Effects of Cryptorchidism on the Semen Quality of Giant Pandas from the Perspective of Seminal Plasma Proteomics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Preparation
2.2. Total Protein Extraction, Quality Control, and Trypsin Treatment
2.3. LC-MS/MS Analysis
2.4. The Identification and Quantitation of Protein
2.5. The Functional Analysis of Protein and DEP
3. Results
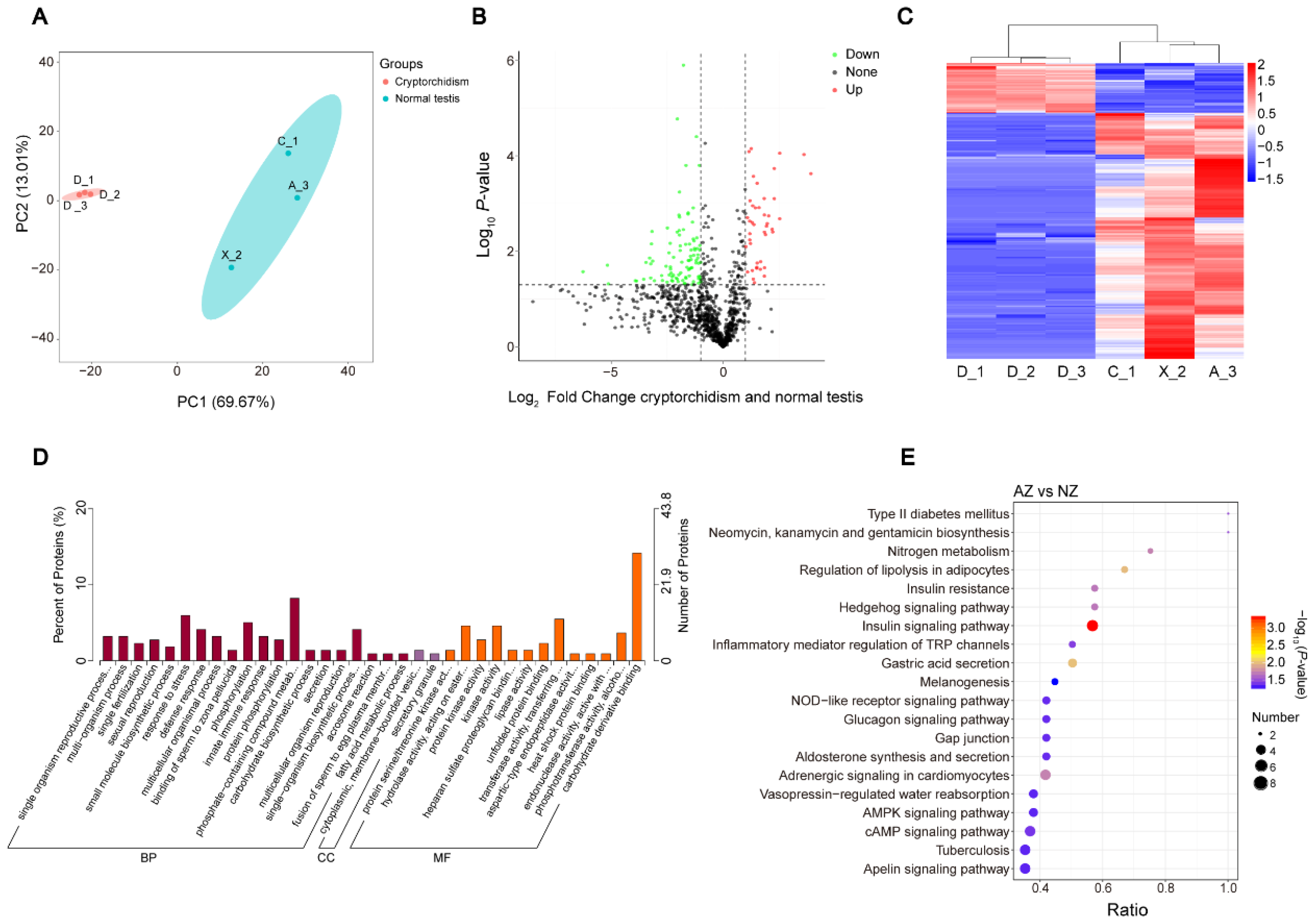
3.1. Seminal Plasma Proteome Profiles of the Giant Panda and One with Cryptorchidism
3.2. Annotation and Analysis of Seminal Plasma Protein
3.3. Identification of the DEPs between Cryptorchid and Normal Testis Group
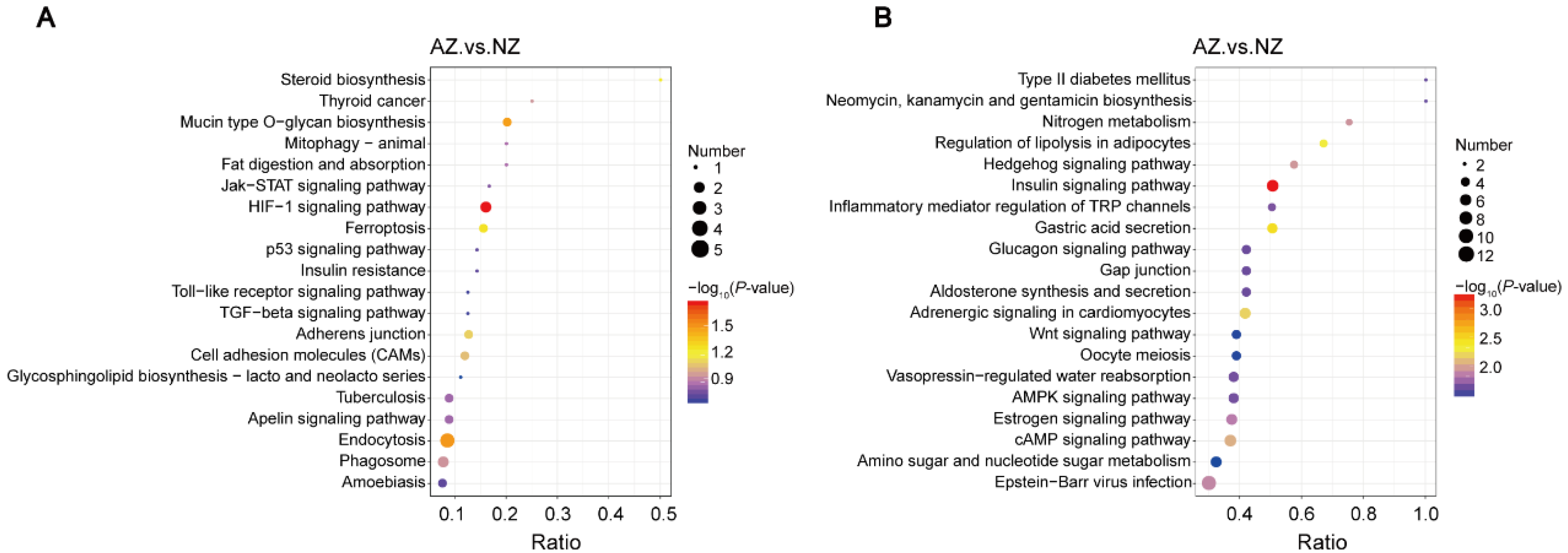
3.4. Comprehensive Functional Analysis of DEPs
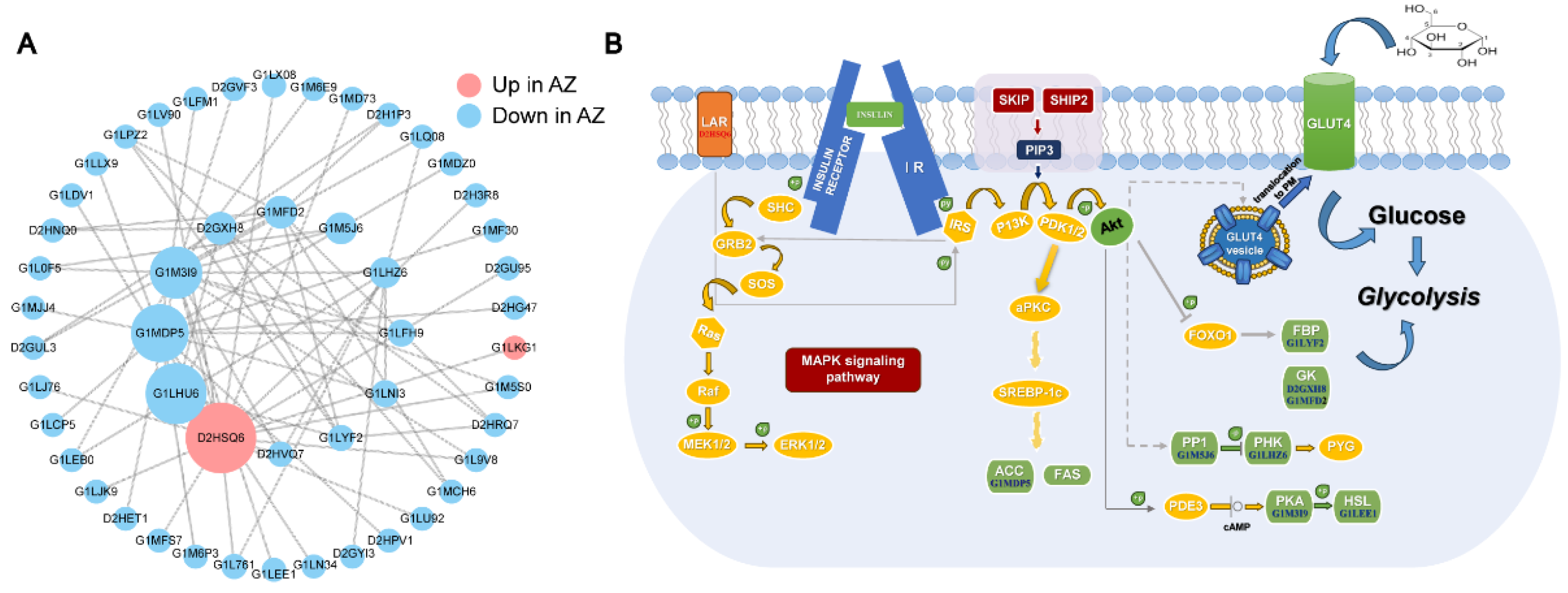
3.5. Network Analysis of the DEPs Involved in Regulating the Semen Quality
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tsutsui, T.; Hori, T.; Nakashige, T.; Narushima, E.; Hara, T.; Akikawa, T.; Nose, N.; Saito, K.; Shichiri, S.; Hashizaki, F.; et al. Semen Quality in a Giant Panda (Ailuropoda melanoleuca) in Relation to Estrus of a Nearby Resident Female Panda. Theriogenology 2006, 66, 1803–1806. [Google Scholar] [CrossRef] [PubMed]
- Martin-Wintle, M.S.; Kersey, D.C.; Wintle, N.J.P.; Aitken-Palmer, C.; Owen, M.A.; Swaisgood, R.R. Comprehensive Breeding Techniques for the Giant Panda. Adv. Exp. Med. Biol. 2019, 1200, 275–308. [Google Scholar] [CrossRef]
- Charlton, B.D.; Owen, M.A.; Zhou, X.; Zhang, H.; Swaisgood, R.R. Influence of Season and Social Context on Male Giant Panda (Ailuropoda melanoleuca) Vocal Behaviour. PLoS ONE 2019, 14, e0225772. [Google Scholar] [CrossRef]
- Zhou, W.; Nie, Y.; Hu, Y.; Swaisgood, R.R.; Zhang, Y.; Liu, D.; Wei, F. Seasonal and Reproductive Variation in Chemical Constituents of Scent Signals in Wild Giant Pandas. Sci. China Life Sci. 2019, 62, 648–660. [Google Scholar] [CrossRef] [PubMed]
- Aitken-Palmer, C.; Hou, R.; Burrell, C.; Zhang, Z.; Wang, C.; Spindler, R.; Wildt, D.E.; Ottinger, M.A.; Howard, J. Protracted Reproductive Seasonality in the Male Giant Panda (Ailuropoda melanoleuca) Reflected by Patterns in Androgen Profiles, Ejaculate Characteristics, and Selected Behaviors. Biol. Reprod. 2012, 86, 195. [Google Scholar] [CrossRef] [PubMed]
