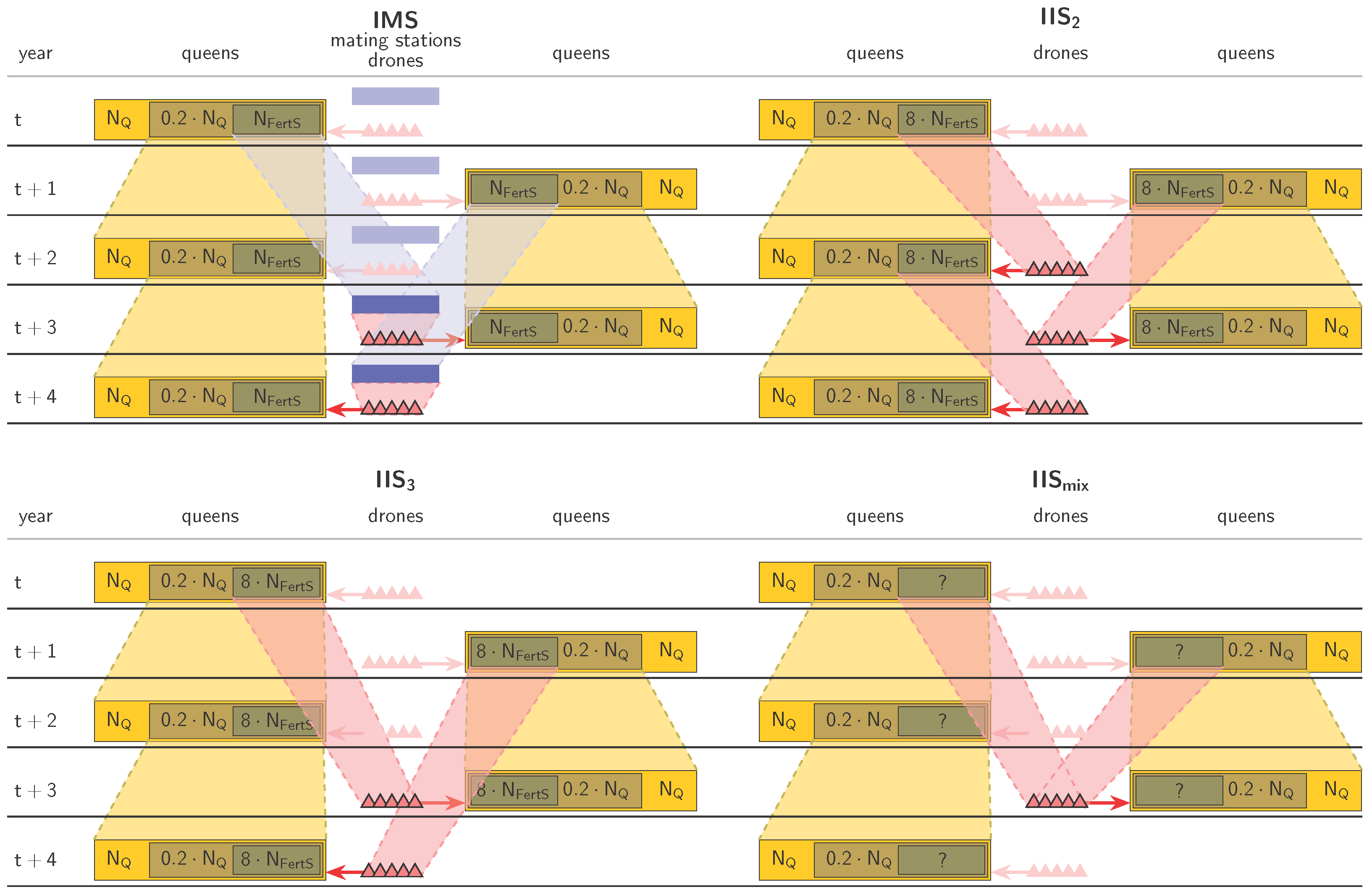
2.3. Controlled Mating
- IMS,
- IIS with a two-year paternal generation interval (called IIS2),
- IIS with a three-year paternal generation interval (called IIS3),
- IIS with a mixed paternal generation interval (called IISmix).
- , representing a realistic number for small-scale honeybee breeding programs (e.g., A. m. mellifera in Switzerland [60]),
- , which was found to be a theoretical minimum number for successful and sustainable breeding with IMS [24],
- , representing the ideal situation of an abundance of fertilization stations.
