GIGANTEA Unveiled: Exploring Its Diverse Roles and Mechanisms
Abstract
1. Introduction
2. Structural Insights into GI
2.1. Structural Conservation and Functional Dynamics
2.2. Subcellular Localization of GI
3. Deciphering the Intricacies of GI Transcription and Post-Transcriptional Regulations
4. Unraveling the Enigmatic Role of GI: A Multifaceted Player in Plant Biology
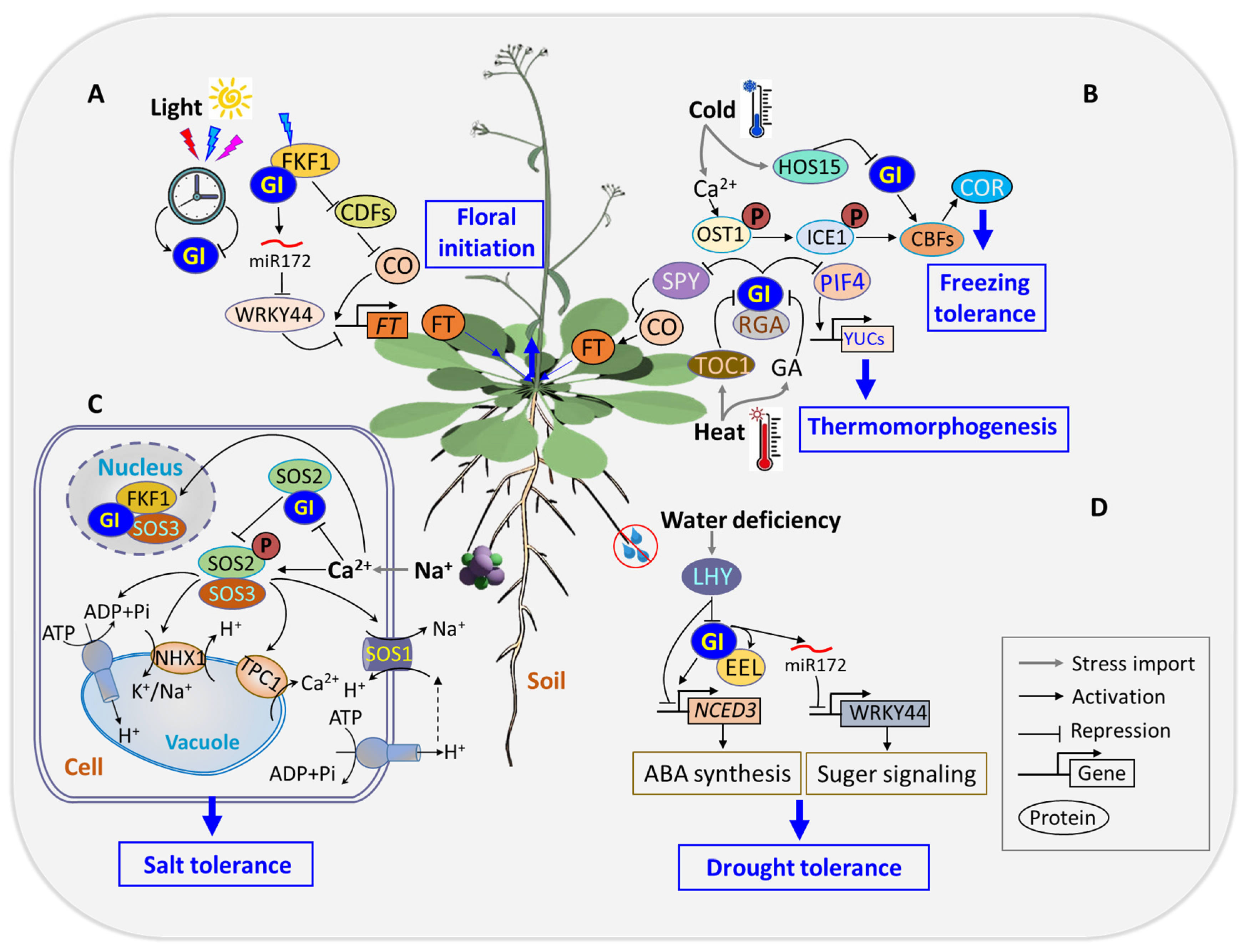
4.1. Stimulus Response
- (1)
- Coordination of red light signaling by GI
- (2)
- Coordination of blue light signaling by GI
- (3)
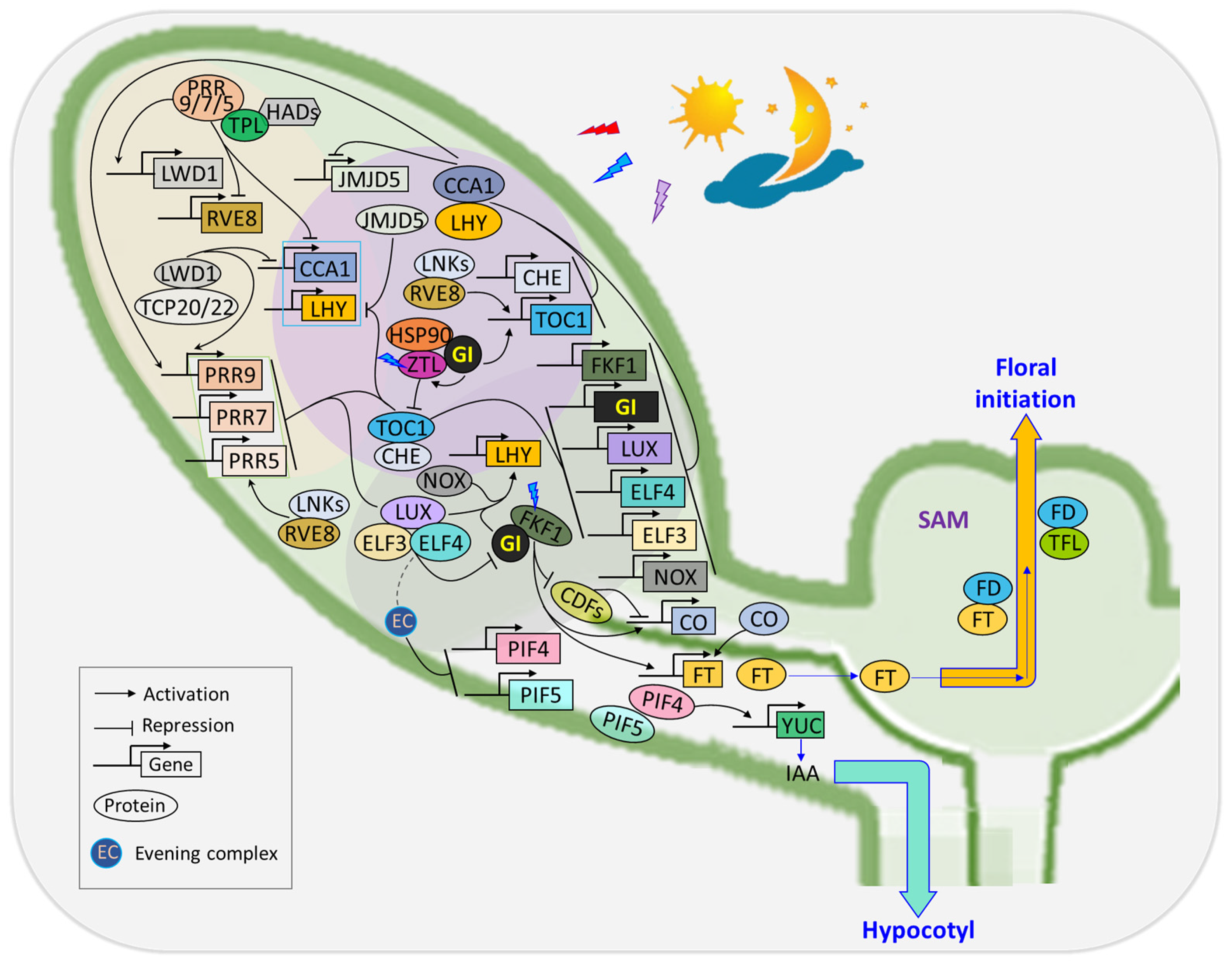
- Circadian clock regulation
4.2. Flowering Time Regulation
- (1)
- Orchestrating floral transition in response to photoperiodic signals
- (2)
- Stress tolerance
4.3. Chlorophyll Accumulation Is Regulated by GI in Plants
4.4. GI Regulates Stomatal Opening in Plants
4.5. GI’s Role in Plant Sugar Signaling
4.6. GI’s Unexplored Role in Anthocyanin Metabolism
4.7. Integrative Role of GI in Hormonal Signaling
- (1)
- The role of GI in ABA-mediated responses to drought stress
- (2)
- GI’s involvement in gibberellin signaling for hypocotyl elongation
- (3)
- GI’s impact on phytohormones in biotic stress response
- (4)
- GI’s role in brassinosteroid signaling pathway
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Savchenko, T.V.; Rolletschek, H.; Dehesh, K. Jasmonates-Mediated Rewiring of Central Metabolism Regulates Adaptive Responses. Plant Cell Physiol. 2019, 60, 2613–2620. [Google Scholar] [CrossRef] [PubMed]
- Zandalinas, S.I.; Mittler, R.; Balfagon, D.; Arbona, V.; Gomez-Cadenas, A. Plant adaptations to the combination of drought and high temperatures. Physiol. Plant 2018, 162, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Badouin, H.; Gouzy, J.; Grassa, C.J.; Murat, F.; Staton, S.E.; Cottret, L.; Lelandais-Brière, C.; Owens, G.L.; Carrère, S.; Mayjonade, B.; et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 2017, 546, 148–152. [Google Scholar] [CrossRef] [PubMed]
- De Frenne, P.; Lenoir, J.; Luoto, M.; Scheffers, B.R.; Zellweger, F.; Aalto, J.; Ashcroft, M.B.; Christiansen, D.M.; Decocq, G.; De Pauw, K.; et al. Forest microclimates and climate change: Importance, drivers and future research agenda. Glob. Chang. Biol. 2021, 27, 2279–2297. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Rico-Medina, A.; Cano-Delgado, A.I. The physiology of plant responses to drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Wallrad, L.; Almutairi, B.O.; Kudla, J. Ca2+ signaling in plant responses to abiotic stresses. J. Integr. Plant Biol. 2022, 64, 287–300. [Google Scholar] [CrossRef] [PubMed]
- Kultz, D.; Fiol, D.; Valkova, N.; Gomez-Jimenez, S.; Chan, S.Y.; Lee, J. Functional genomics and proteomics of the cellular osmotic stress response in ‘non-model’ organisms. J. Exp. Biol. 2007, 210 Pt 9, 1593–1601. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yu, C.; Li, H.; Ouyang, B.; Wang, T.; Zhang, J.; Wang, X.; Ye, Z. Overexpression of ShDHN, a dehydrin gene from Solanum habrochaites enhances tolerance to multiple abiotic stresses in tomato. Plant Sci. 2015, 231, 198–211. [Google Scholar] [CrossRef]
- Hirayama, T.; Shinozaki, K. Research on plant abiotic stress responses in the post-genome era: Past, present and future. Plant J. 2010, 61, 1041–1052. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K.; Seki, M. Regulatory network of gene expression in the drought and cold stress responses. Curr Opin. Plant Biol. 2003, 6, 410–417. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef]
- Khurana, P.; Vishnudasan, D.; Chhibbar, A.K. Genetic approaches towards overcoming water deficit in plants—Special emphasis on LEAs. Physiol. Mol. Biol. Plants 2008, 14, 277–298. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Villagra, J.; Kurepin, L.V.; Reyes-Diaz, M.M. Evaluating the involvement and interaction of abscisic acid and miRNA156 in the induction of anthocyanin biosynthesis in drought-stressed plants. Planta 2017, 246, 299–312. [Google Scholar] [CrossRef]
- Xie, Q.; Lou, P.; Hermand, V.; Aman, R.; Park, H.J.; Yun, D.J.; Kim, W.Y.; Salmela, M.J.; Ewers, B.E.; Weinig, C.; et al. Allelic polymorphism of GIGANTEA is responsible for naturally occurring variation in circadian period in Brassica rapa. Proc. Natl. Acad. Sci. USA 2015, 112, 3829–3834. [Google Scholar] [CrossRef] [PubMed]
- De Montaigu, A.; Toth, R.; Coupland, G. Plant development goes like clockwork. Trends Genet. 2010, 26, 296–306. [Google Scholar] [CrossRef] [PubMed]
- Shu, K.; Yang, W. E3 Ubiquitin Ligases: Ubiquitous Actors in Plant Development and Abiotic Stress Responses. Plant Cell Physiol. 2017, 58, 1461–1476. [Google Scholar] [CrossRef]
- Patnaik, A.; Kumar, A.; Behera, A.; Mishra, G.; Dehery, S.K.; Panigrahy, M.; Das, A.B.; Panigrahi, K.C.S. GIGANTEA supresses wilt disease resistance by down-regulating the jasmonate signaling in Arabidopsis thaliana. Front. Plant Sci. 2023, 14, 1091644. [Google Scholar] [CrossRef]
