Genetics of Calcific Aortic Stenosis: A Systematic Review
Abstract
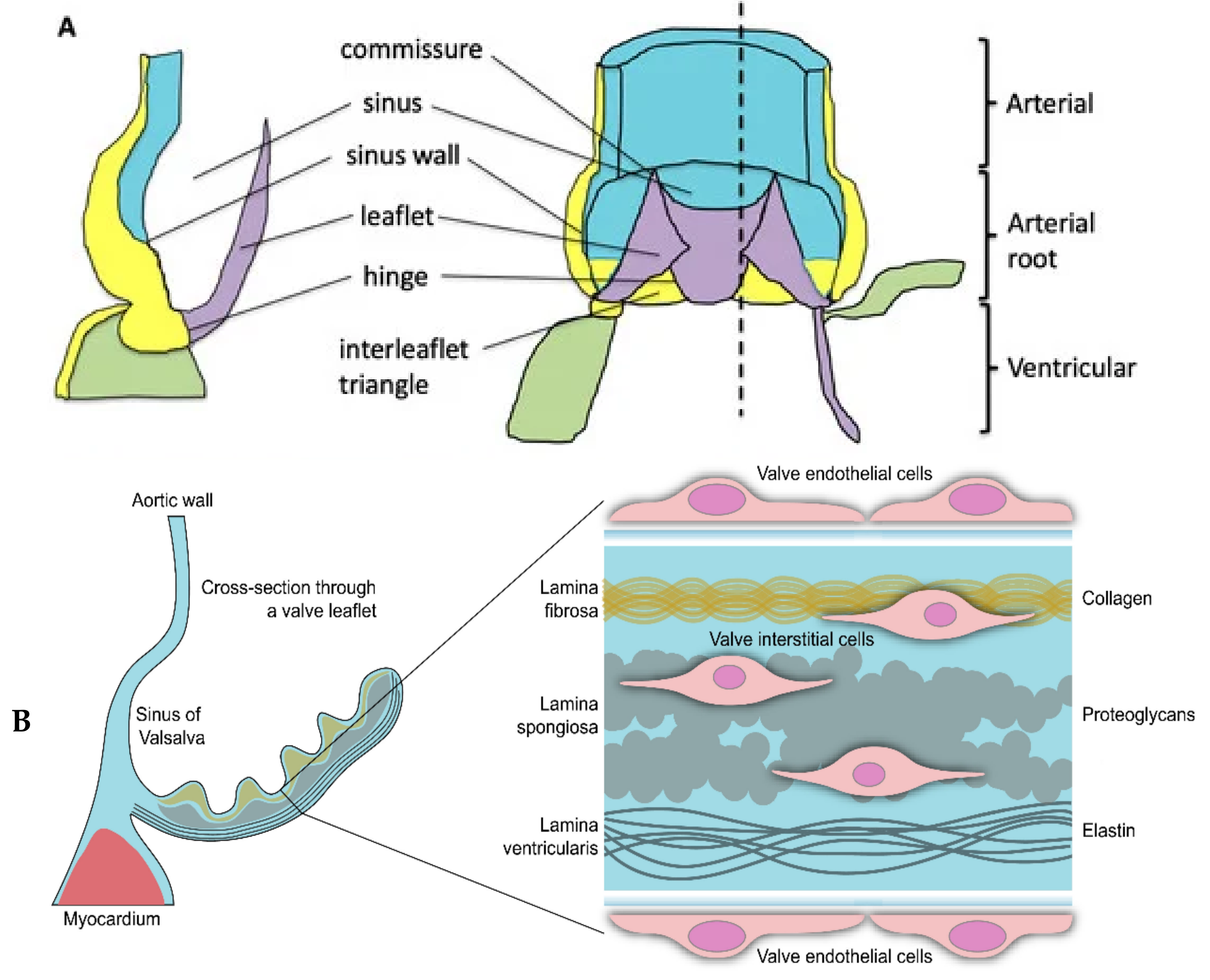
:1. Introduction
