Selection of Reference Genes and HSP17.9A Expression Profiling in Heat-Stressed Grapevine Varieties
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and HS Induction
2.2. Extraction of Total RNA, cDNA Synthesis, and qPCR Assays
2.3. Evaluation and Selection of the Reference Genes for qPCR Assays
2.4. Statistical Analyses
3. Results
3.1. Selection of the Candidate Reference Genes
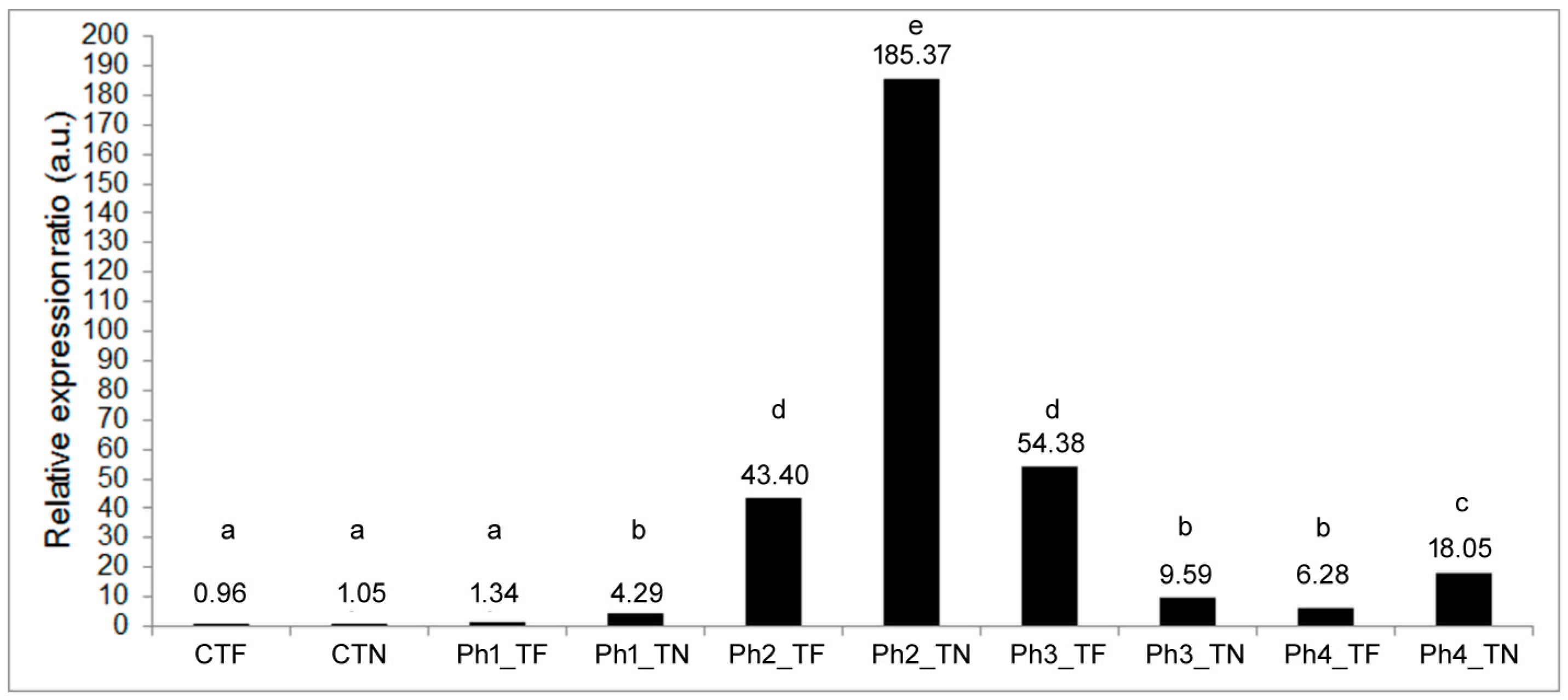
3.2. Expression Profiling of the HSP17.9A Target Gene
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Janni, M.; Maestri, E.; Gullì, M.; Marmiroli, M.; Marmiroli, N. Plant responses to climate change, how global warming may impact on food security: A critical review. Front. Plant Sci. 2024, 14, 1297569. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.V.; White, M.A.; Cooper, O.R.; Storchmann, K. Climate change and global wine quality. Clim. Change 2005, 73, 319–343. [Google Scholar] [CrossRef]
- Fraga, H.; Santos, J.A.; Malheiro, A.C.; Oliveira, A.A.; Moutinho-Pereira, J.; Jones, G.V. Climatic suitability of Portuguese grapevine varieties and climate change adaptation. Int. J. Clim. 2015, 36, 1–12. [Google Scholar] [CrossRef]
- Venios, X.; Korkas, E.; Nisiotou, A.; Banillas, G. Grapevine responses to heat stress and global warming. Plants 2020, 9, 1754. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, L.C.; Amâncio, S. Cutting the Gordian Knot of abiotic stress in grapevine: From the test tube to climate change adaptation. Physiol. Plant. 2018, 165, 330–342. [Google Scholar] [CrossRef]
- Carvalho, A.; Leal, F.; Matos, M.; Lima-Brito, J. Effects of heat stress in the leaf mitotic cell cycle and chromosomes of four wine-producing grapevine varieties. Protoplasma 2018, 255, 1725–1740. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, A.; Leal, F.; Matos, M.; Lima-Brito, J. Heat stress tolerance assayed in four wine-producing grapevine varieties using a cytogenetic approach. Cienc. Tec. Vitiv. 2019, 34, 61–70. [Google Scholar] [CrossRef]
- Nogales, A.; Ribeiro, H.; Nogales-Bueno, J.; Hansen, L.D.; Gonçalves, E.F.; Coito, J.L.; Rato, A.E.; Peixe, A.; Viegas, W.; Cardoso, H. Response of Mycorrhizal ’Touriga Nacional’ Variety Grapevines to High Temperatures Measured by Calorespirometry and Near-Infrared Spectroscopy. Plants 2020, 9, 1499. [Google Scholar] [CrossRef]