- Santiago-Moreno, J.; Blesbois, E. Functional Aspects of Seminal Plasma in Bird Reproduction. Int. J. Mol. Sci. 2020, 21, 5664. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Martínez, H.; Kvist, U.; Ernerudh, J.; Sanz, L.; Calvete, J.J. Seminal Plasma Proteins: What Role Do They Play? Am. J. Reprod. Immunol. 2011, 66 (Suppl. S1), 11–22. [Google Scholar] [CrossRef] [PubMed]
- Duncan, M.W.; Thompson, H.S. Proteomics of Semen and Its Constituents. Proteom. Clin. Appl. 2007, 1, 861–875. [Google Scholar] [CrossRef] [PubMed]
- Carver, D.A.; Ball, B.A. Lipase Activity in Stallion Seminal Plasma and the Effect of Lipase on Stallion Spermatozoa during Storage at 5 Degrees C. Theriogenology 2002, 58, 1587–1595. [Google Scholar] [CrossRef] [PubMed]
- Buchman-Shaked, O.; Kraiem, Z.; Gonen, Y.; Goldman, S. Presence of Matrix Metalloproteinases and Tissue Inhibitor of Matrix Metalloproteinase in Human Sperm. J. Androl. 2002, 23, 702–708. [Google Scholar] [CrossRef]
- Baumgart, E.; Lenk, S.V.; Loening, S.A.; Jung, K. Quantitative Differences in Matrix Metalloproteinase (MMP)-2, but Not in MMP-9, Tissue Inhibitor of Metalloproteinase (TIMP)-1 or TIMP-2, in Seminal Plasma of Normozoospermic and Azoospermic Patients. Hum. Reprod. 2002, 17, 2919–2923. [Google Scholar] [CrossRef] [PubMed]
- Gwathmey, T.M.; Ignotz, G.G.; Mueller, J.L.; Manjunath, P.; Suarez, S.S. Bovine Seminal Plasma Proteins PDC-109, BSP-A3, and BSP-30-kDa Share Functional Roles in Storing Sperm in the Oviduct. Biol. Reprod. 2006, 75, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Gwathmey, T.M.; Ignotz, G.G.; Suarez, S.S. PDC-109 (BSP-A1/A2) Promotes Bull Sperm Binding to Oviductal Epithelium in Vitro and May Be Involved in Forming the Oviductal Sperm Reservoir. Biol. Reprod. 2003, 69, 809–815. [Google Scholar] [CrossRef] [PubMed]
- Kelly, V.C.; Kuy, S.; Palmer, D.J.; Xu, Z.; Davis, S.R.; Cooper, G.J. Characterization of Bovine Seminal Plasma by Proteomics. Proteomics 2006, 6, 5826–5833. [Google Scholar] [CrossRef] [PubMed]
- Moura, A.A.; Chapman, D.A.; Koc, H.; Killian, G.J. A Comprehensive Proteomic Analysis of the Accessory Sex Gland Fluid from Mature Holstein Bulls. Anim. Reprod. Sci. 2007, 98, 169–188. [Google Scholar] [CrossRef]
- Töpfer-Petersen, E.; Romero, A.; Varela, P.F.; Ekhlasi-Hundrieser, M.; Dostàlovà, Z.; Sanz, L.; Calvete, J.J. Spermadhesins: A New Protein Family. Facts, Hypotheses and Perspectives. Andrologia 1998, 30, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Sridharn, T.B.; Rao, K.A. Role of Seminal Plasma Proteins in Effective Zygote Formation—A Success Road to Pregnancy. Protein Pept. Lett. 2019, 26, 238–250. [Google Scholar] [CrossRef]