2.4. Data Analysis
2.5. Effective Number of Sires
3. Results
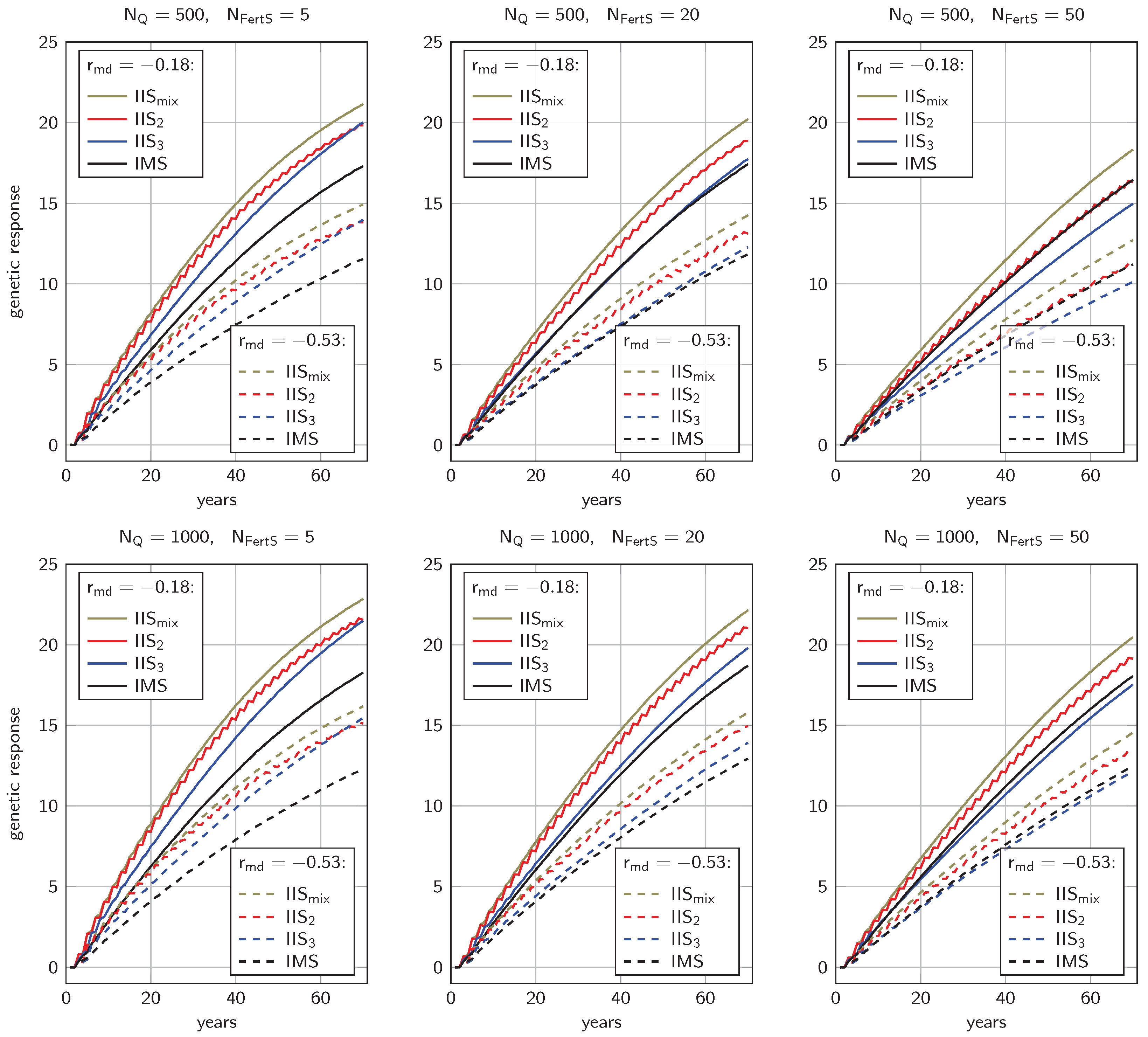
3.1. Genetic Progress
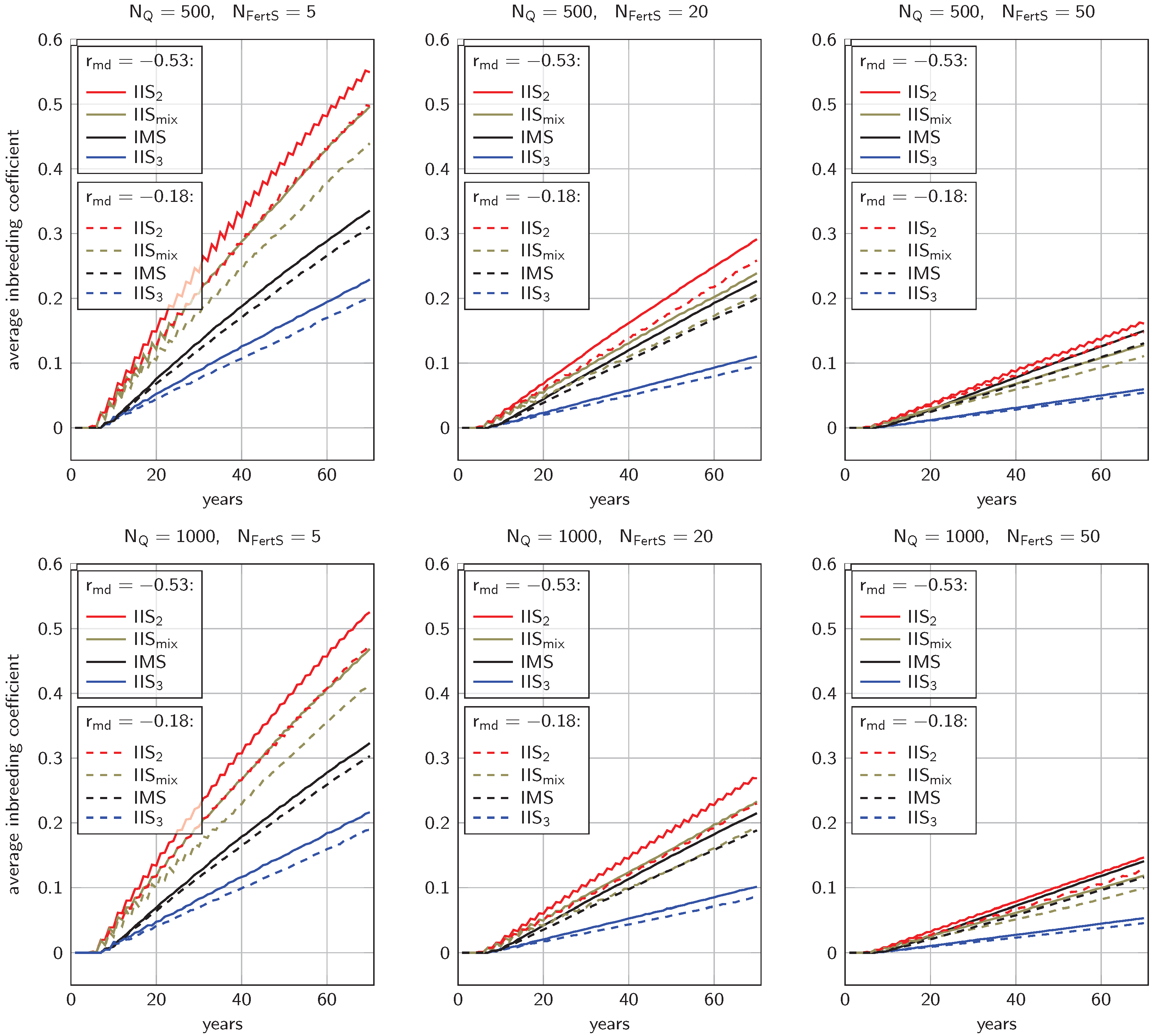
3.2. Inbreeding Coefficients
3.3. Generation Intervals and Inbreeding Rates
4. Discussion
4.1. Explanation of Observations
4.1.1. Genetic Progress
4.1.2. Inbreeding
4.1.3. Paternal Generation Intervals
4.2. Implications for Real Honeybee Breeding Programs
4.2.1. Sustainability
4.2.2. IIS as an Alternative to IMS
4.2.3. Diversity of Drones
4.2.4. Genetic Model
4.3. Limitations of the Study
4.3.1. Reproduction Genetics
4.3.2. Other Biological Factors
4.3.3. Economic Factors
4.3.4. Sociological Factors
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Average relationship between sister queens | |
| BLUP | Best linear unbiased prediction |
| DPQ | Drone producing queen |
| FAO | Food and Agriculture Organization of the United Nations |
| ibd | Identical by descent |
| IIS | Instrumental insemination station |
| IIS2/IIS3/IISmix | IIS with two-year/three-year/mixed paternal generation interval |
| IMS | Isolated mating station |
| (=8) Number of drone producing queens per fertilization station | |
| (=5, 20, or 50) Number of fertilization stations per year | |
| (=500 or 1000) Number of breeding queens per year | |
| Effective number of sires | |
| Probability of paternally inherited alleles to be identical by descent | |
| (= or ) Correlation between maternal and direct effects | |
| / | (=1/=2) Maternal/direct genetic variance |
| (=4) Residual variance |
References
- Bienefeld, K.; Pirchner, F. Heritabilities for several colony traits in the honeybee (Apis mellifera carnica). Apidologie 1990, 21, 175–183. [Google Scholar] [CrossRef]
- Brascamp, E.W.; Willam, A.; Boigenzahn, C.; Bijma, P.; Veerkamp, R.F. Heritabilities and genetic correlations for honey yield, gentleness, calmness and swarming behaviour in Austrian honey bees. Apidologie 2016, 47, 739–748. [Google Scholar] [CrossRef]
- Guichard, M.; Neuditschko, M.; Soland, G.; Fried, P.; Grandjean, M.; Gerster, S.; Dainat, B.; Bijma, P.; Brascamp, E.W. Estimates of genetic parameters for production, behaviour, and health traits in two Swiss honey bee populations. Apidologie 2020, 51, 876–891. [Google Scholar] [CrossRef]
- Hoppe, A.; Du, M.; Bernstein, R.; Tiesler, F.K.; Kärcher, M.; Bienefeld, K. Substantial genetic progress in the international Apis mellifera carnica population since the implementation of genetic evaluation. Insects 2020, 11, 768. [Google Scholar] [CrossRef]
- Jensen, A.B.; Palmer, K.A.; Boomsma, J.J.; Ratnieks, F.L.W. Varying degrees of Apis mellifera ligustica introgression in protected populations of the black honeybee, Apis mellifera mellifera, in northwest Europe. Mol. Ecol. 2005, 14, 93–106. [Google Scholar] [CrossRef]
- De La Rúa, P.; Jaffé, R.; Dall’Olio, R.; Muñoz, I.; Serrano, J. Biodiversity, conservation and current threats to European honey bees. Apidologie 2009, 40, 263–284. [Google Scholar] [CrossRef]
