YELLOW LEAF AND DWARF 7, Encoding a Novel Ankyrin Domain-Containing Protein, Affects Chloroplast Development in Rice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Paraffin Sectioning and Microscopic Analysis
2.3. Measurement of Pigment Content and Photosynthetic Rate
2.4. Transmission Electron Microscopy (TEM)
2.5. Histochemical Staining
2.6. Map-Based Cloning
2.7. Vector Construction
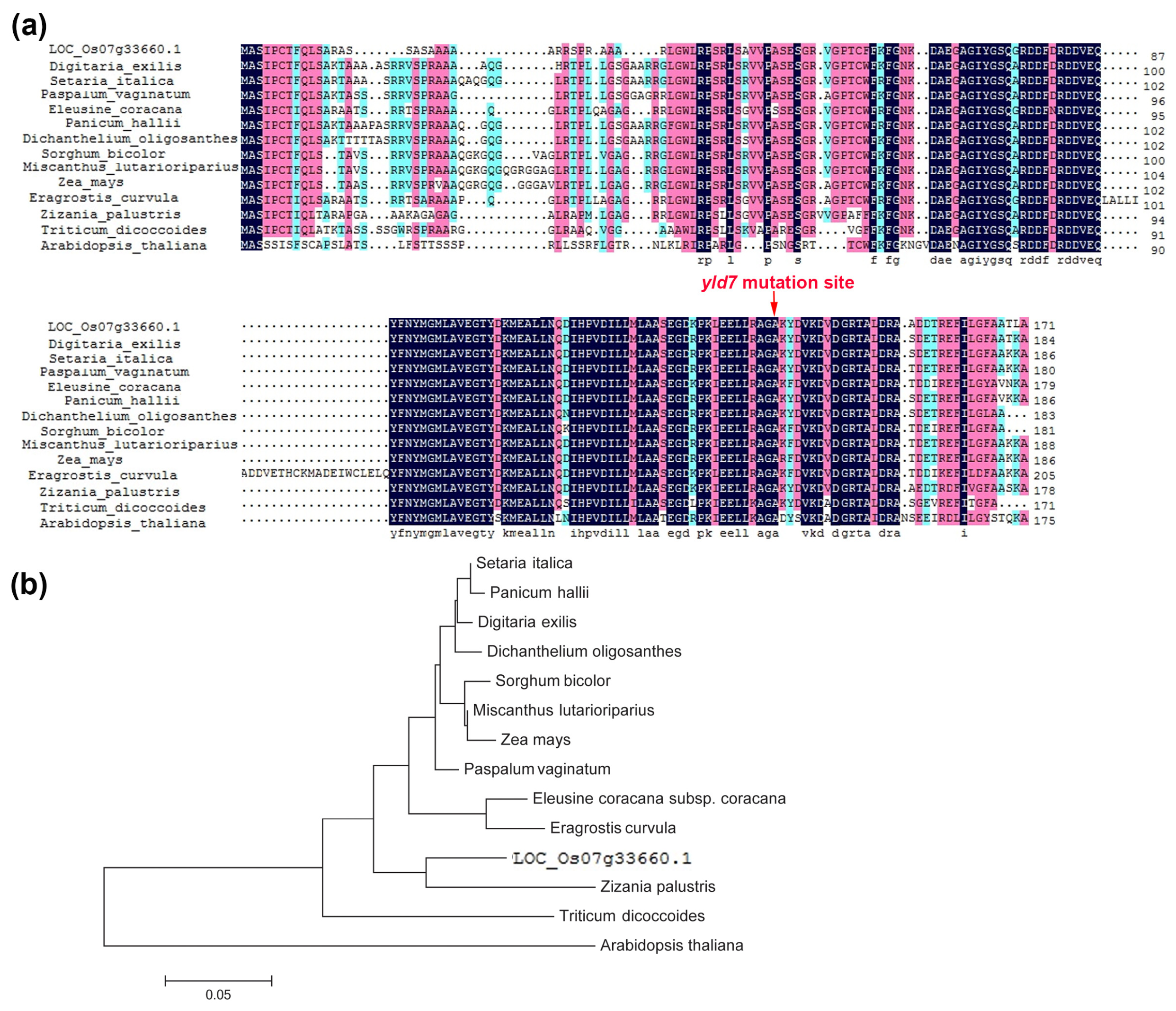
2.8. Phylogenetic Analysis
2.9. Quantitative Real-Time PCR
2.10. Immunoblot Analysis
2.11. RNA-Seq Analysis
2.12. Statistical Analysis
3. Results
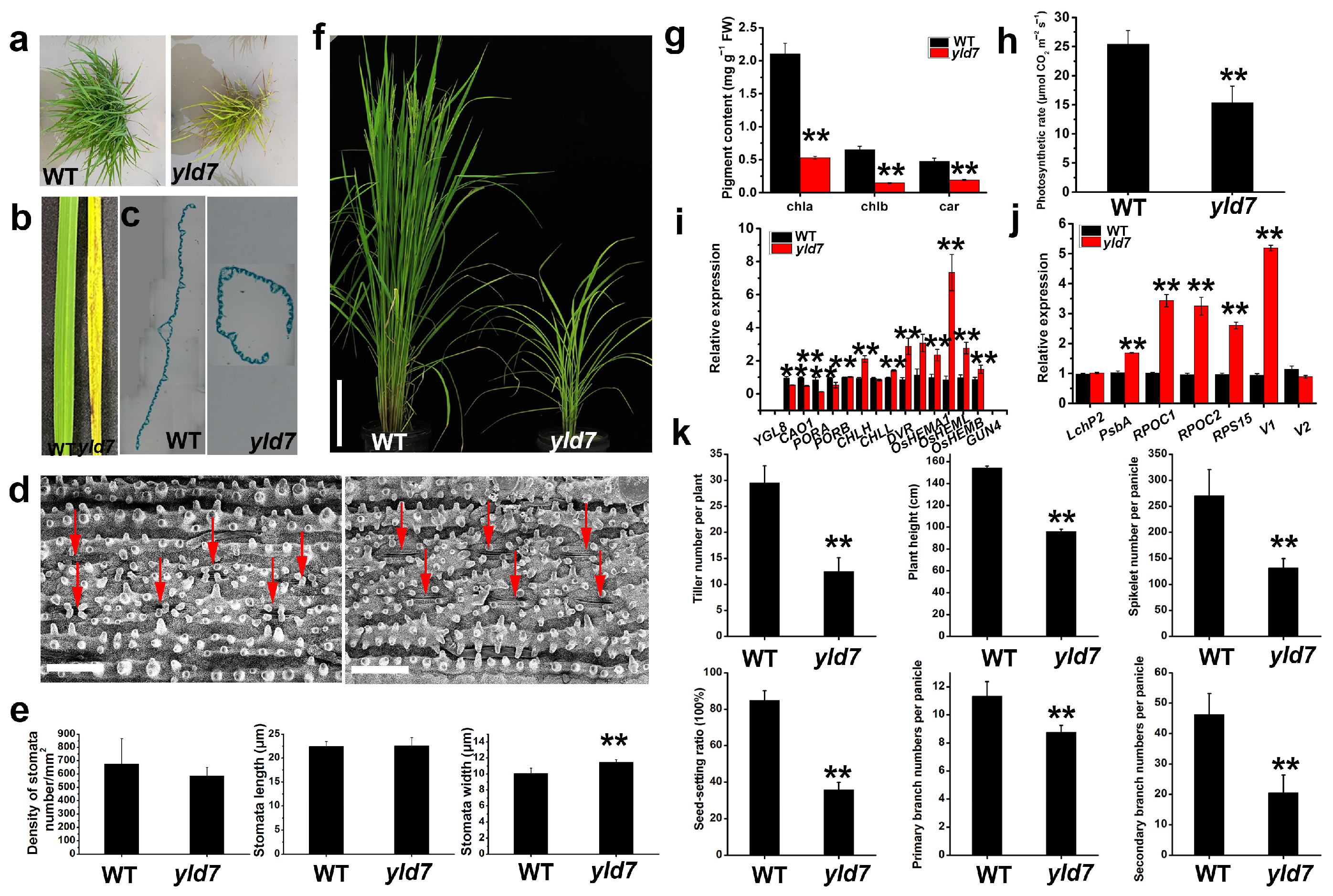
3.1. Phenotypic Analysis Reveals Yellowing and Dwarfism in yld7 Mutants
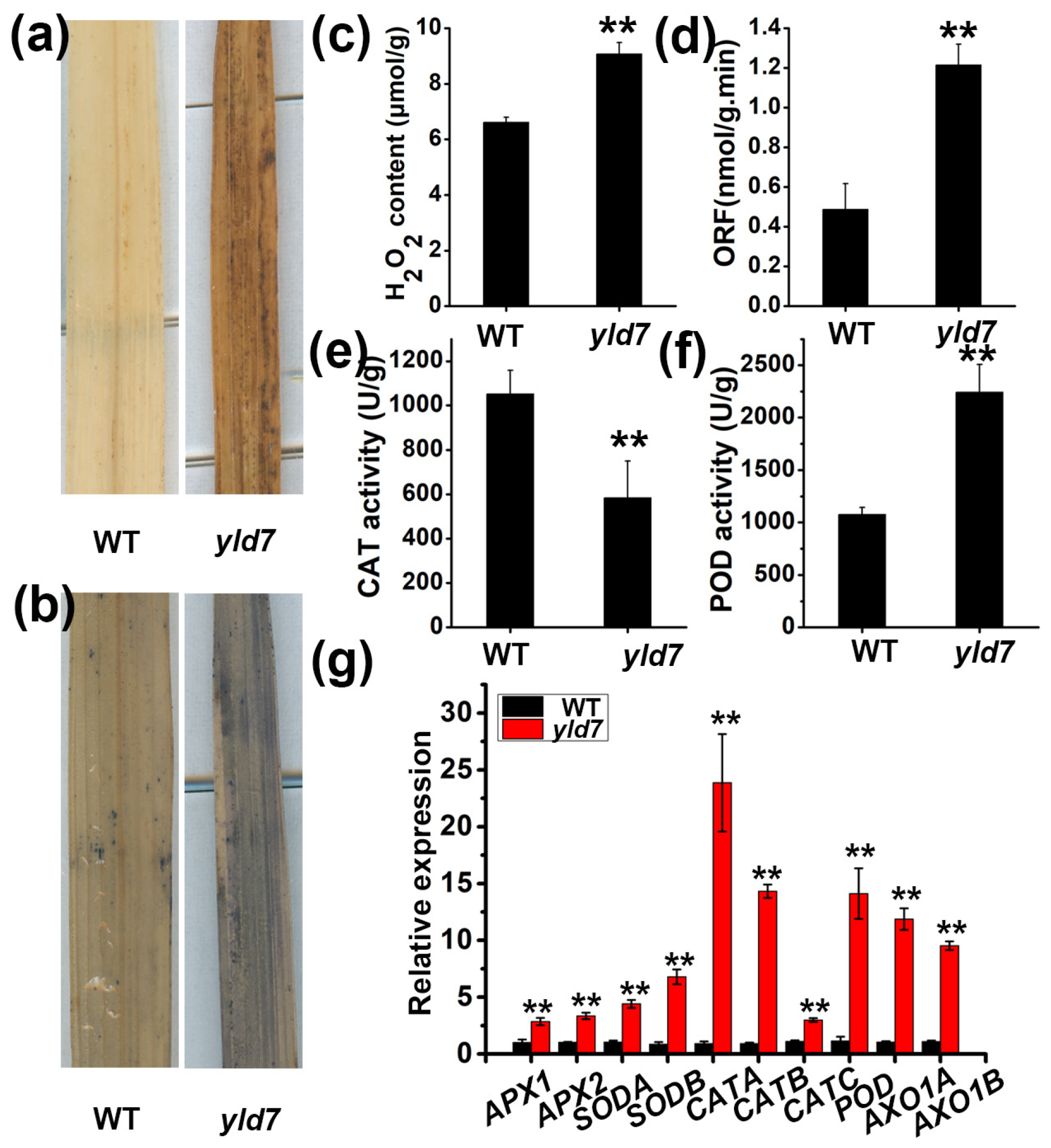
3.2. Premature Cell Death in yld7 Plants
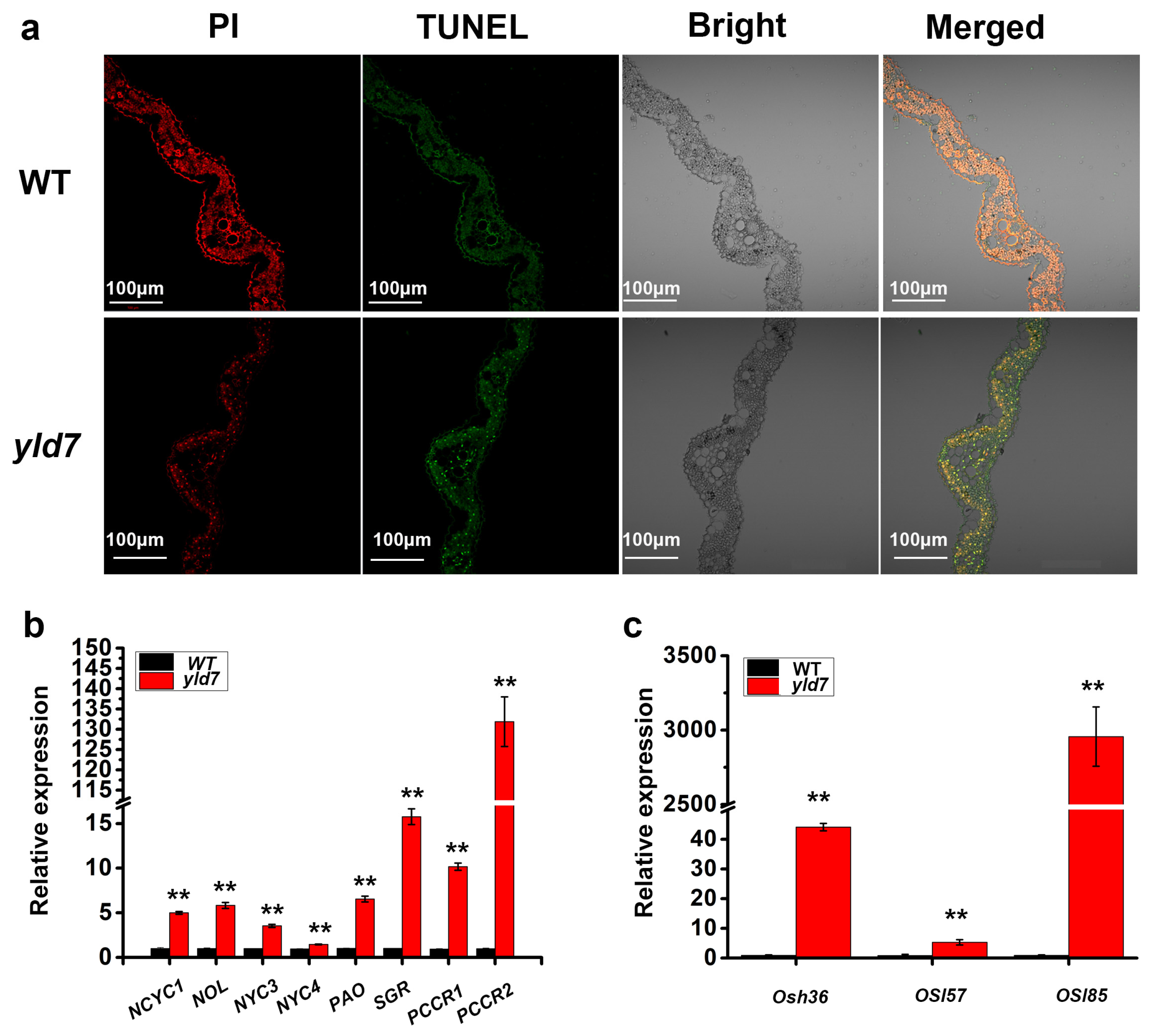
3.3. TUNEL Assay for Cell Death in yld7 Plants
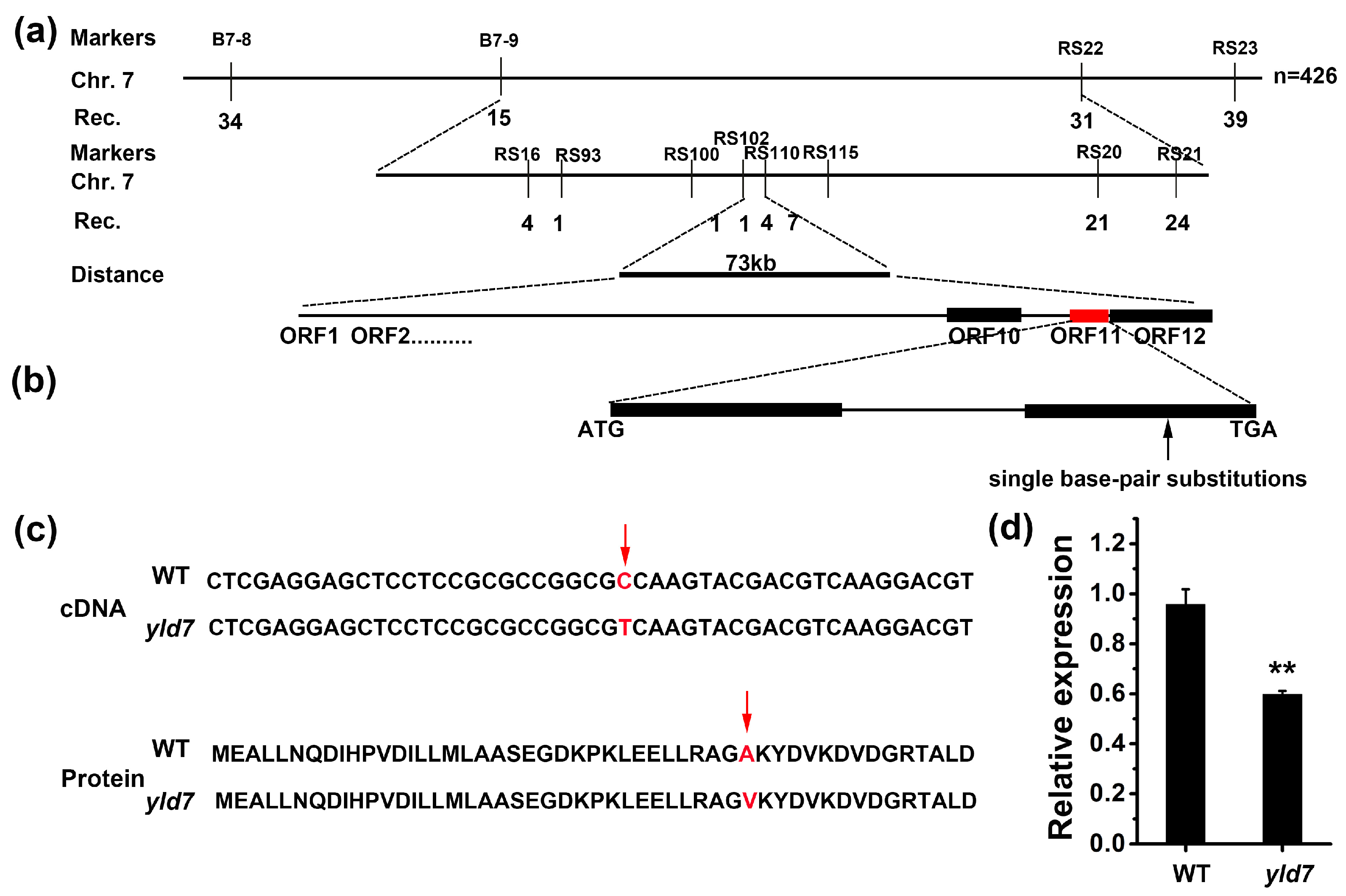
3.4. Map-Based Cloning and Candidate Analysis of the YLD7 Gene
3.5. YLD7 Encodes an Ank-Repeat Containing Protein
3.6. Subcellular Localization of YLD7 Protein
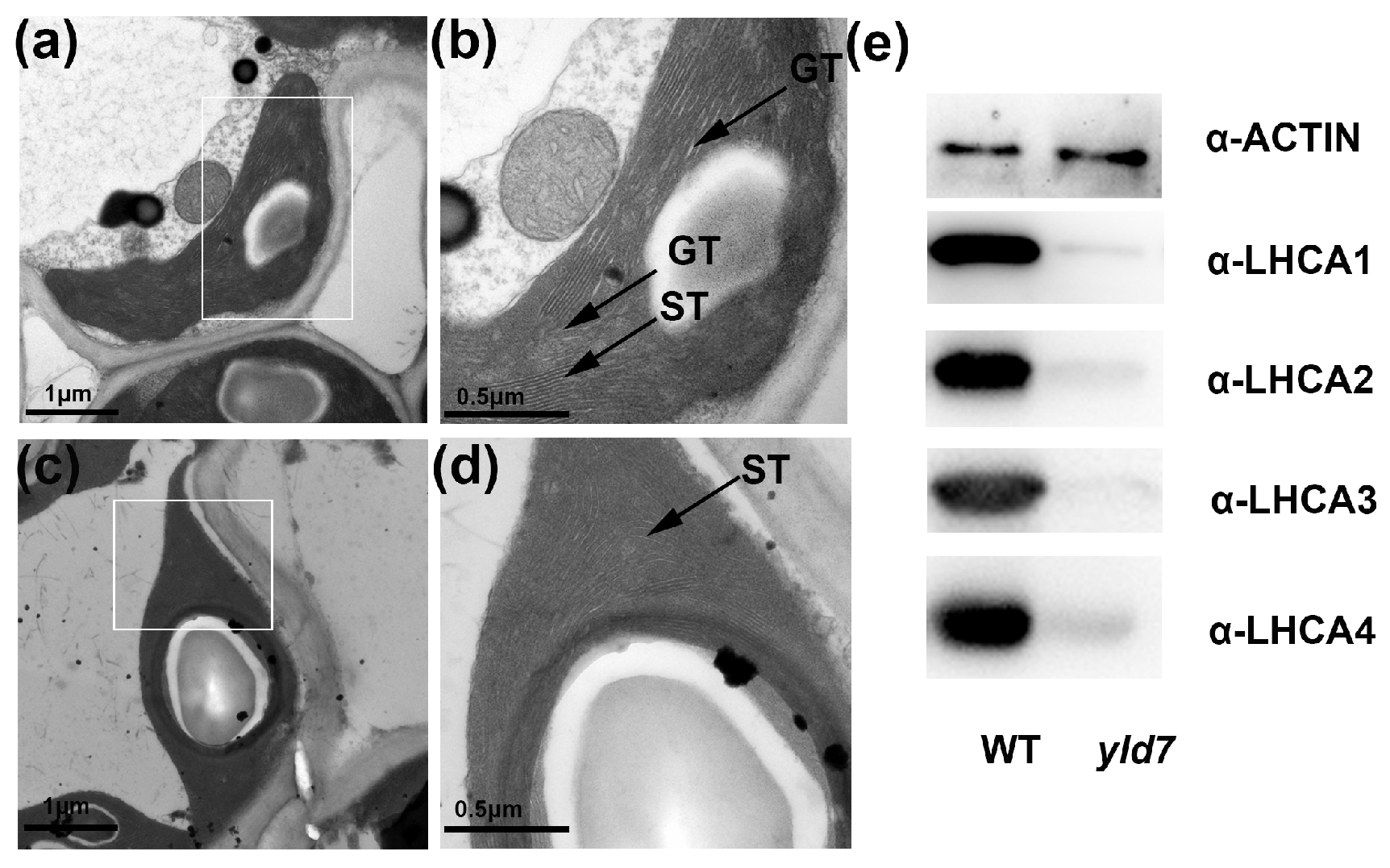
3.7. YLD7 Protein Is Essential for the Formation of Grana and Stroma
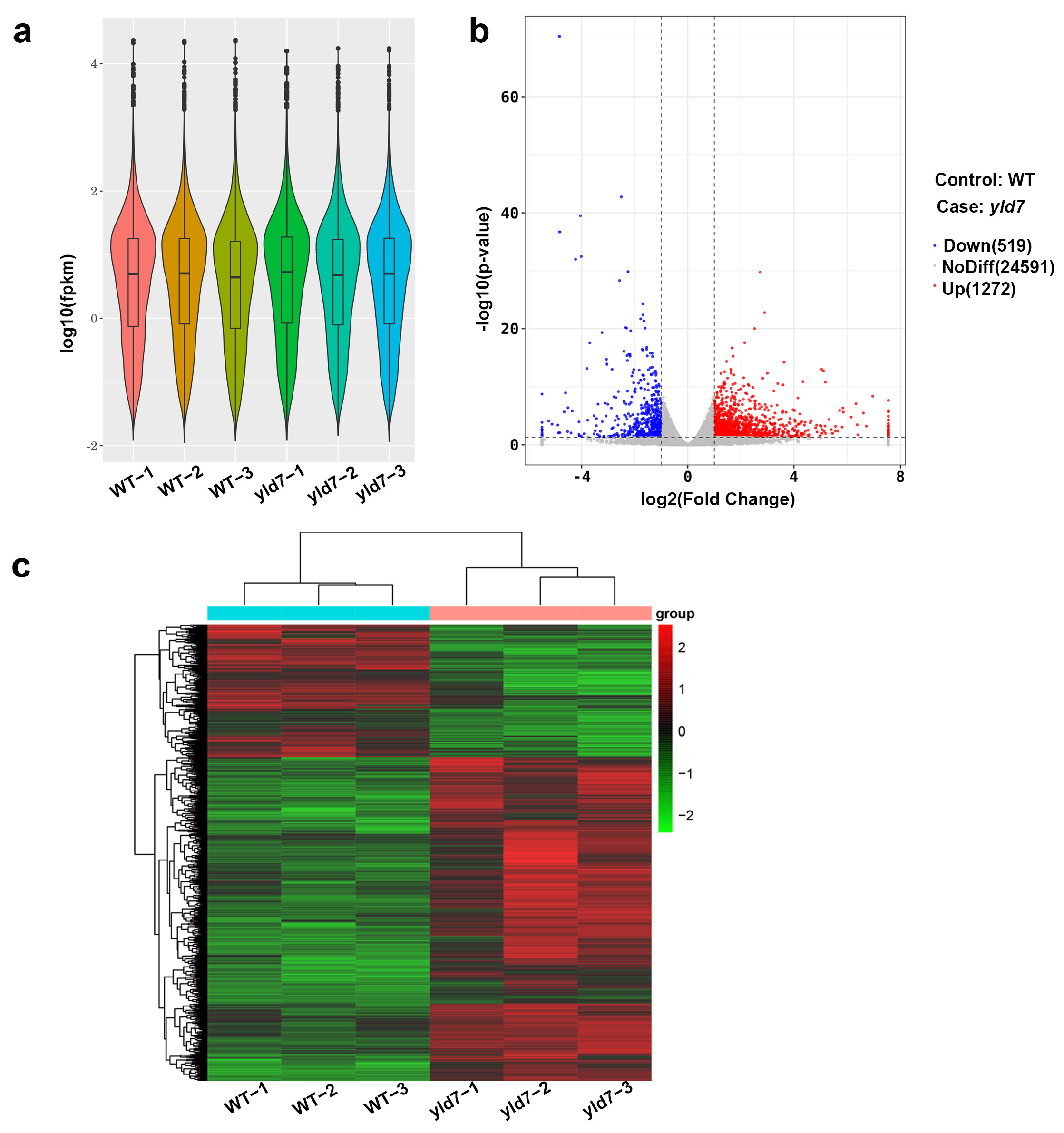
3.8. RNA-Seq Analysis Reveals Altered Gene Expression in yld7 Mutants
3.9. Haplotype Analysis of YLD7
4. Discussion
4.1. YLD7 Act as a Bridge in the Plant Thylakoid Membrane for Grana and Stroma Formation
4.2. yld7 Accelerated Senescence and Cell Death in the yld7 Mutants
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, P.; Hu, H.; Zhang, Y.; Wang, Z.; Dong, G.; Cui, Y.; Qian, Q.; Ren, D.; Guo, L.-b. Genetic analysis and fine-mapping of a new rice mutant, white and lesion mimic leaf1. Plant Growth Regul. 2018, 85, 425–435. [Google Scholar] [CrossRef]