- Park, D.H.; Somers, D.E.; Kim, Y.S.; Choy, Y.H.; Lim, H.K.; Soh, M.S.; Kim, H.J.; Kay, S.A.; Nam, H.G. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 1999, 285, 1579–1582. [Google Scholar] [CrossRef]
- Huq, E.; Tepperman, J.M.; Quail, P.H. GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2000, 97, 9789–9794. [Google Scholar] [CrossRef]
- Kim, W.Y.; Ali, Z.; Park, H.J.; Park, S.J.; Cha, J.Y.; Perez-Hormaeche, J.; Quintero, F.J.; Shin, G.; Kim, M.R.; Qiang, Z.; et al. Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat. Commun. 2013, 4, 1352. [Google Scholar] [CrossRef]
- Sawa, M.; Nusinow, D.A.; Kay, S.A.; Imaizumi, T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 2007, 318, 261–265. [Google Scholar] [CrossRef]
- Patnaik, A.; Alavilli, H.; Rath, J.; Panigrahi, K.C.S.; Panigrahy, M. Variations in Circadian Clock Organization & Function: A Journey from Ancient to Recent. Planta 2022, 256, 91. [Google Scholar]
- Mishra, P.; Panigrahi, K.C. GIGANTEA—An emerging story. Front. Plant Sci. 2015, 6, 8. [Google Scholar] [CrossRef]
- Mizoguchi, T.; Wright, L.; Fujiwara, S.; Cremer, F.; Lee, K.; Onouchi, H.; Mouradov, A.; Fowler, S.; Kamada, H.; Putterill, J.; et al. Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 2005, 17, 2255–2270. [Google Scholar] [CrossRef]
- Filichkin, S.A.; Breton, G.; Priest, H.D.; Dharmawardhana, P.; Jaiswal, P.; Fox, S.E.; Michael, T.P.; Chory, J.; Kay, S.A.; Mockler, T.C. Global profiling of rice and poplar transcriptomes highlights key conserved circadian-controlled pathways and cis-regulatory modules. PLoS ONE 2011, 6, e16907. [Google Scholar] [CrossRef]
- Fowler, S.; Lee, K.; Onouchi, H.; Samach, A.; Richardson, K.; Morris, B.; Coupland, G.; Putterill, J. GIGANTEA: A circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 1999, 18, 4679–4688. [Google Scholar] [CrossRef]
- David, K.M.; Armbruster, U.; Tama, N.; Putterill, J. Arabidopsis GIGANTEA protein is post-transcriptionally regulated by light and dark. FEBS Lett. 2006, 580, 1193–1197. [Google Scholar] [CrossRef]
- Linde, A.M.; Eklund, D.M.; Kubota, A.; Pederson, E.R.A.; Holm, K.; Gyllenstrand, N.; Nishihama, R.; Cronberg, N.; Muranaka, T.; Oyama, T.; et al. Early evolution of the land plant circadian clock. New Phytol. 2017, 216, 576–590. [Google Scholar] [CrossRef]
- Izawa, T.; Mihara, M.; Suzuki, Y.; Gupta, M.; Itoh, H.; Nagano, A.J.; Motoyama, R.; Sawada, Y.; Yano, M.; Hirai, M.Y.; et al. Os- GIGANTEA Confers Robust Diurnal Rhythms on the Global Transcriptome of Rice in the Field. Plant Cell 2011, 23, 1741–1755. [Google Scholar] [CrossRef]
- Holm, K.; Kallman, T.; Gyllenstrand, N.; Hedman, H.; Lagercrantz, U. Does the core circadian clock in the moss Physcomitrella patens (Bryophyta) comprise a single loop? BMC Plant Biol. 2010, 10, 109. [Google Scholar] [CrossRef]
- Brandoli, C.; Petri, C.; Egea-Cortines, M.; Weiss, J. Gigantea: Uncovering New Functions in Flower Development. Genes 2020, 11, 1142. [Google Scholar] [CrossRef]
- Bombarely, A.; Moser, M.; Amrad, A.; Bao, M.; Bapaume, L.; Barry, C.S.; Bliek, M.; Boersma, M.R.; Borghi, L.; Bruggmann, R.; et al. Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida. Nat. Plants 2016, 2, 16074. [Google Scholar] [CrossRef]
- Redei, G.P. Supervital Mutants of Arabidopsis. Genetics 1962, 47, 443–460. [Google Scholar] [CrossRef]
- Mizoguchi, T.; Wheatley, K.; Hanzawa, Y.; Wright, L.; Mizoguchi, M.; Song, H.R.; Carré, I.A.; Coupland, G. LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2002, 2, 629–641. [Google Scholar] [CrossRef]
- Kim, H.; Park, S.J.; Kim, Y.; Nam, H.G. Subcellular Localization of GIGANTEA Regulates the Timing of Leaf Senescence and Flowering in Arabidopsis. Front. Plant. Sci. 2020, 11, 589707. [Google Scholar] [CrossRef]
- Baek, D.; Kim, W.Y.; Cha, J.Y.; Park, H.J.; Shin, G.; Park, J.; Lim, C.J.; Chun, H.J.; Li, N.; Kim, D.H.; et al. The GIGANTEA-ENHANCED EM LEVEL Complex Enhances Drought Tolerance via Regulation of Abscisic Acid Synthesis. Plant Physiol. 2020, 184, 443–458. [Google Scholar] [CrossRef]
- Cha, J.; Shin, G.I.; Ahn, G.; Jeong, S.Y.; Ji, M.G.; Alimzhan, A.; Kim, M.G.; Kim, W.K. Loss-of-function in GIGANTEA confers resistance to PPO-inhibiting herbicide tiafenacil through transcriptional activation of antioxidant genes in Arabidopsis. Appl. Biol. Chem. 2022, 65, 1–10. [Google Scholar] [CrossRef]
- He, Y.; Tang, R.H.; Hao, Y.; Stevens, R.D.; Cook, C.W.; Ahn, S.M.; Jing, L.; Yang, Z.; Chen, L.; Guo, F.; et al. Nitric oxide represses the Arabidopsis floral transition. Science 2004, 305, 1968–1971. [Google Scholar] [CrossRef]
- Abdul-Awal, S.M.; Chen, J.; Xin, Z.; Harmon, F.G. A sorghum gigantea mutant attenuates florigen gene expression and delays flowering time. Plant Direct 2020, 4, e00281. [Google Scholar] [CrossRef]
- Kim, Y.; Lim, J.; Yeom, M.; Kim, H.; Kim, J.; Wang, L.; Kim, W.Y.; Somers, D.E.; Nam, H.G. ELF4 regulates GIGANTEA chromatin access through subnuclear sequestration. Cell Rep. 2013, 3, 671–677. [Google Scholar] [CrossRef]