- Uzunov, A.; Mifsud, D.; Galea, T.; Cutajar, S.; Zammit-Mangion, M.; Meixner, M.D. Development, behaviour, productivity, and health status of the native honey bee Apis mellifera ruttneri vs. the introduced A. m. ligustica in Malta. Apidologie 2023, 54, 34. [Google Scholar] [CrossRef]
- Crawford, M. Conservation by utilisation. Oryx 1972, 11, 427–432. [Google Scholar] [CrossRef]
- Uzunov, A.; Brascamp, E.W.; Büchler, R. The basic concept of honey bee breeding programs. Bee World 2017, 94, 84–87. [Google Scholar] [CrossRef]
- Oxley, P.R.; Olroyd, B.P. The genetic architecture of honeybee breeding. In Advances in Insect Physiology; Simpson, S.J., Casas, J., Eds.; Elsevier: Amsterdam, The Netherlands, 2010; Volume 39, pp. 83–118. [Google Scholar]
- Plate, M.; Bernstein, R.; Hoppe, A.; Bienefeld, K. The importance of controlled mating in honeybee breeding. Genet. Sel. Evol. 2019, 51, 74. [Google Scholar] [CrossRef]
- Du, M.; Bernstein, R.; Hoppe, A.; Bienefeld, K. A theoretical derivation of response to selection with and without controlled mating in honeybees. Genet. Sel. Evol. 2021, 53, 17. [Google Scholar] [CrossRef] [PubMed]
- Uzunov, A.; Brascamp, E.W.; Du, M.; Büchler, R. The relevance of mating control for successful implementation of honey bee breeding programs. Bee World 2022, 99, 94–98. [Google Scholar] [CrossRef]
- Maucourt, S.; Rousseau, A.; Fortin, F.; Robert, C.; Giovenazzo, P. Observation of genetic gain with instrumental insemination of honeybee queens. Insects 2023, 14, 301. [Google Scholar] [CrossRef] [PubMed]
- Jensen, A.B.; Palmer, K.A.; Chaline, N.; Raine, N.E.; Tofilski, A.; Martin, S.J.; Pedersen, B.V.; Boomsma, J.J.; Ratnieks, F.L.W. Quantifying honey bee mating range and isolation in semi-isolated valleys by DNA microsatellite paternity analysis. Conserv. Genet. 2005, 6, 527–537. [Google Scholar] [CrossRef]
- Scharpenberg, H.; Neumann, P.; van Praagh, J.P.; Moritz, R.F.A. Reliability of an island mating apiary under routine management. J. Apic. Res. 2006, 45, 153–154. [Google Scholar] [CrossRef]
- Uzunov, A.; Brascamp, E.W.; Du, M.; Büchler, R. Initiation and implementation of honey bee breeding programs. Bee World 2022, 99, 50–55. [Google Scholar] [CrossRef]
- Uzunov, A.; Brascamp, E.W.; Du, M.; Bijma, P.; Büchler, R. Breeding values in honey bees. Bee World 2023, 100, 9–14. [Google Scholar] [CrossRef]
- Druml, T.; Putz, A.; Rubinigg, M.; Kärcher, M.H.; Neubauer, K.; Boigenzahn, C. Founder gene pool composition and genealogical structure in two populations of Austrian Carniolan honey bees (Apis mellifera carnica) as derived from pedigree analysis. Apidologie 2023, 54, 24. [Google Scholar] [CrossRef]
- Bienefeld, K.; Ehrhardt, K.; Reinhardt, F. Genetic evaluation in the honey bee considering queen and worker effects—A BLUP-Animal Model approach. Apidologie 2007, 38, 77–85. [Google Scholar] [CrossRef]
- Brascamp, E.W.; Bijma, P. Methods to estimate breeding values in honey bees. Genet. Sel. Evol. 2014, 46, 53. [Google Scholar] [CrossRef]
- Neumann, P.; van Praagh, J.P.; Moritz, R.F.A.; Dustmann, J.H. Testing reliability of a potential island mating apiary using DNA microsatellites. Apidologie 1999, 30, 257–276. [Google Scholar] [CrossRef]