- Mao, C.; Lu, S.; Lv, B.; Zhang, B.; Shen, J.; He, J.; Luo, L.; Xi, D.; Chen, X.; Ming, F. A Rice NAC Transcription Factor Promotes Leaf Senescence via ABA Biosynthesis. Plant Physiol. 2017, 174, 1747–1763. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Song, J.; Tang, L.; Xu, X.; Peng, X.; Fan, H.; Wang, J. Genetic Analysis and Fine Mapping of a New Rice Mutant, Leaf Tip Senescence 2. Int. J. Mol Sci. 2024, 25, 7082. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Liu, C.; Zhang, Y.; Cui, Y.; Hu, H.; Jahan, N.; Lv, Y.; Qian, Q.; Guo, L. A 3-bp deletion of WLS5 gene leads to weak growth and early leaf senescence in rice. Rice 2019, 12, 26. [Google Scholar] [CrossRef]
- Kim, S.H.; Yoon, J.; Kim, H.; Lee, S.J.; Paek, N.C. Rice Basic Helix-Loop-Helix 079 (OsbHLH079) Delays Leaf Senescence by Attenuating ABA Signaling. Rice 2023, 16, 60. [Google Scholar] [CrossRef]
- Lee, S.; Masclaux-Daubresse, C. Current Understanding of Leaf Senescence in Rice. Int. J. Mol. Sci. 2021, 22, 4515. [Google Scholar] [CrossRef]
- Chen, D.; Qiu, Z.; He, L.; Hou, L.; Zhu, L. The Rice LRR-Like1 Protein YELLOW AND PREMATURE DWARF 1 is Involved in High Light-Induced Leaf Senescence. J. Exp. Bot. 2020, 72, 1589–1605. [Google Scholar] [CrossRef]
- Sun, L.; Xu, H.; Song, J.; Yang, X.; Wang, X.; Liu, H.; Pang, M.; Hu, Y.; Yang, Q.; Ning, X.; et al. OsNAC103, a NAC Transcription Factor, Positively Regulates Leaf Senescence and Plant Architecture in Rice. Rice 2024, 17, 15. [Google Scholar] [CrossRef]
- Wu, L.; Ren, D.; Hu, S.; Li, G.; Dong, G.; Jiang, L.; Hu, X.; Ye, W.; Cui, Y.; Zhu, L.; et al. Down-Regulation of a Nicotinate Phosphoribosyltransferase Gene, OsNaPRT1, Leads to Withered Leaf Tips. Plant Physiol. 2016, 171, 1085–1098. [Google Scholar] [CrossRef]