- Yu, J.W.; Rubio, V.; Lee, N.Y.; Bai, S.; Lee, S.Y.; Kim, S.S.; Liu, L.; Zhang, Y.; Irigoyen, M.L.; Sullivan, J.A.; et al. COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol. Cell 2008, 32, 617–630. [Google Scholar] [CrossRef]
- Kim, J.; Geng, R.; Gallenstein, R.A.; Somers, D.E. The F-box protein ZEITLUPE controls stability and nucleocytoplasmic partitioning of GIGANTEA. Development 2013, 140, 4060–4069. [Google Scholar] [CrossRef]
- Kim, Y.; Han, S.; Yeom, M.; Kim, H.; Lim, J.; Cha, J.Y.; Kim, W.Y.; Somers, D.E.; Putterill, J.; Nam, H.G.; et al. Balanced nucleocytosolic partitioning defines a spatial network to coordinate circadian physiology in plants. Dev. Cell 2013, 26, 73–85. [Google Scholar] [CrossRef]
- Lu, S.X.; Webb, C.J.; Knowles, S.M.; Kim, S.H.; Wang, Z.; Tobin, E.M. CCA1 and ELF3 Interact in the control of hypocotyl length and flowering time in Arabidopsis. Plant Physiol. 2012, 158, 1079–1088. [Google Scholar] [CrossRef]
- Adams, S.; Manfield, I.; Stockley, P.; Carre, I.A. Revised Morning Loops of the Arabidopsis Circadian Clock Based on Analyses of Direct Regulatory Interactions. PLoS ONE 2015, 10, e0143943. [Google Scholar] [CrossRef]
- Kamioka, M.; Takao, S.; Suzuki, T.; Taki, K.; Higashiyama, T.; Kinoshita, T.; Nakamichi, N. Direct Repression of Evening Genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis Circadian Clock. Plant Cell 2016, 28, 696–711. [Google Scholar] [CrossRef]
- Adams, S.; Grundy, J.; Veflingstad, S.R.; Dyer, N.P.; Hannah, M.A.; Ott, S.; Carré, I.A. Circadian control of abscisic acid biosynthesis and signalling pathways revealed by genome-wide analysis of LHY binding targets. New Phytol. 2018, 220, 893–907. [Google Scholar] [CrossRef]
- Lau, O.S.; Huang, X.; Charron, J.B.; Lee, J.H.; Li, G.; Deng, X.W. Interaction of Arabidopsis DET1 with CCA1 and LHY in mediating transcriptional repression in the plant circadian clock. Mol. Cell 2011, 43, 703–712. [Google Scholar] [CrossRef]
- Nakamichi, N.; Kiba, T.; Henriques, R.; Mizuno, T.; Chua, N.H.; Sakakibara, H. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 2010, 22, 594–605. [Google Scholar] [CrossRef]
- Kim, W.Y.; Hicks, K.A.; Somers, D.E. Independent roles for EARLY FLOWERING 3 and ZEITLUPE in the control of circadian timing, hypocotyl length, and flowering time. Plant Physiol. 2005, 139, 1557–1569. [Google Scholar] [CrossRef]
- Wu, J.F.; Wang, Y.; Wu, S.H. Two new clock proteins, LWD1 and LWD2, regulate Arabidopsis photoperiodic flowering. Plant Physiol. 2008, 148, 948–959. [Google Scholar] [CrossRef]
- Ding, Z.; Millar, A.J.; Davis, A.M.; Davis, S.J. TIME FOR COFFEE Encodes a Nuclear Regulator in the Arabidopsis thaliana Circadian Clock. Plant Cell 2007, 19, 1522–1536. [Google Scholar] [CrossRef]
- Shin, J.; Du, S.; Bujdoso, N.; Hu, Y.; Davis, S.J. Overexpression and loss-of-function at TIME FOR COFFEE results in similar phenotypes in diverse growth and physiological responses. J. Plant Biol. 2013, 56, 152–159. [Google Scholar] [CrossRef]
- Yuan, L.; Hu, Y.; Li, S.; Xie, Q.; Xu, X. PRR9 and PRR7 negatively regulate the expression of EC components under warm temperature in roots. Plant Signal Behav. 2021, 16, 1855384. [Google Scholar] [CrossRef]
- Malapeira, J.; Khaitova, L.C.; Mas, P. Ordered changes in histone modifications at the core of the Arabidopsis circadian clock. Proc. Natl. Acad. Sci. USA 2012, 109, 21540–21545. [Google Scholar] [CrossRef]
- Park, H.J.; Baek, D.; Cha, J.Y.; Liao, X.; Kang, S.H.; McClung, C.R.; Lee, S.Y.; Yun, D.J.; Kim, W.Y. HOS15 Interacts with the Histone Deacetylase HDA9 and the Evening Complex to Epigenetically Regulate the Floral Activator GIGANTEA. Plant Cell 2019, 31, 37–51. [Google Scholar] [CrossRef]
- Lee, H.G.; Jeong, Y.Y.; Lee, H.; Seo, P.J. Arabidopsis HISTONE DEACETYLASE 9 Stimulates Hypocotyl Cell Elongation by Repressing GIGANTEA Expression Under Short Day Photoperiod. Front. Plant Sci. 2022, 13, 950378. [Google Scholar] [CrossRef]
- Keene, J.D. Minireview: Global regulation and dynamics of ribonucleic Acid. Endocrinology 2010, 151, 1391–1397. [Google Scholar] [CrossRef]
- Jang, K.; Gil Lee, H.; Jung, S.-J.; Paek, N.-C.; Joon Seo, P. The E3 Ubiquitin Ligase COP1 Regulates Thermosensory Flowering by Triggering GI Degradation in Arabidopsis. Sci. Rep. 2015, 5, 12071. [Google Scholar] [CrossRef]
- Ahn, G.; Park, H.J.; Jeong, S.Y.; Shin, G.I.; Ji, M.G.; Cha, J.Y.; Kim, J.; Kim, M.G.; Yun, D.J.; Kim, W.Y. HOS15 represses flowering by promoting GIGANTEA degradation in response to low temperature in Arabidopsis. Plant Commun. 2023, 4, 100570. [Google Scholar] [CrossRef]
- Kim, W.Y.; Fujiwara, S.; Suh, S.S.; Kim, J.; Kim, Y.; Han, L.; David, K.; Putterill, J.; Nam, H.G.; Somers, D.E. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 2007, 449, 356–360. [Google Scholar] [CrossRef]