- Musin, E.; Bienefeld, K.; Skerka, H.; Wegener, J. Delayed flight time of drones and queens as a method for mating control in small-scale honey bee breeding. J. Apic. Res. 2021, 62, 450–458. [Google Scholar] [CrossRef]
- Plate, M.; Bernstein, R.; Hoppe, A.; Bienefeld, K. Long-term evaluation of breeding scheme alternatives for endangered honeybee subspecies. Insects 2020, 11, 404. [Google Scholar] [CrossRef] [PubMed]
- Jager, F.; Howard, C.W. The artificial fertilization of queen bees. Science 1914, 40, 720. [Google Scholar] [CrossRef]
- Cobey, S.W.; Tarpy, D.R.; Woyke, J. Standard methods for instrumental insemination of Apis mellifera queens. J. Apic. Res. 2013, 52, 1–18. [Google Scholar] [CrossRef]
- Cobey, S.; Bieńkowska, M.; Wilde, J.; Gąbka, J. Poland, the only country where instrumentally inseminated queens are routinely used in commercial production colonies. Am. Bee J. 2019, 159, 275–280. [Google Scholar]
- Büchler, R.; Andonov, S.; Bienefeld, K.; Costa, C.; Hatjina, F.; Kezic, N.; Kryger, P.; Spivak, M.; Uzunov, A.; Wilde, J. Standard methods for rearing and selection of Apis mellifera queens. J. Apic. Res. 2013, 52, 1–30. [Google Scholar] [CrossRef]
- Moškrič, A.; Formato, G.; Smodiš Škerl, M.I.; Prešern, J. Chapter 14—Reproductive technologies in the honeybee (Apis mellifera). In Reproductive Technologies in Animals; Presicce, G.A., Ed.; Academic Press: London, UK, 2020; pp. 229–243. [Google Scholar]
- Uzunov, A.; Andonov, S.; Dahle, B.; Kovačić, M.; Prešern, J.; Aleksovski, G.; Jaman, F.; Jovanovska, M.; Pavlov, B.; Puškadija, Z.; et al. Evaluating the potential for mating control in honey bee breeding in three SE European countries (preliminary results). In Proceedings of the 12th World Congress on Genetics Applied to Livestock Production, Rotterdam, The Netherlands, 3–8 July 2022. [Google Scholar]
- Meyer-Rochow, V.B.; Jung, C. Remote honey bee breeding centre: A case study of Heligoland Island in Germany. J. Apic. 2019, 34, 285–293. [Google Scholar] [CrossRef]
- Frunze, O.; Akongte, P.N.; Kim, D.; Kang, E.J.; Kim, K.; Park, B.S.; Choi, Y.S. Remote honey bee breeding center in the Wido Island, the Republic of Korea. J. Apic. 2022, 37, 175–183. [Google Scholar] [CrossRef]
- Bernstein, R.; Plate, M.; Hoppe, A.; Bienefeld, K. Computing inbreeding coefficients and the inverse numerator relationship matrix in large populations of honey bees. J. Anim. Breed. Genet. 2018, 135, 323–332. [Google Scholar] [CrossRef]
- Gerula, D.; Węgrzynowicz, P.; Panasiuk, B.; Bieńkowska, M.; Skowronek, W. Performance of Bee Colonies Headed by Queens Instrumentally Inseminated with Semen of Drones Who Come from a Single Colony or Many Colonies. J. Apic. Sci. 2014, 58, 87–97. [Google Scholar] [CrossRef]
- Kistler, T.; Basso, B.; Phocas, F. A simulation study of a honeybee breeding scheme accounting for polyandry, direct and maternal effects on colony performance. Genet. Sel. Evol. 2021, 53, 71. [Google Scholar] [CrossRef]