- Ge, C.; Wang, L.; Ye, W.; Wu, L.; Cui, Y.; Chen, P.; Pan, J.; Zhang, D.; Hu, J.; Zeng, D.; et al. Single-point Mutation of an Histidine-aspartic Domain-containing Gene involving in Chloroplast Ribosome Biogenesis Leads to White Fine Stripe Leaf in Rice. Sci. Rep. 2017, 7, 3298. [Google Scholar] [CrossRef]
- Allred, D.R.; Staehelin, L.A. Lateral Distribution of the Cytochrome b 6/f and Coupling Factor ATP Synthetase Complexes of Chloroplast Thylakoid Membranes 1. Plant Physiol. 1985, 78, 199–202. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.R.; Staehelin, L.A. Analysis of the thylakoid outer surface. Coupling factor is limited to unstacked membrane regions. J. Cell Biol. 1976, 68, 30–47. [Google Scholar] [CrossRef] [PubMed]
- Wood, W.H.J.; Johnson, M.P. Modeling the Role of LHCII-LHCII, PSII-LHCII, and PSI-LHCII Interactions in State Transitions. Biophys J 2020, 119, 287–299. [Google Scholar] [CrossRef]
- Hasan, S.S.; Yamashita, E.; Ryan, C.M.; Whitelegge, J.P.; Cramer, W.A. Conservation of Lipid Functions in Cytochrome bc Complexes. J. Mol. Biol. 2011, 414, 145–162. [Google Scholar] [CrossRef]
- Bai, T.; Guo, L.; Xu, M.; Tian, L. Structural Diversity of Photosystem I and Its Light-Harvesting System in Eukaryotic Algae and Plants. Front. Plant Sci. 2021, 12, 781035. [Google Scholar] [CrossRef]
- Hiroshi, K.; Masahiko, I. Targeted disruption of psbX and biochemical characterization of photosystem II complex in the thermophilic cyanobacterium Synechococcus elongatus. Plant Cell Physiol. 2001, 42, 179–188. [Google Scholar]
- Kamiya, N.; Shen, J.R. Crystal structure of oxygen-evolving photosystem II from Thermosynechococcus vulcanus at 3.7-A resolution. Proc. Natl. Acad. Sci. USA 2003, 100, 98–103. [Google Scholar] [CrossRef]