- Castro, C.; Carvalho, A.; Pavia, I.; Bacelar, E.; Lima-Brito, J. Development of grapevine plants under hydroponic Copper-enriched solutions induced morpho-histological, biochemical and cytogenetic changes. Plant Physiol. Biochem. 2021, 166, 887–901. [Google Scholar] [CrossRef]
- Castro, C.; Carvalho, A.; Pavia, I.; Bacelar, E.; Lima-Brito, J. Grapevine varieties with differential tolerance to Zinc analysed by morpho-histological and cytogenetic approaches. Sci. Hortic. 2021, 288, 110386. [Google Scholar] [CrossRef]
- Mittler, R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 2006, 11, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Portaria No. 380/2012, Diário da República, 1a Série—N.º 226—22 de Novembro de 2012; Ministério da Agricultura, do Mar, do Ambiente e do Ordenamento do Território: Lisboa, Portugal, 2012; pp. 6712–6715. (In Portuguese)
- Portaria No. 383/2017, Diário da República, 1.ª série–N.º 243–20 de Dezembro de 2017; Ministério da Agricultura, Florestas e Desenvolvimento Rural: Lisboa, Portugal, 2017; pp. 6659–6660. (In Portuguese)
- Touriga Nacional—Vine and Wine Cluster; COLAB Vines & Wines: Vila Real, Portugal; 7p, Available online: https://www.advid.pt/uploads/TourigaNacional_Final_25.10.pdf (accessed on 27 May 2024). (In Portuguese)
- Touriga Franca—Vine and Wine Cluster; COLAB Vines & Wines: Vila Real, Portugal; 6p, Available online: https://www.advid.pt/uploads/TourigaFranca_Final_25.10.pdf (accessed on 27 May 2024). (In Portuguese)
- Carvalho, L.C.; Ramos, M.J.N.; Faísca-Silva, D.; van der Kellen, D.; Fernandes, J.C.; Egipto, R.; Lopes, C.M.; Amâncio, S. Developmental regulation of transcription in Touriga Nacional berries under deficit irrigation. Plants 2022, 11, 827. [Google Scholar] [CrossRef] [PubMed]
- Cabral, I.L.; Teixeira, A.; Lanoue, A.; Unlubayir, M.; Munsch, T.; Valente, J.; Alves, F.; da Costa, P.L.; Rogerson, F.S.; Carvalho, S.M.P.; et al. Impact of Deficit Irrigation on Grapevine cv. ‘Touriga Nacional’ during Three Seasons in Douro Region: An Agronomical and Metabolomics Approach. Plants 2022, 11, 732. [Google Scholar] [CrossRef]
- Chaves, M.M.; Pereira, J.S.; Maroco, J.; Rodrigues, M.L.; Ricardo, C.P.; Osório, M.L.; Carvalho, I.; Faria, T.; Pinheiro, C. How plants cope with water stress in the field. Photosynthesis and Growth. Ann. Bot. 2002, 89, 907–916. [Google Scholar] [CrossRef]