- Kistler, T.; Brascamp, E.W.; Basso, B.; Bijma, P.; Phocas, F. Assessment of uncertainty in direct and maternal genetic parameters estimates for honeybee colony phenotypes. In Proceedings of the 12th World Congress on Genetics Applied to Livestock Production, Rotterdam, The Netherlands, 3–8 July 2022. [Google Scholar]
- Guichard, M.; Phocas, F.; Neuditschko, M.; Basso, B.; Dainat, B. An overview of selection concepts applied to honey bees. Bee World 2023, 100, 2–8. [Google Scholar] [CrossRef]
- Mattila, H.R.; Seeley, T.D. Genetic diversity in honey bee colonies enhances productivity and fitness. Science 2007, 317, 362–364. [Google Scholar] [CrossRef] [PubMed]
- Tarpy, D.R.; vanEngelsdorp, D.; Pettis, J.S. Genetic diversity affects colony survivorship in commercial honey bee colonies. Naturwissenschaften 2013, 100, 723–728. [Google Scholar] [CrossRef]
- Delaplane, K.S.; Pietravalle, S.; Brown, M.A.; Budge, G.E. Honey bee colonies headed by hyperpolyandrous queens have improved brood rearing efficiency and lower infestation rates of parasitic Varroa mites. PLoS ONE 2015, 10, e0142985. [Google Scholar] [CrossRef]
- Page, R.E.; Laidlaw, H.H. Closed population honeybee breeding. Bee World 1985, 66, 63–72. [Google Scholar] [CrossRef]
- Moritz, R.F.A. Comparison of within-family and mass selection in honeybee populations. J. Apic. Res. 1986, 25, 146–153. [Google Scholar] [CrossRef]
- Du, M.; Bernstein, R.; Hoppe, A.; Bienefeld, K. Consequences of incorrect genetic parameter estimates for single-trait and multi-trait genetic evaluations in honeybees. J. Anim. Breed. Genet. 2022, 139, 666–678. [Google Scholar] [CrossRef]
- Gupta, P.; Reinsch, N.; Spötter, A.; Conrad, T.; Bienefeld, K. Accuracy of the unified approach in maternally influenced traits—Illustrated by a simulation study in the honey bee (Apis mellifera). BMC Genet. 2013, 14, 36. [Google Scholar] [CrossRef]
- Brascamp, E.W.; Wanders, T.H.V.; Wientjes, Y.C.J.; Bijma, P. Prospects for genomic selection in honey-bee breeding. In Proceedings of the 11th World Congress on Genetics Applied to Livestock Production, Auckland, New Zealand, 11–16 February 2018. [Google Scholar]
- Bernstein, R.; Du, M.; Hoppe, A.; Bienefeld, K. Simulation studies to optimize genomic selection in honey bees. Genet. Sel. Evol. 2021, 53, 64. [Google Scholar] [CrossRef] [PubMed]
- Plate, M.; Bernstein, R.; Hoppe, A.; Bienefeld, K. Comparison of infinitesimal and finite locus models for long-term breeding simulations with direct and maternal effects at the example of honeybees. PLoS ONE 2019, 14, e0213270. [Google Scholar] [CrossRef] [PubMed]
- Muir, W.M. Genetic selection strategies: Computer modeling. Poult. Sci. 1997, 76, 1066–1070. [Google Scholar] [CrossRef] [PubMed]
- Wright, S. Evolution in Mendelian populations. Genetics 1931, 16, 97–159. [Google Scholar] [CrossRef] [PubMed]
- Hill, W.G.; Goddard, M.E.; Visscher, P.M. Data and Theory Point to Mainly Additive Genetic Variance for Complex Traits. PLoS Genet. 2008, 4, e1000008. [Google Scholar] [CrossRef]