- Stefano, C.; Tania, T.; Robert, J.; Stefano, S. A Comparison between Plant Photosystem I and Photosystem II Architecture and Functioning. Curr. Protein Pept. Sci. 2014, 15, 296–331. [Google Scholar]
- Kato, Y.; Kuroda, H.; Ozawa, S.I.; Saito, K.; Dogra, V.; Scholz, M.; Zhang, G.; de Vitry, C.; Ishikita, H.; Kim, C.; et al. Characterization of tryptophan oxidation affecting D1 degradation by FtsH in the photosystem II quality control of chloroplasts. eLife 2023, 12, RP88822. [Google Scholar] [CrossRef]
- Zhao, Y.; Deng, L.; Last, R.L.; Hua, W.; Liu, J. Psb28 protein is indispensable for stable accumulation of PSII core complexes in Arabidopsis. Plant J. 2024, 119, 1226–1238. [Google Scholar] [CrossRef]
- Nelson, N. Plant Photosystem I—The Most Efficient Nano-Photochemical Machine. J. Nanosci. Nanotechnol. 2009, 9, 1709–1713. [Google Scholar] [CrossRef] [PubMed]
- Croce, R.; Amerongen, H.V. Light-harvesting in photosystem I. Photosynth. Res. 2013, 116, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Chen, S.; Wang, C.; Lin, W.; Huang, C.; Fan, C.; Han, D.; Lu, D.; Xu, X.; Sui, S.; et al. Regulatory dynamics of the higher-plant PSI-LHCI supercomplex during state transitions. Mol. Plant. 2023, 16, 1937–1950. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Xia, H.; Li, Q.; Li, Z.; Zhai, C.; Weng, L.; Mi, H.; Yan, S.; Datla, R.; Wang, H.; et al. PALE-GREEN LEAF 1, a rice cpSRP54 protein, is essential for the assembly of the PSI-LHCI supercomplex. Plant Direct. 2022, 6, e436. [Google Scholar] [CrossRef]
- Cui, Y.L.; Jia, Q.S.; Yin, Q.Q.; Lin, G.N.; Kong, M.M.; Yang, Z.N. The GDC1 gene encodes a novel ankyrin domain-containing protein that is essential for grana formation in Arabidopsis. Plant Physiol. 2011, 155, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Mustárdy, L.; Garab, G. Granum revisited. A three-dimensional model—Where things fall into place. Trends Plant Sci. 2003, 8, 117–122. [Google Scholar] [CrossRef]