- Moutinho-Pereira, J.M.; Correia, C.M.; Gonçalves, B.M.; Bacelar, E.A.; Torres-Pereira, J.M. Leaf gas exchange and water relations of grapevines grown in three different conditions. Photosynthetica 2004, 42, 81–86. [Google Scholar] [CrossRef]
- Carvalho, A.; Dinis, L.-T.; Luzio, A.; Bernardo, S.; Moutinho-Pereira, J.; Lima-Brito, J. Cytogenetic and molecular effects of kaolin’s foliar application in grapevine (Vitis vinifera L.) under summer’s stressful growing conditions. Genes 2024, 15, 747. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, S.; Dinis, L.-T.; Machado, N.; Moutinho-Pereira, J. Grapevine abiotic stress assessment and search for sustainable adaptation strategies in Mediterranean-like climates. A review. Agron. Sustain. Dev. 2018, 38, 66. [Google Scholar] [CrossRef]
- Carvalho, L.C.; Coito, J.L.; Colaço, S.; Sangiogo, M.; Amâncio, S. Heat stress in grapevine: The pros and cons of acclimation. Plant Cell Environ. 2015, 38, 777–789. [Google Scholar] [CrossRef]
- Chen, Z.; Galli, M.; Gallavotti, A. Mechanisms of temperature-regulated growth and thermotolerance in crop species. Curr. Opin. Plant Biol. 2022, 65, 102134. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Euring, D.; Cha, J.Y.; Lin, Z.; Lu, M.; Huang, L.-J.; Kim, W.Y. Plant hormone-mediated regulation of heat tolerance in response to global climate change. Front. Plant Sci. 2021, 11, 627969. [Google Scholar] [CrossRef]
- Banilas, G.; Korkas, E.; Englezos, V.; Nisiotou, A.A.; Hatzopoulos, P. Genome-wide analysis of the heat shock protein 90 gene family in grapevine (Vitis vinifera L.). Aust. J. Grape Wine Res. 2012, 18, 29–38. [Google Scholar] [CrossRef]
- Wu, H.; Wang, L.; Wang, H.; Lua, Z. Characterization of Hsp17, a novel small heat shock protein, in Sphingomonas melonis TY under heat stress. Microbiol. Spectr. 2023, 11, e01360-23. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, A.; Graça, C.; Carocha, V.; Pêra, S.; Lousada, J.L.; Lima-Brito, J.; Paiva, J.A.P. An improved total RNA isolation from secondary tissues of Woody species for coding and non-coding gene expression analyses. Wood Sci. Technol. 2015, 49, 647–658. [Google Scholar] [CrossRef]