- Ji, H.; Pardo, J.M.; Batelli, G.; Van Oosten, M.J.; Bressan, R.A.; Li, X. The Salt Overly Sensitive (SOS) Pathway: Established and Emerging Roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef]
- Cha, J.-Y.; Kim, J.; Jeong, S.Y.; Shin, G.-I.; Ji, M.G.; Hwang, J.-W.; Khaleda, L.; Liao, X.; Ahn, G.; Park, H.J.; et al. The Na+/H+ antiporter SALT OVERLY SENSITIVE 1 regulates salt compensation of circadian rhythms by stabilizing GIGANTEA in Arabidopsis. Proc. Natl. Acad. Sci. USA 2022, 119, e2207275119. [Google Scholar] [CrossRef]
- Park, H.J.; Gamez-Arjona, F.M.; Lindahl, M.; Aman, R.; Villalta, I.; Cha, J.Y.; Carranco, R.; Lim, C.J.; García, E.; Bressan, R.A.; et al. S-acylated and nucleus-localized SALT OVERLY SENSITIVE3/CALCINEURIN B-LIKE4 stabilizes GIGANTEA to regulate Arabidopsis flowering time under salt stress. Plant Cell 2023, 35, 298–317. [Google Scholar] [CrossRef]
- Bader, Z.E.; Bae, M.J.; Ali, A.; Park, J.; Baek, D.; Yun, D.J. GIGANTEA-ENHANCED EM LEVEL complex initiates drought escape response via dual function of ABA synthesis and flowering promotion. Plant Signal Behav. 2023, 18, 2180056. [Google Scholar] [CrossRef]
- Goyal, A.; Szarzynska, B.; Fankhauser, C. Phototropism: At the crossroads of light-signaling pathways. Trends Plant Sci. 2013, 18, 393–401. [Google Scholar] [CrossRef]
- Ballare, C.L.; Pierik, R. The shade-avoidance syndrome: Multiple signals and ecological consequences. Plant Cell Environ. 2017, 40, 2530–2543. [Google Scholar] [CrossRef]
- Lu, T.; Meng, Z.; Zhang, G.; Qi, M.; Sun, Z.; Liu, Y.; Li, T. Sub-high Temperature and High Light Intensity Induced Irreversible Inhibition on Photosynthesis System of Tomato Plant (Solanum lycopersicum L.). Front. Plant Sci. 2017, 8, 365. [Google Scholar] [CrossRef]
- Virsile, A.; Brazaityte, A.; Vastakaite-Kairiene, V.; Miliauskiene, J.; Jankauskiene, J.; Novickovas, A.; Samuolienė, G. Lighting intensity and photoperiod serves tailoring nitrate assimilation indices in red and green baby leaf lettuce. J. Sci. Food Agric. 2019, 99, 6608–6619. [Google Scholar] [CrossRef]
- Yu, X.; Liu, H.; Klejnot, J.; Lin, C. The Cryptochrome Blue Light Receptors. Arab. Book 2010, 8, e0135. [Google Scholar] [CrossRef]
- Suetsugu, N.; Wada, M. Evolution of three LOV blue light receptor families in green plants and photosynthetic stramenopiles: Phototropin, ZTL/FKF1/LKP2 and aureochrome. Plant Cell Physiol. 2013, 54, 8–23. [Google Scholar] [CrossRef]
- Moglich, A.; Yang, X.; Ayers, R.A.; Moffat, K. Structure and function of plant photoreceptors. Annu. Rev. Plant Biol. 2010, 61, 21–47. [Google Scholar] [CrossRef]
- Xu, X.; Paik, I.; Zhu, L.; Huq, E. Illuminating Progress in Phytochrome-Mediated Light Signaling Pathways. Trends Plant Sci. 2015, 20, 641–650. [Google Scholar] [CrossRef]
- Leivar, P.; Monte, E. PIFs: Systems integrators in plant development. Plant Cell 2014, 26, 56–78. [Google Scholar] [CrossRef]
- Nohales, M.A.; Liu, W.; Duffy, T.; Nozue, K.; Sawa, M.; Pruneda-Paz, J.L.; Maloof, J.N.; Jacobsen, S.E.; Kay, S.A. Multi-level Modulation of Light Signaling by GIGANTEA Regulates Both the Output and Pace of the Circadian Clock. Dev. Cell 2019, 49, 840–851.e8. [Google Scholar] [CrossRef]
- Fornara, F.; de Montaigu, A.; Sanchez-Villarreal, A.; Takahashi, Y.; Ver Loren van Themaat, E.; Huettel, B.; Davis, S.J.; Coupland, G. The GI-CDF module of Arabidopsis affects freezing tolerance and growth as well as flowering. Plant J. 2015, 81, 695–706. [Google Scholar] [CrossRef]
- De Montaigu, A.; Giakountis, A.; Rubin, M.; Toth, R.; Cremer, F.; Sokolova, V.; Porri, A.; Reymond, M.; Weinig, C.; Coupland, G. Natural diversity in daily rhythms of gene expression contributes to phenotypic variation. Proc. Natl. Acad. Sci. USA 2015, 112, 905–910. [Google Scholar] [CrossRef]
- Martinez-Vasallo, C.; Cole, B.; Gallego-Bartolome, J.; Chory, J.; Kay, S.A.; Nohales, M.A. Epidermal GIGANTEA adjusts the response to shade at dusk by directly impinging on PHYTOCHROME INTERACTING FACTOR 7 function. bioRxiv 2023. [Google Scholar] [CrossRef]
- Heyes, D.J.; Khara, B.; Sakuma, M.; Hardman, S.J.; O’Cualain, R.; Rigby, S.E.; Scrutton, N.S. Ultrafast red light activation of Synechocystis phytochrome Cph1 triggers major structural change to form the Pfr signalling-competent state. PLoS ONE 2012, 7, e52418. [Google Scholar] [CrossRef]
- Casal, J.J.; Luccioni, L.G.; Oliverio, K.A.; Boccalandro, H.E. Light, phytochrome signalling and photomorphogenesis in Arabidopsis. Photochem. Photobiol. Sci. 2003, 2, 625–636. [Google Scholar] [CrossRef]
- Molas, M.L.; Kiss, J.Z.; Correll, M.J. Gene profiling of the red light signalling pathways in roots. J. Exp. Bot. 2006, 57, 3217–3229. [Google Scholar] [CrossRef]
- Singh, A. GIGANTEA regulates lateral root formation by modulating auxin signaling in Arabidopsis thaliana. Plant Signal Behav. 2022, 17, 2096780. [Google Scholar] [CrossRef]
- Wang, H.; Deng, X.W. Phytochrome signaling mechanism. Arab. Book 2004, 3, e0074.1. [Google Scholar] [CrossRef]