- Dekker, J.P.; Boekema, E.J. Supramolecular organization of thylakoid membrane proteins in green plants. Biochim. Biophys. Acta 2005, 1706, 12–39. [Google Scholar] [CrossRef]
- Anderson, J.M.; Aro, E.M. Grana stacking and protection of Photosystem II in thylakoid membranes of higher plant leaves under sustained high irradiance: An hypothesis. Photosynth. Res. 1994, 41, 315–326. [Google Scholar] [CrossRef]
- Allen, J.F.; Forsberg, J. Molecular recognition in thylakoid structure and function. Trends Plant Sci. 2001, 6, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Haldrup, A.; Jensen, P.E.; Lunde, C.; Scheller, H.V. Balance of power: A view of the mechanism of photosynthetic state transitions. Trends Plant Sci. 2001, 6, 301–305. [Google Scholar] [CrossRef]
- Li, L.; Yang, J.; Zhang, Q.; Xue, Q.; Li, M.; Xue, Q.; Liu, W.; Niu, Z.; Ding, X. Genome-wide identification of Ankyrin (ANK) repeat gene families in three Dendrobium species and the expression of ANK genes in D. officinale under gibberellin and abscisic acid treatments. BMC Plant Biol. 2024, 24, 762. [Google Scholar] [CrossRef] [PubMed]
- Arnon, D.I. Copper enzymes in isolated chloroplasts. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Wellburn, A.R. The spectral determination of chlorophyll a and chlorophyll b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J. Plant Physiol. 1994, 144, 307–313. [Google Scholar] [CrossRef]
- Peterhänsel, C.; Freialdenhoven, A.; Kurth, J.; Kolsch, R.; Schulzelefert, P. Interaction Analyses of Genes Required for Resistance Responses to Powdery Mildew in Barley Reveal Distinct Pathways Leading to Leaf Cell Death. Plant Cell 1997, 9, 1397–1409. [Google Scholar] [CrossRef]