- Borges, A.F.; Fonseca, C.; Ferreira, R.B.; Lourenço, A.M.; Monteiro, S. Reference gene validation for quantitative RT-PCR during biotic and abiotic stresses in Vitis vinifera. PLoS ONE 2014, 9, e111399. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Sepeleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Pfaffl, M.; Tichopad, A.; Prgomet, C.; Neuvians, T. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef]
- Droulia, F.; Charalampopoulos, I. Future Climate Change Impacts on European Viticulture: A Review on Recent Scientific Advances. Atmosphere 2021, 12, 495. [Google Scholar] [CrossRef]
- Durall, C.; Sandesh Kanchugal, P.; Selmer, M.; Lindblad, P. Oligomerization and characteristics of phosphoenolpyruvate carboxylase in Synechococcus PCC 7002. Sci. Rep. 2020, 10, 3607. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wang, Y.; Xin, H.; Li, S.; Liang, Z. Involvement of ubiquitin-conjugating enzyme (e2 gene family) in ripening process and response to cold and heat stress of Vitis vinifera. Sci. Rep. 2017, 7, 13290. [Google Scholar] [CrossRef]
- Seidel, T. The plant V-ATPase. Front. Plant Sci. 2022, 13, 931777. [Google Scholar] [CrossRef]
- Huang, L.-Z.; Zhou, M.; Ding, Y.-F.; Zhu, C. Gene networks involved in plant heat stress response and tolerance. Int. J. Mol. Sci. 2022, 23, 11970. [Google Scholar] [CrossRef]
- Liu, X.; Chen, H.; Li, S.; Lecourieux, D.; Duan, W.; Fan, P.; Liang, Z.; Wan, L. Natural variations of HSFA2 enhance thermotolerance in grapevine. Hortic. Res. 2023, 10, uhac250. [Google Scholar] [CrossRef]
- Jung, K.; Friede, T.; Beißbarth, T. Reporting FDR analogous confidence intervals for the log fold change of differentially expressed genes. BMC Bioinform. 2011, 12, 288. [Google Scholar] [CrossRef]
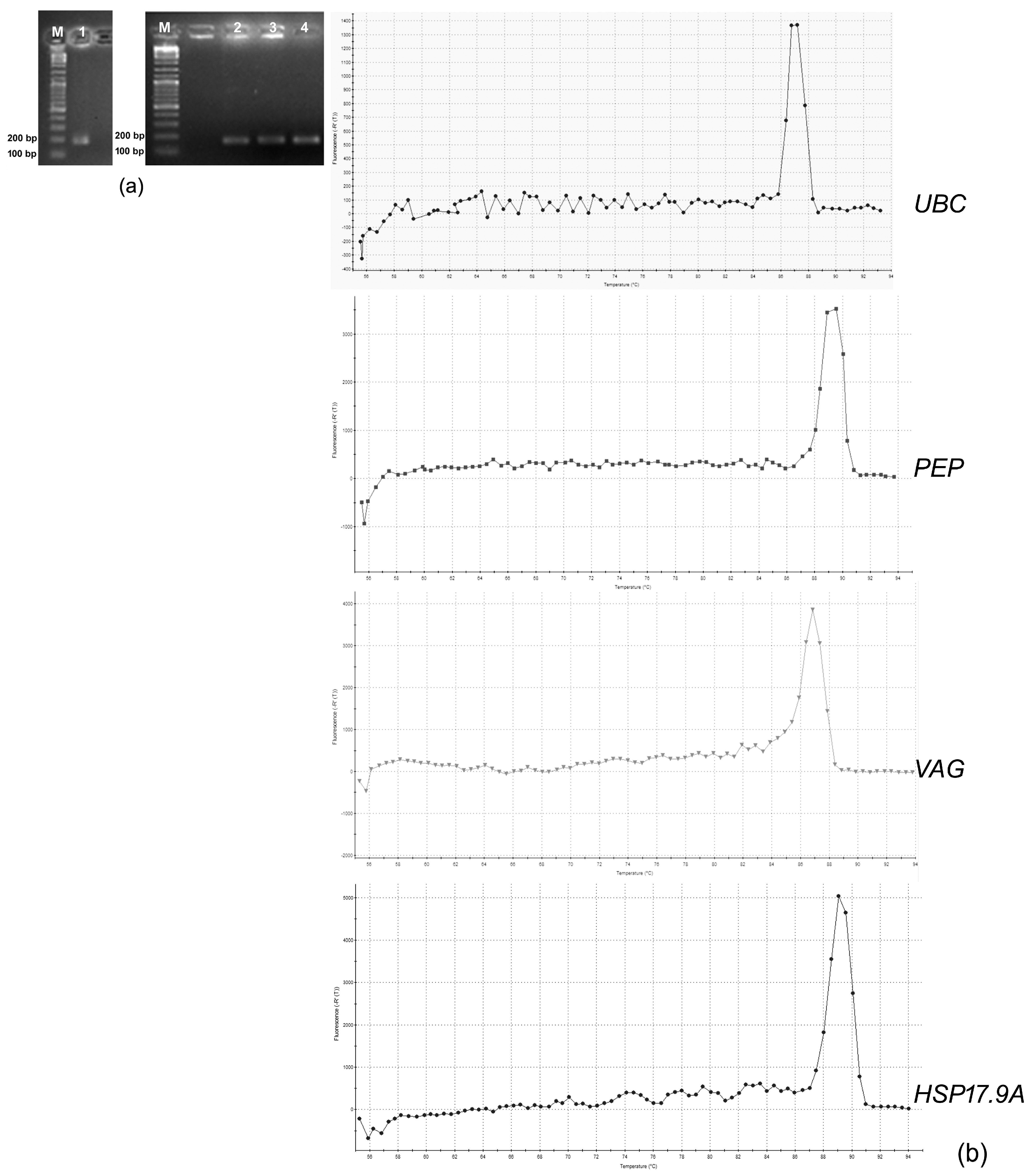
| Candidate Reference Genes | * NIH—NCBI Reference Sequence | Sequence (5′→3′) | Expected Amplicon Size (bp) | Reference |
|---|---|---|---|---|
| Phosphoenolpyruvate carboxylase (PEP) | AF236126.1 | F: CCTCCTCCTCCAGATTGC R: GGCTTGCTTGATTCCATTATC | 198 | [30] |
| Vacuolar ATPase subunit G (VAG) | XM_002281110.1 | F: TTGCCTGTGTCTCTTGTTC R: TCAATGCTGCCAGAAGTG | 174 | [30] |
| Ubiquitin-conjugating enzyme (UBC) | EE253706 GenBank: EE253706.1 | F: CATAAGGGCTATCAGGAGGAC R: TGGCGGTCGGAGTTAGG | 161 | [30] |
| Target gene | * NIH—NCBI Reference Sequence | Sequence (5′→3′) | Expected amplicon size (bp) | Reference |