- Mas, P.; Kim, W.Y.; Somers, D.E.; Kay, S.A. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 2003, 426, 567–570. [Google Scholar] [CrossRef]
- Cha, J.; Kim, J.; Kim, T.S.; Zeng, Q.; Wang, L.; Lee, S.Y.; Kim, W.Y.; Somers, D.E. GIGANTEA is a co-chaperone which facilitates maturation of ZEITLUPE in the Arabidopsis circadian clock. Nat. Commun. 2017, 8, 3. [Google Scholar] [CrossRef]
- Kiba, T.; Henriques, R.; Sakakibara, H.; Chua, N.-H. Targeted Degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL Complex Regulates Clock Function and Photomorphogenesis in Arabidopsis thaliana. Plant Cell 2007, 19, 2516–2530. [Google Scholar] [CrossRef]
- Martin-Tryon, E.L.; Kreps, J.A.; Harmer, S.L. GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiol. 2007, 143, 473–486. [Google Scholar] [CrossRef]
- Kwon, E.; Pathak, D.; Dahal, P.; Tandukar, S.; Jung, H.S.; Kim, W.Y.; Kim, D.Y. Structural analysis of the regulation of blue-light receptors by GIGANTEA. Cell Rep. 2022, 39, 110700. [Google Scholar] [CrossRef]
- Venkat, A.; Muneer, S. Role of Circadian Rhythms in Major Plant Metabolic and Signaling Pathways. Front. Plant Sci. 2022, 13, 836244. [Google Scholar] [CrossRef]
- Kielbowicz-Matuk, A.; Rey, P.; Rorat, T. Interplay between circadian rhythm, time of the day and osmotic stress constraints in the regulation of the expression of a Solanum Double B-box gene. Ann. Bot. 2014, 113, 831–842. [Google Scholar] [CrossRef]
- Inoue, K.; Araki, T.; Endo, M. Integration of input signals into the gene network in the plant circadian clock. Plant Cell Physiol. 2017, 58, 977–982. [Google Scholar] [CrossRef]
- Yan, J.; Kim, Y.J.; Somers, D.E. Post-Translational Mechanisms of Plant Circadian Regulation. Genes 2021, 12, 325. [Google Scholar] [CrossRef]
- Tataroglu, O.; Emery, P. The molecular ticks of the Drosophila circadian clock. Curr. Opin. Insect Sci. 2015, 7, 51–57. [Google Scholar] [CrossRef]
- Pokhilko, A.; Fernandez, A.P.; Edwards, K.D.; Southern, M.M.; Halliday, K.J.; Millar, A.J. The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol. Syst. Biol. 2012, 8, 574. [Google Scholar] [CrossRef]
- Staiger, D.; Shin, J.; Johansson, M.; Davis, S.J. The circadian clock goes genomic. Genome Biol. 2013, 14, 208. [Google Scholar] [CrossRef]
- Gray, J.A.; Shalit-Kaneh, A.; Chu, D.N.; Hsu, P.Y.; Harmer, S.L. The REVEILLE Clock Genes Inhibit Growth of Juvenile and Adult Plants by Control of Cell Size. Plant Physiol. 2017, 173, 2308–2322. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, J.F.; Nakamichi, N.; Sakakibara, H.; Nam, H.G.; Wu, S.H. LIGHT-REGULATED WD1 and PSEUDO-RESPONSE REGULATOR9 form a positive feedback regulatory loop in the Arabidopsis circadian clock. Plant Cell 2011, 23, 486–498. [Google Scholar] [CrossRef]
- Green, R.M.; Tobin, E.M. The role of CCA1 and LHY in the plant circadian clock. Dev. Cell 2002, 2, 516–518. [Google Scholar] [CrossRef]
- McClung, C.R. The Plant Circadian Oscillator. Biology 2019, 8, 14. [Google Scholar] [CrossRef]
- McClung, C.R.; Gutierrez, R.A. Network news: Prime time for systems biology of the plant circadian clock. Curr. Opin. Genet. Dev. 2010, 20, 588–598. [Google Scholar] [CrossRef]
- Nagel, D.H.; Doherty, C.J.; Pruneda-Paz, J.L.; Schmitz, R.J.; Ecker, J.R.; Kay, S.A. Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc. Natl. Acad. Sci. USA 2015, 112, E4802–E4810. [Google Scholar] [CrossRef]
- Harmer, S.L.; Kay, S.A. Positive and negative factors confer phase-specific circadian regulation of transcription in Arabidopsis. Plant Cell 2005, 17, 1926–1940. [Google Scholar] [CrossRef]
- Hayama, R.; Izawa, T.; Shimamoto, K. Isolation of rice genes possibly involved in the photoperiodic control of flowering by a fluorescent differential display method. Plant Cell Physiol. 2002, 43, 494–504. [Google Scholar] [CrossRef]
- Valverde, F. CONSTANS and the evolutionary origin of photoperiodic timing of flowering. J. Exp. Bot. 2011, 62, 2453–2463. [Google Scholar] [CrossRef]
- Valverde, F.; Mouradov, A.; Soppe, W.; Ravenscroft, D.; Samach, A.; Coupland, G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 2004, 303, 1003–1006. [Google Scholar] [CrossRef]
- Song, Y.H.; Smith, R.W.; To, B.J.; Millar, A.J.; Imaizumi, T. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 2012, 336, 1045–1049. [Google Scholar] [CrossRef]
- Suarez-Lopez, P.; Wheatley, K.; Robson, F.; Onouchi, H.; Valverde, F.; Coupland, G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 2001, 410, 1116–1120. [Google Scholar] [CrossRef]
- Jung, J.H.; Seo, Y.H.; Seo, P.J.; Reyes, J.L.; Yun, J.; Chua, N.H.; Park, C.M. The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 2007, 19, 2736–2748. [Google Scholar] [CrossRef]
- Aukerman, M.J.; Sakai, H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 2003, 15, 2730–2741. [Google Scholar] [CrossRef]
- Tseng, T.S.; Salome, P.A.; McClung, C.R.; Olszewski, N.E. SPINDLY and GIGANTEA interact and act in Arabidopsis thaliana pathways involved in light responses, flowering, and rhythms in cotyledon movements. Plant Cell 2004, 16, 1550–1563. [Google Scholar] [CrossRef]