- Rozeboom, K.J.; Troedsson, M.H.; Hodson, H.H.; Shurson, G.C.; Crabo, B.G. The Importance of Seminal Plasma on the Fertility of Subsequent Artificial Inseminations in Swine. J. Anim. Sci. 2000, 78, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Rath, D.; Knorr, C.; Taylor, U. Communication Requested: Boar Semen Transport through the Uterus and Possible Consequences for Insemination. Theriogenology 2016, 85, 94–104. [Google Scholar] [CrossRef]
- Kraus, M.; Tichá, M.; Zelezná, B.; Peknicová, J.; Jonáková, V. Characterization of Human Seminal Plasma Proteins Homologous to Boar AQN Spermadhesins. J. Reprod. Immunol. 2005, 65, 33–46. [Google Scholar] [CrossRef]
- Maxwell, W.M.C.; de Graaf, S.P.; Ghaoui, R.E.-H.; Evans, G. Seminal Plasma Effects on Sperm Handling and Female Fertility. Soc. Reprod. Fertil. Suppl. 2007, 64, 13–38. [Google Scholar] [CrossRef] [PubMed]
- Menezes, E.B.; Velho, A.L.C.; Santos, F.; Dinh, T.; Kaya, A.; Topper, E.; Moura, A.A.; Memili, E. Uncovering Sperm Metabolome to Discover Biomarkers for Bull Fertility. BMC Genom. 2019, 20, 714. [Google Scholar] [CrossRef]
- Rego, J.P.A.; Crisp, J.M.; Moura, A.A.; Nouwens, A.S.; Li, Y.; Venus, B.; Corbet, N.J.; Corbet, D.H.; Burns, B.M.; Boe-Hansen, G.B.; et al. Seminal Plasma Proteome of Electroejaculated Bos Indicus Bulls. Anim. Reprod. Sci. 2014, 148, 1–17. [Google Scholar] [CrossRef]
- De Lazari, F.L.; Sontag, E.R.; Schneider, A.; Moura, A.A.A.; Vasconcelos, F.R.; Nagano, C.S.; Mattos, R.C.; Jobim, M.I.M.; Bustamante-Filho, I.C. Seminal Plasma Proteins and Their Relationship with Sperm Motility and Morphology in Boars. Andrologia 2019, 51, e13222. [Google Scholar] [CrossRef]
- Qu, F.; Ying, X.; Guo, W.; Guo, Q.; Chen, G.; Liu, Y.; Ding, Z. The Role of Zn-Alpha2 Glycoprotein in Sperm Motility Is Mediated by Changes in Cyclic AMP. Reproduction 2007, 134, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Russell, S.T.; Zimmerman, T.P.; Domin, B.A.; Tisdale, M.J. Induction of Lipolysis in Vitro and Loss of Body Fat in Vivo by Zinc-Alpha2-Glycoprotein. Biochim. Biophys. Acta 2004, 1636, 59–68. [Google Scholar] [CrossRef]
- Tisdale, M.J. Tumor-Host Interactions. J. Cell Biochem. 2004, 93, 871–877. [Google Scholar] [CrossRef]