- Daetwyler, H.D.; Capitan, A.; Pausch, H.; Stothard, P.; van Binsbergen, R.; Brøndum, R.F.; Liao, X.; Djari, A.; Rodriguez, S.C.; Grohs, C.; et al. Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat. Genet. 2014, 46, 858–865. [Google Scholar] [CrossRef]
- Hayes, B.J.; Goddard, M.E. The distribution of the effects of genes affecting quantitative traits in livestock. Genet. Sel. Evol. 2001, 33, 209–229. [Google Scholar] [CrossRef]
- Sun, C.; VanRaden, P.M. Increasing long-term response by selecting for favorable minor alleles. PLoS ONE 2014, 9, e88510. [Google Scholar] [CrossRef]
- Esfandyari, H.; Henryon, M.; Berg, P.; Thomasen, J.R.; Bijma, P.; Sørensen, A.C. Response to selection in finite locus models with nonadditive effects. J. Hered. 2017, 108, 318–327. [Google Scholar]
- Fernando, R.L.; Grossmann, M. Genetic evaluation with autosomal and X-chromosomal inheritance. Theor. Appl. Genet. 1990, 80, 75–80. [Google Scholar] [CrossRef]
- Obšteter, J.; Strachan, L.K.; Bubnič, J.; Prešern, J.; Gorjanc, G. SIMplyBee: An R package to simulate honeybee populations and breeding programs. Genet. Sel. Evol. 2023, 55, 31. [Google Scholar] [CrossRef]
- Misztal, I.; Tsuruta, S.; Strabel, T.; Auvray, B.; Druet, T.; Lee, D.H. BLUPF90 and related programs (BGF90). In Proceedings of the 7th World Congress on Genetics Applied to Livestock Production, Montpellier, France, 19–23 August 2002. [Google Scholar]
- Brascamp, E.W.; Bijma, P. A note on genetic parameters and accuracy of estimated breeding values in honey bees. Genet. Sel. Evol. 2019, 51, 71. [Google Scholar] [CrossRef]
- Du, M.; Bernstein, R.; Hoppe, A.; Bienefeld, K. Short-term effects of controlled mating and selection on the genetic variance of honeybee populations. Heredity 2021, 162, 733–747. [Google Scholar] [CrossRef]
- Liste der A- und B-Belegstationen. mellifera.ch magazin. February 2023, pp. 32–34. Available online: https://mellifera.clicdesign.ch/downloads/ (accessed on 8 September 2023).
- Tarpy, D.R.; Nielsen, D.I. Sampling Error, Effective Paternity, and Estimating the Genetic Structure of Honey Bee Colonies (Hymenoptera: Apidae). Ann. Entomol. Soc. Am. 2002, 95, 513–528. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; D’Agostino McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Welch, B.L. The generalization of ‘Student’s’ problem when several different population variances are involved. Biometrika 1947, 34, 28–35. [Google Scholar] [CrossRef]
- Bienefeld, K.; Reinhardt, F.; Pirchner, F. Inbreeding effects of queen and workers on colony traits in the honey bee. Apidologie 1989, 20, 439–450. [Google Scholar] [CrossRef]
- Bernstein, R.; Du, M.; Du, Z.G.; Strauss, A.S.; Hoppe, A.; Bienefeld, K. First large-scale genomic prediction in the honey bee. Heredity 2023, 130, 320–328. [Google Scholar] [CrossRef]
- Xu, S. Quantitative Genetics, 1st ed.; Springer: Cham, Switzerland, 2022. [Google Scholar]
- Draft Guidelines on in Vivo Conservation of Animal Genetic Resources; Number 14 in FAO Animal Production and Health Guidelines; Food and Agriculture Organization of the United Nations (FAO): Rome, Italy, 2013.