- Huang, T.; Böhlenius, H.; Eriksson, S.; Parcy, F.; Nilsson, O. The mRNA of the Arabidopsis Gene FT Moves from Leaf to Shoot Apex and Induces Flowering. Science 2005, 309, 1694–1696. [Google Scholar] [CrossRef]
- Corbesier, L.; Vincent, C.; Jang, S.; Fornara, F.; Fan, Q.; Searle, I.; Giakountis, A.; Farrona, S.; Gissot, L.; Turnbull, C.; et al. FT Protein Movement Contributes to Long-Distance Signaling in Floral Induction of Arabidopsis. Science 2007, 316, 1030–1033. [Google Scholar] [CrossRef]
- Khoso, M.A.; Hussain, A.; Ritonga, F.N.; Ali, Q.; Channa, M.M.; Alshegaihi, R.M.; Meng, Q.; Ali, M.; Zaman, W.; Brohi, R.D.; et al. WRKY transcription factors (TFs): Molecular switches to regulate drought, temperature, and salinity stresses in plants. Front. Plant Sci. 2022, 13, 1039329. [Google Scholar] [CrossRef]
- Ivey, C.T.; Carr, D.E. Tests for the joint evolution of mating system and drought escape in Mimulus. Ann. Bot. 2012, 109, 583–598. [Google Scholar] [CrossRef]
- Franks, S.J. Plasticity and evolution in drought avoidance and escape in the annual plant Brassica rapa. New Phytol. 2011, 190, 249–257. [Google Scholar] [CrossRef]
- Riboni, M.; Galbiati, M.; Tonelli, C.; Conti, L. GIGANTEA enables drought escape response via abscisic acid-dependent activation of the florigens and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS. Plant Physiol. 2013, 162, 1706–1719. [Google Scholar] [CrossRef]
- Han, Y.; Zhang, X.; Wang, W.; Wang, Y.; Ming, F. Correction: The suppression of WRKY44 by GIGANTEA-miR172 pathway is involved in drought response of Arabidopsis thaliana. PLoS ONE 2015, 10, e0124854. [Google Scholar] [CrossRef]
- Fahad, S.; Bajwa, A.A.; Nazir, U.; Anjum, S.A.; Farooq, A.; Zohaib, A.; Sadia, S.; Nasim, W.; Adkins, S.; Saud, S.; et al. Crop Production under Drought and Heat Stress: Plant Responses and Management Options. Front. Plant Sci. 2017, 8, 1147. [Google Scholar] [CrossRef]
- Grover, A.; Mittal, D.; Negi, M.; Lavania, D. Generating high temperature tolerant transgenic plants: Achievements and challenges. Plant Sci. 2013, 205–206, 38–47. [Google Scholar] [CrossRef]
- Sanghera, G.S.; Wani, S.H.; Hussain, W.; Singh, N.B. Engineering cold stress tolerance in crop plants. Curr. Genom. 2011, 12, 30–43. [Google Scholar] [CrossRef]
- Fowler, S.; Thomashow, M.F. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 2002, 14, 1675–1690. [Google Scholar] [CrossRef]
- Cao, S.; Ye, M.; Jiang, S. Involvement of GIGANTEA gene in the regulation of the cold stress response in Arabidopsis. Plant Cell Rep. 2005, 24, 683–690. [Google Scholar] [CrossRef]
- Mizuno, T.; Nomoto, Y.; Oka, H.; Kitayama, M.; Takeuchi, A.; Tsubouchi, M.; Yamashino, T. Ambient Temperature Signal Feeds into the Circadian Clock Transcriptional Circuitry Through the EC Night-Time Repressor in Arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 958–976. [Google Scholar] [CrossRef]
- Park, Y.-J.; Kim, J.Y.; Lee, J.-H.; Lee, B.-D.; Paek, N.-C.; Park, C.-M. GIGANTEA Shapes the Photoperiodic Rhythms of Thermomorphogenic Growth in Arabidopsis. Mol. Plant 2020, 13, 459–470. [Google Scholar] [CrossRef]
- Zhou, H.; Lin, H.; Chen, S.; Becker, K.; Yang, Y.; Zhao, J.; Kudla, J.; Schumaker, K.S.; Guo, Y. Inhibition of the Arabidopsis salt overly sensitive pathway by 14-3-3 proteins. Plant Cell 2014, 26, 1166–1182. [Google Scholar] [CrossRef]
- Qiu, Q.S.; Guo, Y.; Quintero, F.J.; Pardo, J.M.; Schumaker, K.S.; Zhu, J.K. Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the salt-overly-sensitive (SOS) pathway. J. Biol. Chem. 2004, 279, 207–215. [Google Scholar] [CrossRef]
- Ali, A.; Petrov, V.; Yun, D.J.; Gechev, T. Revisiting plant salt tolerance: Novel components of the SOS pathway. Trends Plant Sci. 2023, 28, 1060–1069. [Google Scholar] [CrossRef]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Almeida, D.M.; Oliveira, M.M.; Saibo, N.J.M. Regulation of Na+ and K+ homeostasis in plants: Towards improved salt stress tolerance in crop plants. Genet. Mol. Biol. 2017, 40 (Suppl. S1), 326–345. [Google Scholar] [CrossRef]
- Ke, Q.; Kim, H.S.; Wang, Z.; Ji, C.Y.; Jeong, J.C.; Lee, H.S.; Choi, Y.I.; Xu, B.; Deng, X.; Yun, D.J.; et al. Down-regulation of GIGANTEA-like genes increases plant growth and salt stress tolerance in poplar. Plant Biotechnol. J. 2017, 15, 331–343. [Google Scholar] [CrossRef]
- Cao, S.; Jiang, S.; Zhang, R. The Role of GIGANTEA Gene in Mediating the Oxidative Stress Response and in Arabidopsis. Plant Growth Regul. 2006, 48, 261–270. [Google Scholar] [CrossRef]
- Cha, J.Y.; Lee, D.Y.; Ali, I.; Jeong, S.Y.; Shin, B.; Ji, H.; Kim, J.S.; Kim, M.G.; Kim, W.Y. Arabidopsis GIGANTEA negatively regulates chloroplast biogenesis and resistance to herbicide butafenacil. Plant Cell Rep. 2019, 38, 793–801. [Google Scholar] [CrossRef] [PubMed]