- McDermott, L.C.; Freel, J.A.; West, A.P.; Bjorkman, P.J.; Kennedy, M.W. Zn-Alpha2-Glycoprotein, an MHC Class I-Related Glycoprotein Regulator of Adipose Tissues: Modification or Abrogation of Ligand Binding by Site-Directed Mutagenesis. Biochemistry 2006, 45, 2035–2041. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Qu, F.; Guo, W.; Ying, X.; Wu, M.; Zhang, Y. Identification of Sperm Forward Motility-Related Proteins in Human Seminal Plasma. Mol. Reprod. Dev. 2007, 74, 1124–1131. [Google Scholar] [CrossRef] [PubMed]
- Bezerra, M.J.B.; Arruda-Alencar, J.M.; Martins, J.A.M.; Viana, A.G.A.; Viana Neto, A.M.; Rêgo, J.P.A.; Oliveira, R.V.; Lobo, M.; Moreira, A.C.O.; Moreira, R.A.; et al. Major Seminal Plasma Proteome of Rabbits and Associations with Sperm Quality. Theriogenology 2019, 128, 156–166. [Google Scholar] [CrossRef]
- Li, Y.; Sun, Y.; Ni, A.; Shi, L.; Wang, P.; Isa, A.M.; Ge, P.; Jiang, L.; Fan, J.; Ma, H.; et al. Seminal Plasma Proteome as an Indicator of Sperm Dysfunction and Low Sperm Motility in Chickens. Mol. Cell Proteom. 2020, 19, 1035–1046. [Google Scholar] [CrossRef] [PubMed]
- Dunbar, M.R.; Cunningham, M.W.; Wooding, J.B.; Roth, R.P. Cryptorchidism and Delayed Testicular Descent in Florida Black Bears. J. Wildl. Dis. 1996, 32, 661–664. [Google Scholar] [CrossRef]
- Rozynek, J.; Nowacka-Woszuk, J.; Stachowiak, M.; Sowinska, N.; Lukomska, A.; Gruss, M.; Switonski, M.; Szczerbal, I. Lack of Causative Mutation in the AMH and AMHR2 Genes in a Cat (38,XY) with Persistent Mullerian Duct Syndrome (PMDS). Reprod. Domest. Anim. 2024, 59, e14635. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.A.; Gartley, C.J.; Khanam, A. Canine Cryptorchidism: An Update. Reprod. Domest. Anim. 2018, 53, 1263–1270. [Google Scholar] [CrossRef] [PubMed]
- Foresta, C.; Zuccarello, D.; Garolla, A.; Ferlin, A. Role of Hormones, Genes, and Environment in Human Cryptorchidism. Endocr. Rev. 2008, 29, 560–580. [Google Scholar] [CrossRef] [PubMed]
- Trabert, B.; Zugna, D.; Richiardi, L.; McGlynn, K.A.; Akre, O. Congenital Malformations and Testicular Germ Cell Tumors. Int. J. Cancer 2013, 133, 1900–1904. [Google Scholar] [CrossRef] [PubMed]
- Rodprasert, W.; Virtanen, H.E.; Mäkelä, J.-A.; Toppari, J. Hypogonadism and Cryptorchidism. Front. Endocrinol. 2019, 10, 906. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.A.; Coughlin, M.T. Fertility after Bilateral Cryptorchidism. Evaluation by Paternity, Hormone, and Semen Data. Horm. Res. 2001, 55, 28–32. [Google Scholar] [CrossRef]