- Bowling, S.A. The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 1997, 9, 1573–1584. [Google Scholar]
- Chun, W.; Lan, S.; Yaping, F.; Changjie, Y.; Kejian, W. A Simple CRISPR/Cas9 System for Multiplex Genome Editing in Rice. J. Genet. Genom. 2015, 2, 703–706. [Google Scholar]
- Cui, Y.; Peng, Y.; Zhang, Q.; Xia, S.; Ruan, B.; Xu, Q.; Yu, X.; Zhou, T.; Liu, H.; Zeng, D.; et al. Disruption of EARLY LESION LEAF 1, encoding a cytochrome P450 monooxygenase, induces ROS accumulation and cell death in rice. Plant J. Cell Mol. Biol. 2021, 105, 942–956. [Google Scholar] [CrossRef] [PubMed]
- Roy, N.; Bagchi, S.; Raychaudhuri, P. Damaged DNA Binding Protein 2 in Reactive Oxygen Species (ROS) Regulation and Premature Senescence. Int. J. Mol. Sci. 2012, 13, 11012–11026. [Google Scholar] [CrossRef]
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef]
- Tan, J. A Novel Chloroplast-Localized Pentatricopeptide Repeat Protein Involved in Splicing Affects Chloroplast Development and Abiotic Stress Response in Rice. Mol. Plant. 2014, 7, 1329–1349. [Google Scholar] [CrossRef]
- Leng, Y.; Yang, Y.; Ren, D.; Huang, L.; Dai, L.; Wang, Y.; Chen, L.; Tu, Z.; Gao, Y.; Li, X. A Rice PECTATE LYASE-LIKE Gene Is Required for Plant Growth and Leaf Senescence. Plant Physiol. 2017, 174, 1151–1166. [Google Scholar] [CrossRef] [PubMed]
- Kirchhoff, H.A.-O. Chloroplast ultrastructure in plants. New Phytol. 2019, 223, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, N.C.; Smillie, R.M.; Henningsen, K.W.; Von Wettstein, D.; French, C.S. Composition and Function of Thylakoid Membranes from Grana-rich and Grana-deficient Chloroplast Mutants of Barley. Plant Physiol. 1979, 63, 174–182. [Google Scholar] [CrossRef]