- Bürger, R.; Lynch, M. Evolution and extinction in a changing environment: A quantitative-genetic analysis. Evolution 1995, 49, 151–163. [Google Scholar] [PubMed]
- Panziera, D.; Requier, F.; Chantawannakul, P.; Pirk, C.W.W.; Blacquière, T. The diversity decline in wild and managed honey bee populations urges for an integrated conservation approach. Front. Ecol. Evol. 2022, 10, 767950. [Google Scholar] [CrossRef]
- Kristensen, T.N.; Sørensen, P. Inbreeding—Lessons from animal breeding, evolutionary biology and conservation genetics. Anim. Sci. 2005, 80, 121–133. [Google Scholar] [CrossRef]
- Franklin, I.R. Evolutionary change in small populations. In Conservation Biology: An Evolutionary-Ecological Perspective; Soulé, M.E., Wilcox, B.A., Eds.; Sinauer: Sunderland, MA, USA, 1980; pp. 135–150. [Google Scholar]
- Soulé, M.E. Thresholds for survival: Maintaining fitness and evolutionary potential. In Conservation Biology: An Evolutionary-Ecological Perspective; Soulé, M.E., Wilcox, B.A., Eds.; Sinauer: Sunderland, MA, USA, 1980; pp. 151–170. [Google Scholar]
- Welsh, C.S.; Stewart, T.S.; Schwab, C.; Blackburn, H.D. Pedigree analysis of 5 swine breeds in the United States and the implications for genetic conservation. J. Anim. Sci. 2010, 88, 1610–1618. [Google Scholar] [CrossRef] [PubMed]
- Pham, M.H.; Tran, X.H.; Berthouly-Salazar, C.; Tixier-Boichard, M.; Chen, C.F.; Lee, Y.P. Monitoring of genetic diversity in Taiwan conserved chickens assessed by pedigree and molecular data. Livest. Sci. 2016, 184, 85–91. [Google Scholar] [CrossRef]
- Mc Parland, S.; Kearney, J.F.; Rath, M.; Berry, D.P. Inbreeding trends and pedigree analysis of Irish dairy and beef cattle populations. J. Anim. Sci. 2007, 85, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Heimpel, G.E.; de Boer, J.G. Sex determination in the Hymenoptera. Annu. Rev. Entomol. 2008, 50, 209–230. [Google Scholar] [CrossRef] [PubMed]
- Mackensen, O. Viability and sex determination in the honey bee (Apis mellifera L.). Genetics 1951, 36, 500–509. [Google Scholar] [CrossRef]
- Zayed, A. Bee genetics and conservation. Apidologie 2009, 40, 237–262. [Google Scholar] [CrossRef]
- Tien, N.S.H.; Sabelis, M.W.; Egas, M. Inbreeding depression and purging in a haplodiploid: Gender-related effects. Heredity 2015, 114, 327–332. [Google Scholar] [CrossRef]
- Xia, S. Exploring the Potential of Genetic Improvement of Insects: A Case Study Using the Haplodiploid Parasitoid Wasp Nasonia vitripennis. Ph.D. Thesis, Wageningen University, Wageningen, The Netherlands, 2020. [Google Scholar]
- Meuwissen, T.H.E. Maximizing the response of selection with a predefined rate of inbreeding. J. Anim. Sci. 1997, 75, 934–940. [Google Scholar] [CrossRef]
- Seeley, T.D.; Tarpy, D.R. Queen promiscuity lowers disease within honeybee colonies. Proc. R. Soc. B 2007, 274, 67–72. [Google Scholar] [CrossRef]
- Mattila, H.R.; Seeley, T.D. Extreme polyandry improves a honey bee colony’s ability to track dynamic foraging opportunities via greater activity of inspecting bees. Apidologie 2014, 45, 347–363. [Google Scholar] [CrossRef]
- Neumann, P.; Moritz, R.F.A. Testing genetic variance hypotheses for the evolution of polyandry in the honeybee (Apis mellifera L.). Insectes Soc. 2000, 47, 271–279. [Google Scholar] [CrossRef]
- Baumdicker, F.; Bisschop, G.; Goldstein, D.; Gower, G.; Ragsdale, A.P.; Tsambos, G.; Zhu, S.; Eldon, B.; Ellerman, E.C.; Galloway, J.G.; et al. Efficient ancestry and mutation simulation with msprime 1.0. Genetics 2022, 220, iyab229. [Google Scholar] [CrossRef] [PubMed]
- Lauterbur, M.E.; Cavassim, M.I.A.; Gladstein, A.L.; Gower, G.; Pope, N.S.; Tsambos, G.; Adrion, J.; Belsare, S.; Biddanda, A.; Caudill, V.; et al. Expanding the stdpopsim species catalog, and lessons learned for realistic genome simulations. eLife 2023, 12, RP84874. [Google Scholar] [CrossRef]
- Faux, A.M.; Gorjanc, G.; Gaynor, R.C.; Battagin, M.; Edwards, S.M.; Wilson, D.L.; Hearne, S.J.; Gonen, S.; Hickey, J.M. AlphaSim: Software for breeding program simulation. Plant Genome 2016, 9, 3. [Google Scholar] [CrossRef]