| Class II Heat Shock Protein 17.9A (HSP17.9A) | XM_002280644.4 Replaced by: Gene Id: 100268056; LOC 100268056: 17.3 kDa class II heat shock protein V. vinifera (grapevine) | F: CGTCAAGGAGTACCCCAATTC R: AACTTCCCCACCCTCCTCT | 177 | [23] |
| Candidate Reference Gene | Delta-CT (S.D.) | Bestkeeper | GeNorm (M-Value) | NormFinder (Stability Value ± S.E.) | Comprehensive Ranking (Geomean of S.D. Values) |
|---|---|---|---|---|---|
| PEP | 2.926 | S.D. = 2.610 %C.V. = 7.81 r = 0.869 (p = 0.001) | - | 0.117 ± 0.019 | 0.562 |
| VAG | 1.126 | S.D. = 0.940 %C.V. = 4.13 r = 0.336 (p = 0.075) | - | 0.094 ± 0.018 | 0.293 |
| UBC | 0.716 | S.D. = 0.520 %C.V. = 2.59 r = 0.618 (p = 0.001) | - | 0.045 ± 0.028 | 0.227 |
| Pairwise analyses | |||||
| PEP + VAG | 2.382 | r = 0.900 | 2.698 | - | - |
| VAG + UBC | 0.807 | r = 0.929 | 1.391 | - | - |
| PEP + UBC | 2.605 | r = 0.791 | 2.723 | - | - |
| ‘Grapevine Variety × Experimental Phase’ | Mean Relative Expression Ratio of the HSP17.9A Gene ± S.E. | S.D. | 95% C.I. |
|---|---|---|---|
| CTF | 0.995 ± 0.095 a | 0.136 | 0.99–1.00 |
| CTN | 1.085 ± 0.065 a | 0.086 | 1.08–1.09 |
| Ph1_TF | 1.345 ± 0.135 a | 0.190 | 1.34–1.36 |
| Ph1_TN | 4.310 ± 0.280 b | 0.399 | 4.29–4.33 |
| Ph2_TF | 43.815 ± 5.995 d | 8.479 | 43.34–44.29 |
| Ph2_TN | 185.405 ± 3.485 e | 4.923 | 185.13–185.68 |
| Ph3_TF | 54.265 ± 2.545 d | 3.600 | 54.07–54.47 |
| Ph3_TN | 9.590 ± 0.240 b | 0.339 | 9.58–9.61 |
| Ph4_TF | 6.280 ± 0.080 b | 0.111 | 6.27–6.29 |
| Ph4_TN | 18.110 ± 0.170 c | 0.240 | 18.10–18.13 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carvalho, A.; Crisóstomo, C.; Leal, F.; Lima-Brito, J. Selection of Reference Genes and HSP17.9A Expression Profiling in Heat-Stressed Grapevine Varieties. Genes 2024, 15, 1283. https://doi.org/10.3390/genes15101283
Carvalho A, Crisóstomo C, Leal F, Lima-Brito J. Selection of Reference Genes and HSP17.9A Expression Profiling in Heat-Stressed Grapevine Varieties. Genes. 2024; 15(10):1283. https://doi.org/10.3390/genes15101283
Chicago/Turabian StyleCarvalho, Ana, Christina Crisóstomo, Fernanda Leal, and José Lima-Brito. 2024. "Selection of Reference Genes and HSP17.9A Expression Profiling in Heat-Stressed Grapevine Varieties" Genes 15, no. 10: 1283. https://doi.org/10.3390/genes15101283