- Kusaba, M.; Ito, H.; Morita, R.; Iida, S.; Sato, Y.; Fujimoto, M.; Kawasaki, S.; Tanaka, R.; Hirochika, H.; Nishimura, M. Rice NON-YELLOW COLORING1 Is Involved in Light-Harvesting Complex II and Grana Degradation during Leaf Senescence. Plant Cell 2007, 19, 1362–1375. [Google Scholar] [CrossRef]
- Ki-Hong, J.; Junghe, H.; Choong-Hwan, R.; Youngju, C.; Yong-Yoon, C.; Akio, M.; Hirohiko, H.; Gynheung, A. Characterization of a Rice Chlorophyll-Deficient Mutant Using the T-DNA Gene-Trap System. Plant Cell Physiol. 2003, 44, 463–472. [Google Scholar]
- Wu, Z.; Zhang, X.; He, B.; Diao, L.; Wan, J. A Chlorophyll-Deficient Rice Mutant with Impaired Chlorophyllide Esterification in Chlorophyll Biosynthesis. Plant Physiol. 2007, 145, 29–40. [Google Scholar] [CrossRef]
- Yang, Y.; Xu, J.; Huang, L.; Yujia, L.; Dai, L.; Rao, Y.; Chen, L.; Wang, Y.; Tu, Z.; Hu, J. PGL, encoding chlorophyllide a oxygenase 1, impacts leaf senescence and indirectly affects grain yield and quality in rice. J. Exp. Bot. 2016, 67, 1297–1310. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Cheng, Z.; Ma, X.; Wu, H.; Liu, Y.; Zhou, K.; Chen, Y.; Ma, W.; Bi, J.; Zhang, X. A knockdown mutation of YELLOW-GREEN LEAF2 blocks chlorophyll biosynthesis in rice. Plant Cell Rep. 2013, 32, 1855–1867. [Google Scholar] [CrossRef]
- Xu, J.; Yang, J.; Wu, Z.; Liu, H.; Huang, F.; Wu, Y.; Carrie, C.; Narsai, R.; Murcha, M.; Whelan, J.; et al. Identification of a Dual-Targeted Protein Belonging to the Mitochondrial Carrier Family That Is Required for Early Leaf Development in Rice; American Society of Plant Biologists: Rockville, MD, USA, 2013. [Google Scholar]