- Bay, K.; Matthiesson, K.L.; McLachlan, R.I.; Andersson, A.-M. The Effects of Gonadotropin Suppression and Selective Replacement on Insulin-like Factor 3 Secretion in Normal Adult Men. J. Clin. Endocrinol. Metab. 2006, 91, 1108–1111. [Google Scholar] [CrossRef] [PubMed]
- Bay, K.; Hartung, S.; Ivell, R.; Schumacher, M.; Jürgensen, D.; Jorgensen, N.; Holm, M.; Skakkebaek, N.E.; Andersson, A.-M. Insulin-like Factor 3 Serum Levels in 135 Normal Men and 85 Men with Testicular Disorders: Relationship to the Luteinizing Hormone-Testosterone Axis. J. Clin. Endocrinol. Metab. 2005, 90, 3410–3418. [Google Scholar] [CrossRef]
- Ferlin, A.; Arredi, B.; Zuccarello, D.; Garolla, A.; Selice, R.; Foresta, C. Paracrine and Endocrine Roles of Insulin-like Factor 3. J. Endocrinol. Invest. 2006, 29, 657–664. [Google Scholar] [CrossRef] [PubMed]
- Foresta, C.; Bettella, A.; Vinanzi, C.; Dabrilli, P.; Meriggiola, M.C.; Garolla, A.; Ferlin, A. A Novel Circulating Hormone of Testis Origin in Humans. J. Clin. Endocrinol. Metab. 2004, 89, 5952–5958. [Google Scholar] [CrossRef]
- Ferlin, A.; Garolla, A.; Rigon, F.; Rasi Caldogno, L.; Lenzi, A.; Foresta, C. Changes in Serum Insulin-like Factor 3 during Normal Male Puberty. J. Clin. Endocrinol. Metab. 2006, 91, 3426–3431. [Google Scholar] [CrossRef] [PubMed]
- Ping, D.U.; Yu-Wei, Y.; Yan, S.; Zheng, G.U.; Jian, W. Uchl1 and Its Associated Proteins Were Involved in Spermatocyte Apoptosis in Mouse Experimental Cryptorchidism. Sheng Li Xue Bao Acta Physiol. Sin. 2014, 66, 528–536. [Google Scholar] [CrossRef]
- Krzeminska, P.; Stachowiak, M.; Skrzypski, M.; Nowak, T.; Maslak, A.; Switonski, M. Altered Expression of CYP17A1 and CYP19A1 in Undescended Testes of Dogs with Unilateral Cryptorchidism. Anim. Genet. 2020, 51, 763–771. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.G.; An, J.H.; Liu, Y.L.; Yie, S.M.; Zhang, Y.; Li, F.P.; Chen, J.S.; Wang, X.; Morrell, J.M.; Hou, R. Single Layer Centrifugation Improves the Quality of Frozen-Thawed Sperm of Giant Panda (Ailuropoda melanoleuca). Anim. Reprod. Sci. 2018, 195, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.-H.; Liu, Y.-L.; Cai, Z.-G.; An, J.-H.; Lan, J.-C.; Chen, J.-S.; Li, Y.; He, L.; Zhang, Y.; He, P.; et al. Effects of Extender Type on the Quality of Post-Thaw Giant Panda (Ailuropoda melanoleuca) Semen. Cryobiology 2020, 94, 95–99. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-Scale Protein Function Classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics Enrichment Tools: Paths toward the Comprehensive Functional Analysis of Large Gene Lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; von Mering, C.; et al. STRING v9.1: Protein-Protein Interaction Networks, with Increased Coverage and Integration. Nucleic Acids Res. 2013, 41, D808–D815. [Google Scholar] [CrossRef] [PubMed]
- Martín, H.; Flández, M.; Nombela, C.; Molina, M. Protein Phosphatases in MAPK Signalling: We Keep Learning from Yeast. Mol. Microbiol. 2005, 58, 6–16. [Google Scholar] [CrossRef]
- Hutson, J.M.; Hasthorpe, S. Testicular Descent and Cryptorchidism: The State of the Art in 2004. J. Pediatr. Surg. 2005, 40, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Overbeek, P.A.; Gorlov, I.P.; Sutherland, R.W.; Houston, J.B.; Harrison, W.R.; Boettger-Tong, H.L.; Bishop, C.E.; Agoulnik, A.I. A Transgenic Insertion Causing Cryptorchidism in Mice. Genesis 2001, 30, 26–35. [Google Scholar] [CrossRef]
- Tomiyama, H.; Hutson, J.M.; Truong, A.; Agoulnik, A.I. Transabdominal Testicular Descent Is Disrupted in Mice with Deletion of Insulinlike Factor 3 Receptor. J. Pediatr. Surg. 2003, 38, 1793–1798. [Google Scholar] [CrossRef] [PubMed]
- Kubota, Y.; Nef, S.; Farmer, P.J.; Temelcos, C.; Parada, L.F.; Hutson, J.M. Leydig Insulin-like Hormone, Gubernacular Development and Testicular Descent. J. Urol. 2001, 165, 1673–1675. [Google Scholar] [CrossRef] [PubMed]
- Klonisch, T.; Fowler, P.A.; Hombach-Klonisch, S. Molecular and Genetic Regulation of Testis Descent and External Genitalia Development. Dev. Biol. 2004, 270, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Dias, T.R.; Samanta, L.; Agarwal, A.; Pushparaj, P.N.; Panner Selvam, M.K.; Sharma, R. Proteomic Signatures Reveal Differences in Stress Response, Antioxidant Defense and Proteasomal Activity in Fertile Men with High Seminal ROS Levels. Int. J. Mol. Sci. 2019, 20, 203. [Google Scholar] [CrossRef] [PubMed]
- Recuero, S.; Fernandez-Fuertes, B.; Bonet, S.; Barranco, I.; Yeste, M. Potential of Seminal Plasma to Improve the Fertility of Frozen-Thawed Boar Spermatozoa. Theriogenology 2019, 137, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Sun, X.; Zheng, C.; Xue, C.; Jin, Y.; Zhou, N.; Sun, S. The Evolutionarily Conserved Hif-1/Bnip3 Pathway Promotes Mitophagy and Mitochondrial Fission in Crustacean Testes during Hypoxia. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2023, 324, R128–R142. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, J.; Zhao, T.; Chen, J.; Kang, L.; Wei, Y.; Han, L.; Shen, L.; Long, C.; Wu, S.; et al. Di-(2-Ethylhexyl) Phthalate Exposure Leads to Ferroptosis via the HIF-1α/HO-1 Signaling Pathway in Mouse Testes. J. Hazard. Mater. 2022, 426, 127807. [Google Scholar] [CrossRef] [PubMed]
- Adams, V.; Griffin, L.D.; Gelb, B.D.; McCabe, E.R. Protein Kinase Activity of Rat Brain Hexokinase. Biochem. Biophys. Res. Commun. 1991, 177, 1101–1106. [Google Scholar] [CrossRef]
- Wysocki, P.; Strzezek, J. Purification and Characterization of a Protein Tyrosine Acid Phosphatase from Boar Seminal Vesicle Glands. Theriogenology 2003, 59, 1011–1025. [Google Scholar] [CrossRef]
- Tomes, C.N.; Roggero, C.M.; De Blas, G.; Saling, P.M.; Mayorga, L.S. Requirement of Protein Tyrosine Kinase and Phosphatase Activities for Human Sperm Exocytosis. Dev. Biol. 2004, 265, 399–415. [Google Scholar] [CrossRef]
- Sidhu, K.S.; Guraya, S.S. Calmodulin-like Protein in Buffalo (Bubalus bubalis) Seminal Plasma and Its Effect on Sperm Ca++, Mg++-ATPase. Int. J. Androl. 1989, 12, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Boonsaeng, V.; Techa-Udomtaworn, W. Prostatic Origin of Pyruvate Kinase Activity in Human Seminal Plasma. Andrologia 1980, 12, 559–563. [Google Scholar] [CrossRef]
- Feiden, S.; Wolfrum, U.; Wegener, G.; Kamp, G. Expression and Compartmentalisation of the Glycolytic Enzymes GAPDH and Pyruvate Kinase in Boar Spermatogenesis. Reprod. Fertil. Dev. 2008, 20, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Lahnsteiner, F.; Berger, B.; Weismann, T.; Patzner, R.A. Motility of Spermatozoa of Alburnus alburnus (Cyprinidae) and Its Relationship to Seminal Plasma Composition and Sperm Metabolism. Fish. Physiol. Biochem. 1996, 15, 167–179. [Google Scholar] [CrossRef]
- Li, P.; Du, H.; Qiao, X.M.; Liu, Z.G.; Zhou, Q.; Wei, Q. Protein Profile of Dabry’s Sturgeon (Acipenser dabryanus) Spermatozoa and Relationship to Sperm Quality. Anim. Reprod. Sci. 2019, 201, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Vijayvargiya, R.; Schwark, W.S.; Singhal, R.L. Metabolic Control Mechanisms in Mammalian Systems. XI. Pyruvate Kinase Modulation in the Rat Prostate and Seminal Vesicles. Can. J. Biochem. 1970, 48, 1268–1277. [Google Scholar] [CrossRef] [PubMed]