- Gaynor, R.C.; Gorjanc, G.; Hickey, J.M. AlphaSimR: An R package for breeding program simulations. G3 2021, 11, jkaa017. [Google Scholar] [CrossRef]
- Spötter, A.; Gupta, P.; Mayer, M.; Reinsch, N.; Bienefeld, K. Genome-wide association study of a Varroa-specific defense behavior in honeybees (Apis mellifera). J. Hered. 2016, 107, 220–227. [Google Scholar] [CrossRef]
- Conlon, B.H.; Aurori, A.; Giurgiu, A.; Kefuss, J.; Dezmirean, D.S.; Moritz, R.F.A.; Routtu, J. A gene for resistance to the Varroa mite (Acari) in honey bee (Apis mellifera) pupae. Mol. Ecol. 2019, 28, 2958–2966. [Google Scholar] [CrossRef]
- Guichard, M.; Dainat, B.; Eynard, S.; Vignal, A.; Servin, B.; the Beestrong Consortium; Neuditschko, M. Identification of quantitative trait loci associated with calmness and gentleness in honey bees using whole-genome sequences. Anim. Genet. 2021, 52, 472–481. [Google Scholar] [CrossRef]
- Du, M.; Bernstein, R.; Hoppe, A.; Bienefeld, K. Influence of model selection and data structure on the estimation of genetic parameters in honeybee populations. G3 2022, 12, jkab450. [Google Scholar] [CrossRef]
- Koeniger, N.; Koeniger, G.; Gries, M.; Tingek, S. Drone competition at drone congregation areas in four Apis species. Apidologie 2005, 36, 211–221. [Google Scholar] [CrossRef]
- Neumann, P.; Blacquière, T. The Darwin cure for apiculture? Natural selection and managed honey bee health. Evol. Appl. 2017, 10, 226–230. [Google Scholar] [CrossRef] [PubMed]
- Slater, G.P.; Harpur, B.A. Using genomics to predict drone quality: Why are there so many ‘dud’ male honey bees. In Proceedings of the 12th World Congress on Genetics Applied to Livestock Production, Rotterdam, The Netherlands, 3–8 July 2022. [Google Scholar]
- Cobey, S.W. Comparison studies of instrumental inseminated and naturally mated honey bee queens and factors affecting their performance. Apidologie 2007, 38, 390–410. [Google Scholar] [CrossRef]
- Pritsch, G.; Bienefeld, K. Performance of colonies with artificially inseminated and naturally mated queens. J. Apic. Sci. 2003, 47, 81–86. [Google Scholar]
- Buechler, R.; Uzunov, A.; Costa, C.; Meixner, M.; Le Conte, Y.; Mondet, F.; Kovacic, M.; Andonov, S.; Carreck, N.L.; Dimitrov, L.; et al. EurBeST—A pilot study testing varroa-resistant bees under commercial beekeeping conditions. Am. Bee J. 2022, 162, 213–215. [Google Scholar]
- Gerula, D. Observations of body injuries of artificially inseminated honeybee queens inflicted in the subsequent stages of rearing and during their introduction into colonies. J. Apic. Sci. 2007, 51, 5–17. [Google Scholar]
- Alonso, A.D.; Kok, S.K.; O’Shea, M. Perceived contributory leisure in the context of hobby beekeeping: A multi-country comparison. Leis. Stud. 2021, 40, 243–260. [Google Scholar] [CrossRef]

| IMS | IIS2 | IIS3 | IISmix | |||
|---|---|---|---|---|---|---|
| 500 | 5 | 1.34 | 1.97 | 0.81 | 1.77 | |
| 500 | 20 | 0.80 | 0.86 | 0.37 | 0.72 | |
| 500 | 50 | 0.51 | 0.46 | 0.20 | 0.37 | |
| 500 | 5 | 1.47 | 2.28 | 0.94 | 2.10 | |
| 500 | 20 | 0.92 | 0.99 | 0.42 | 0.85 | |
| 500 | 50 | 0.59 | 0.51 | 0.22 | 0.43 | |
| 1000 | 5 | 1.30 | 1.86 | 0.76 | 1.64 | |
| 1000 | 20 | 0.75 | 0.75 | 0.32 | 0.66 | |
| 1000 | 50 | 0.45 | 0.39 | 0.17 | 0.33 | |
| 1000 | 5 | 1.40 | 2.14 | 0.88 | 1.93 | |
| 1000 | 20 | 0.87 | 0.90 | 0.39 | 0.82 | |
| 1000 | 50 | 0.55 | 0.46 | 0.20 | 0.39 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, M.; Bernstein, R.; Hoppe, A. The Potential of Instrumental Insemination for Sustainable Honeybee Breeding. Genes 2023, 14, 1799. https://doi.org/10.3390/genes14091799
Du M, Bernstein R, Hoppe A. The Potential of Instrumental Insemination for Sustainable Honeybee Breeding. Genes. 2023; 14(9):1799. https://doi.org/10.3390/genes14091799
Chicago/Turabian StyleDu, Manuel, Richard Bernstein, and Andreas Hoppe. 2023. "The Potential of Instrumental Insemination for Sustainable Honeybee Breeding" Genes 14, no. 9: 1799. https://doi.org/10.3390/genes14091799
APA StyleDu, M., Bernstein, R., & Hoppe, A. (2023). The Potential of Instrumental Insemination for Sustainable Honeybee Breeding. Genes, 14(9), 1799. https://doi.org/10.3390/genes14091799