- Ohmiya, A.; Hirashima, M.; Yagi, M.; Tanase, K.; Yamamizo, C. Identification of genes associated with chlorophyll accumulation in flower petals. PLoS ONE 2014, 9, e113738. [Google Scholar] [CrossRef] [PubMed]
- Cortleven, A.; Schmulling, T. Regulation of chloroplast development and function by cytokinin. J. Exp. Bot. 2015, 66, 4999–5013. [Google Scholar] [CrossRef] [PubMed]
- Kurepa, J.; Smalle, J.; Van Montagu, M.; Inze, D. Oxidative stress tolerance and longevity in Arabidopsis: The late-flowering mutant gigantea is tolerant to paraquat. Plant J. 1998, 14, 759–764. [Google Scholar] [CrossRef]
- Shimazaki, K.; Doi, M.; Assmann, S.M.; Kinoshita, T. Light regulation of stomatal movement. Annu. Rev. Plant Biol. 2007, 58, 219–247. [Google Scholar] [CrossRef] [PubMed]
- Jezek, M.; Blatt, M.R. The Membrane Transport System of the Guard Cell and Its Integration for Stomatal Dynamics. Plant Physiol. 2017, 174, 487–519. [Google Scholar] [CrossRef]
- Inoue, S.I.; Kinoshita, T. Blue Light Regulation of Stomatal Opening and the Plasma Membrane H+-ATPase. Plant Physiol. 2017, 174, 531–538. [Google Scholar] [CrossRef]
- Kinoshita, T.; Doi, M.; Suetsugu, N.; Kagawa, T.; Wada, M.; Shimazaki, K. Phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 2001, 414, 656–660. [Google Scholar] [CrossRef]
- Kinoshita, T.; Ono, N.; Hayashi, Y.; Morimoto, S.; Nakamura, S.; Soda, M.; Kato, Y.; Ohnishi, M.; Nakano, T.; Inoue, S.; et al. FLOWERING LOCUS T regulates stomatal opening. Curr. Biol. 2011, 21, 1232–1238. [Google Scholar] [CrossRef]
- Kimura, Y.; Aoki, S.; Ando, E.; Kitatsuji, A.; Watanabe, A.; Ohnishi, M.; Takahashi, K.; Inoue, S.; Nakamichi, N.; Tamada, Y.; et al. A flowering integrator, SOC1, affects stomatal opening in Arabidopsis thaliana. Plant Cell Physiol. 2015, 56, 640–649. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.W.; Wen, X.H.; Fu, J.X.; Dai, S.L. ClCRY2 facilitates floral transition in Chrysanthemum lavandulifolium by affecting the transcription of circadian clock-related genes under short-day photoperiods. Hortic. Res. 2018, 5, 58. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.J.; Zhang, Y.C.; Li, Q.H.; Sang, Y.; Mao, J.; Lian, H.L.; Wang, L.; Yang, H.Q. COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 2008, 20, 292–306. [Google Scholar] [CrossRef] [PubMed]
- Ando, E.; Ohnishi, M.; Wang, Y.; Matsushita, T.; Watanabe, A.; Hayashi, Y.; Fujii, M.; Ma, J.F.; Inoue, S.; Kinoshita, T. TWIN SISTER OF FT, GIGANTEA, and CONSTANS have a positive but indirect effect on blue light-induced stomatal opening in Arabidopsis. Plant Physiol. 2013, 162, 1529–1538. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.M.; Lo, S.F.; Ho, T.D. Source-Sink Communication: Regulated by Hormone, Nutrient, and Stress Cross-Signaling. Trends Plant Sci. 2015, 20, 844–857. [Google Scholar] [CrossRef]
- Bolouri Moghaddam, M.R.; Van den Ende, W. Sugars, the clock and transition to flowering. Front. Plant Sci. 2013, 4, 22. [Google Scholar]
- Rolland, F.; Baena-Gonzalez, E.; Sheen, J. Sugar sensing and signaling in plants: Conserved and novel mechanisms. Annu. Rev. Plant Biol. 2006, 57, 675–709. [Google Scholar] [CrossRef]
- Bolouri Moghaddam, M.R.; Van den Ende, W. Sugars and plant innate immunity. J. Exp. Bot. 2012, 63, 3989–3998. [Google Scholar] [CrossRef]
- Shin, J.; Sanchez-Villarreal, A.; Davis, A.M.; Du, S.X.; Berendzen, K.W.; Koncz, C.; Ding, Z.; Li, C.; Davis, S.J. The metabolic sensor AKIN10 modulates the Arabidopsis circadian clock in a light-dependent manner. Plant Cell Environ. 2017, 40, 997–1008. [Google Scholar] [CrossRef]
- Frank, A.; Matiolli, C.C.; Viana, A.J.C.; Hearn, T.J.; Kusakina, J.; Belbin, F.E.; Wells Newman, D.; Yochikawa, A.; Cano-Ramirez, D.L.; Chembath, A.; et al. Circadian Entrainment in Arabidopsis by the Sugar-Responsive Transcription Factor bZIP63. Curr. Biol. 2018, 28, 2597–2606.e6. [Google Scholar] [CrossRef]
- Haydon, M.J.; Mielczarek, O.; Frank, A.; Roman, A.; Webb, A.A.R. Sucrose and Ethylene Signaling Interact to Modulate the Circadian Clock. Plant Physiol. 2017, 175, 947–958. [Google Scholar] [CrossRef] [PubMed]
- Dalchau, N.; Baek, S.J.; Briggs, H.M.; Robertson, F.C.; Dodd, A.N.; Gardner, M.J.; Stancombe, M.A.; Haydon, M.J.; Stan, G.B.; Gonçalves, J.M.; et al. The circadian oscillator gene GIGANTEA mediates a long-term response of the Arabidopsis thaliana circadian clock to sucrose. Proc. Natl. Acad. Sci. USA 2011, 108, 5104–5109. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.Q.; Song, Y.Q.; Su, L. Freezing sensitivity in the gigantea mutant of Arabidopsis is associated with sugar deficiency. Biol. Plant 2007, 51, 359–362. [Google Scholar] [CrossRef]
- Eimert, K.; Wang, S.M.; Lue, W.I.; Chen, J. Monogenic Recessive Mutations Causing Both Late Floral Initiation and Excess Starch Accumulation in Arabidopsis. Plant Cell 1995, 7, 1703–1712. [Google Scholar] [CrossRef] [PubMed]
- Krahmer, J.; Goralogia, G.S.; Kubota, A.; Zardilis, A.; Johnson, R.S.; Song, Y.H.; MacCoss, M.J.; Le Bihan, T.; Halliday, K.J.; Imaizumi, T.; et al. Time-resolved interaction proteomics of the GIGANTEA protein under diurnal cycles in Arabidopsis. FEBS Lett. 2019, 593, 319–338. [Google Scholar] [CrossRef]