- Yildirim, O.G.; Guney, C.; Alcigir, M.E.; Akar, F. High-Fructose Consumption Suppresses Insulin Signaling Pathway Accompanied by Activation of Macrophage and Apoptotic Markers in Rat Testis. Reprod. Biol. 2023, 23, 100815. [Google Scholar] [CrossRef] [PubMed]
- Paloma Álvarez-Rendón, J.; Manuel Murillo-Maldonado, J.; Rafael Riesgo-Escovar, J. The Insulin Signaling Pathway a Century after Its Discovery: Sexual Dimorphism in Insulin Signaling. Gen. Comp. Endocrinol. 2023, 330, 114146. [Google Scholar] [CrossRef] [PubMed]
- Zabolotny, J.M.; Kim, Y.B.; Peroni, O.D.; Kim, J.K.; Pani, M.A.; Boss, O.; Klaman, L.D.; Kamatkar, S.; Shulman, G.I.; Kahn, B.B.; et al. Overexpression of the LAR (Leukocyte Antigen-Related) Protein-Tyrosine Phosphatase in Muscle Causes Insulin Resistance. Proc. Natl. Acad. Sci. USA 2001, 98, 5187–5192. [Google Scholar] [CrossRef] [PubMed]
- Gurney, J.K.; McGlynn, K.A.; Stanley, J.; Merriman, T.; Signal, V.; Shaw, C.; Edwards, R.; Richiardi, L.; Hutson, J.; Sarfati, D. Risk Factors for Cryptorchidism. Nat. Rev. Urol. 2017, 14, 534–548. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Ma, J.; Liu, Y.; Chen, Z.; Xiao, N.; Lu, Y.; Fu, Y.; Yang, C.; Li, M.; Wu, S.; et al. iProX in 2021: Connecting Proteomics Data Sharing with Big Data. Nucleic Acids Res. 2022, 50, D1522–D1527. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Chen, T.; Wu, S.; Yang, C.; Bai, M.; Shu, K.; Li, K.; Zhang, G.; Jin, Z.; He, F.; et al. iProX: An Integrated Proteome Resource. Nucleic Acids Res. 2019, 47, D1211–D1217. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qian, Y.; Liu, Y.; Wang, T.; Wang, S.; Chen, J.; Li, F.; Zhang, M.; Hu, X.; Wang, J.; Li, Y.; et al. Effects of Cryptorchidism on the Semen Quality of Giant Pandas from the Perspective of Seminal Plasma Proteomics. Genes 2024, 15, 1288. https://doi.org/10.3390/genes15101288
Qian Y, Liu Y, Wang T, Wang S, Chen J, Li F, Zhang M, Hu X, Wang J, Li Y, et al. Effects of Cryptorchidism on the Semen Quality of Giant Pandas from the Perspective of Seminal Plasma Proteomics. Genes. 2024; 15(10):1288. https://doi.org/10.3390/genes15101288
Chicago/Turabian StyleQian, Yicheng, Yuliang Liu, Tao Wang, Shenfei Wang, Jiasong Chen, Feiping Li, Mengshi Zhang, Xianbiao Hu, Juan Wang, Yan Li, and et al. 2024. "Effects of Cryptorchidism on the Semen Quality of Giant Pandas from the Perspective of Seminal Plasma Proteomics" Genes 15, no. 10: 1288. https://doi.org/10.3390/genes15101288