- Apel, K.; Hirt, H. REACTIVE OXYGEN SPECIES: Metabolism, Oxidative Stress, and Signal Transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Smerdon, S.S.J. The ankyrin repeat: A diversity of interactions on a common structural framework. Trends Biochem. Sci. 1999, 24, 311–316. [Google Scholar]
- Bennett, V.A. Adaptors between diverse plasma membrane proteins and the cytoplasm. J. Biol. Chem. 1992, 267, 8703–8706. [Google Scholar] [CrossRef] [PubMed]
- Breeden, L.; Nasmyth, K. Similarity between cell-cycle genes of budding yeast and fission yeast and the Notch gene of Drosophila. Nature 1987, 329, 651–654. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zeng, F.; Song, P.; Sun, B.; Wang, Q.; Wang, J. Effects of reduced chlorophyll content on photosystem functions and photosynthetic electron transport rate in rice leaves. J. Plant Physiol. 2022, 272, 153669. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, Y.; Song, J.; Tang, L.; Wang, J. YELLOW LEAF AND DWARF 7, Encoding a Novel Ankyrin Domain-Containing Protein, Affects Chloroplast Development in Rice. Genes 2024, 15, 1267. https://doi.org/10.3390/genes15101267
Cui Y, Song J, Tang L, Wang J. YELLOW LEAF AND DWARF 7, Encoding a Novel Ankyrin Domain-Containing Protein, Affects Chloroplast Development in Rice. Genes. 2024; 15(10):1267. https://doi.org/10.3390/genes15101267
Chicago/Turabian StyleCui, Yongtao, Jian Song, Liqun Tang, and Jianjun Wang. 2024. "YELLOW LEAF AND DWARF 7, Encoding a Novel Ankyrin Domain-Containing Protein, Affects Chloroplast Development in Rice" Genes 15, no. 10: 1267. https://doi.org/10.3390/genes15101267
APA StyleCui, Y., Song, J., Tang, L., & Wang, J. (2024). YELLOW LEAF AND DWARF 7, Encoding a Novel Ankyrin Domain-Containing Protein, Affects Chloroplast Development in Rice. Genes, 15(10), 1267. https://doi.org/10.3390/genes15101267