- Chen, Z.H.; Soltis, D.E. Evolution of environmental stress responses in plants. Plant Cell Environ. 2020, 43, 2827–2831. [Google Scholar] [CrossRef]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef]
- Odgerel, K.; Jose, J.; Karsai-Rektenwald, F.; Ficzek, G.; Simon, G.; Vegvari, G.; Bánfalvi, Z. Effects of the repression of GIGANTEA gene StGI.04 on the potato leaf transcriptome and the anthocyanin content of tuber skin. BMC Plant Biol. 2022, 22, 249. [Google Scholar] [CrossRef]
- Tal, L.; Gil, M.X.A.; Guercio, A.M.; Shabek, N. Structural Aspects of Plant Hormone Signal Perception and Regulation by Ubiquitin Ligases. Plant Physiol. 2020, 182, 1537–1544. [Google Scholar] [CrossRef]
- Singh, M.; Mas, P. A Functional Connection between the Circadian Clock and Hormonal Timing in Arabidopsis. Genes 2018, 9, 567. [Google Scholar] [CrossRef]
- Hanano, S.; Domagalska, M.A.; Nagy, F.; Davis, S.J. Multiple phytohormones influence distinct parameters of the plant circadian clock. Genes Cells 2006, 11, 1381–1392. [Google Scholar] [CrossRef]
- Siemiatkowska, B.; Chiara, M.; Badiger, B.G.; Riboni, M.; D’Avila, F.; Braga, D.; Salem, M.A.A.; Martignago, D.; Colanero, S.; Galbiati, M.; et al. GIGANTEA Is a Negative Regulator of Abscisic Acid Transcriptional Responses and Sensitivity in Arabidopsis. Plant Cell Physiol. 2022, 63, 1285–1297. [Google Scholar] [CrossRef] [PubMed]
- Riboni, M.; Robustelli Test, A.; Galbiati, M.; Tonelli, C.; Conti, L. ABA-dependent control of GIGANTEA signalling enables drought escape via up-regulation of FLOWERING LOCUS T in Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 6309–6322. [Google Scholar] [CrossRef]
- Arana, M.V.; Marín-de la Rosa, N.; Maloof, J.N.; Blázquez, M.A.; Alabadí, D. Circadian oscillation of gibberellin signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 9292–9297. [Google Scholar] [CrossRef] [PubMed]
- Nohales, M.A.; Kay, S.A. GIGANTEA gates gibberellin signaling through stabilization of the DELLA proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 21893–21899. [Google Scholar] [CrossRef] [PubMed]
- Zentella, R.; Sui, N.; Barnhill, B.; Hsieh, W.-P.; Hu, J.; Shabanowitz, J.; Boyce, M.; Olszewski, N.E.; Zhou, P.; Hunt, D.F.; et al. The Arabidopsis O-fucosyltransferase SPINDLY activates nuclear growth repressor DELLA. Nat. Chem. Biol. 2017, 13, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Hedden, P.; Thomas, S.G. Gibberellin biosynthesis and its regulation. Biochem. J. 2012, 444, 11–25. [Google Scholar] [CrossRef]
- Oh, E.; Yamaguchi, S.; Hu, J.; Yusuke, J.; Jung, B.; Paik, I.; Lee, H.S.; Sun, T.P.; Kamiya, Y.; Choi, G. PIL5, a Phytochrome-Interacting bHLH Protein, Regulates Gibberellin Responsiveness by Binding Directly to theGAIandRGAPromoters inArabidopsisSeeds. Plant Cell 2007, 19, 1192–1208. [Google Scholar] [CrossRef]
- Kundu, P.; Sahu, R. GIGANTEA confers susceptibility to plants during spot blotch attack by regulating salicylic acid signalling pathway. Plant Physiol. Biochem. 2021, 167, 349–357. [Google Scholar] [CrossRef]
- Lyons, R.R.A.; Stiller, J.; Powell, J.; Manners, J.M.; Kazan, K. Investigating the Association between Flowering Time and Defense in the Arabidopsis thaliana-Fusarium oxysporum Interaction. PLoS ONE 2015, 10, e0127699. [Google Scholar] [CrossRef]
- Li, Z.; He, Y. Roles of Brassinosteroids in Plant Reproduction. Int. J. Mol. Sci. 2020, 21, 872. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.; Park, J.; Lee, B.; Cheong, H. Loss of Function in GIGANTEA Gene is Involved in Brassinosteroid Signaling. J. Chosun Nat. Sci. 2011, 4, 113–120. [Google Scholar]
- Park, S.-H.; Jeong, J.S.; Zhou, Y.; Binte Mustafa, N.F.; Chua, N.-H. Deubiquitination of BES1 by UBP12/UBP13 promotes brassinosteroid signaling and plant growth. Plant Commun. 2022, 3, 100348. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.M.; Li, M.W.; Feke, A.; Liu, W.; Saffer, A.M.; Gendron, J.M. GIGANTEA recruits the UBP12 and UBP13 deubiquitylases to regulate accumulation of the ZTL photoreceptor complex. Nat. Commun. 2019, 10, 3750. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Xie, Y.; Yahaya, B.S.; Wu, F. GIGANTEA Unveiled: Exploring Its Diverse Roles and Mechanisms. Genes 2024, 15, 94. https://doi.org/10.3390/genes15010094
Liu L, Xie Y, Yahaya BS, Wu F. GIGANTEA Unveiled: Exploring Its Diverse Roles and Mechanisms. Genes. 2024; 15(1):94. https://doi.org/10.3390/genes15010094
Chicago/Turabian StyleLiu, Ling, Yuxin Xie, Baba Salifu Yahaya, and Fengkai Wu. 2024. "GIGANTEA Unveiled: Exploring Its Diverse Roles and Mechanisms" Genes 15, no. 1: 94. https://doi.org/10.3390/genes15010094
APA StyleLiu, L., Xie, Y., Yahaya, B. S., & Wu, F. (2024). GIGANTEA Unveiled: Exploring Its Diverse Roles and Mechanisms. Genes, 15(1), 94. https://doi.org/10.3390/genes15010094