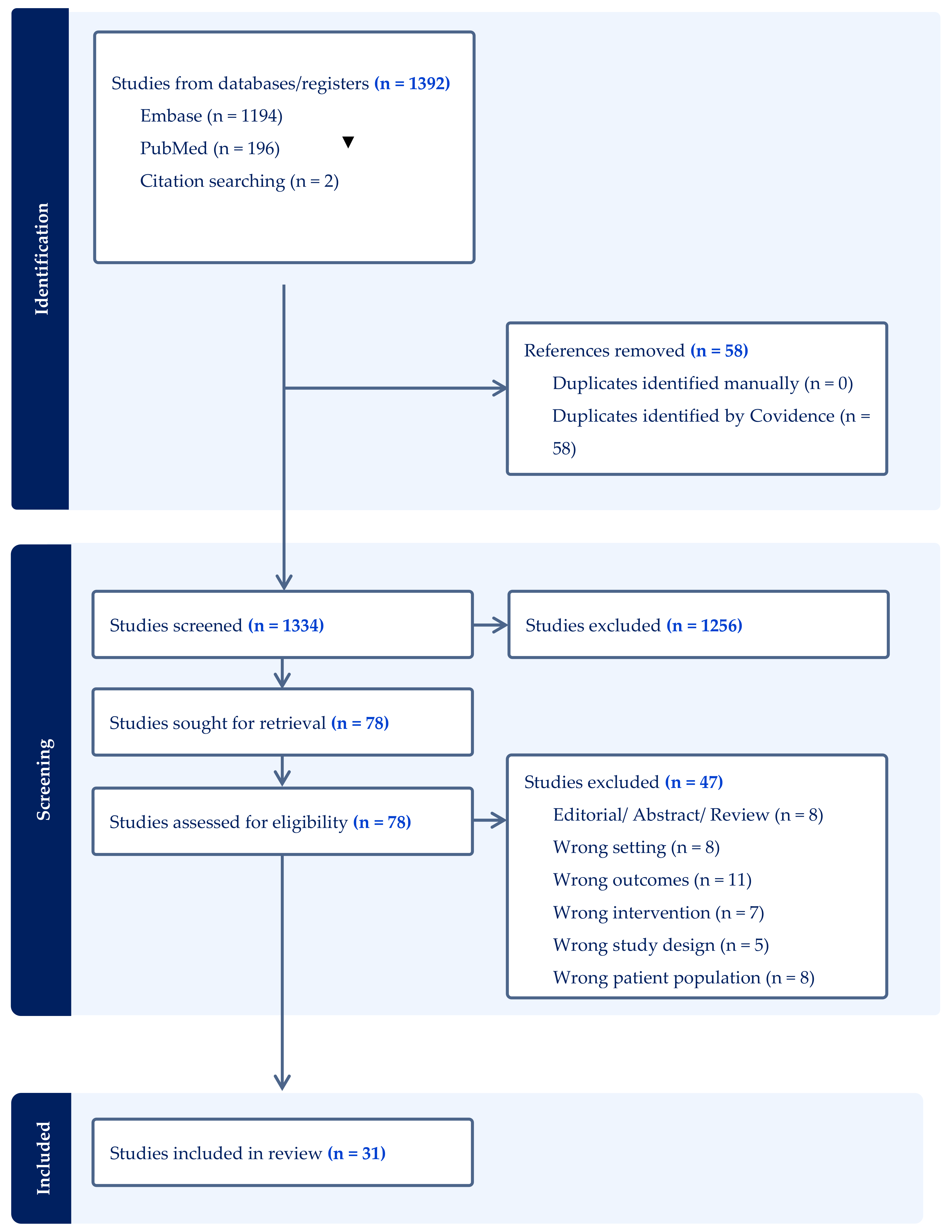
2. Materials and Methods
3. Results
3.1. Lipid Metabolism Genes and CAS
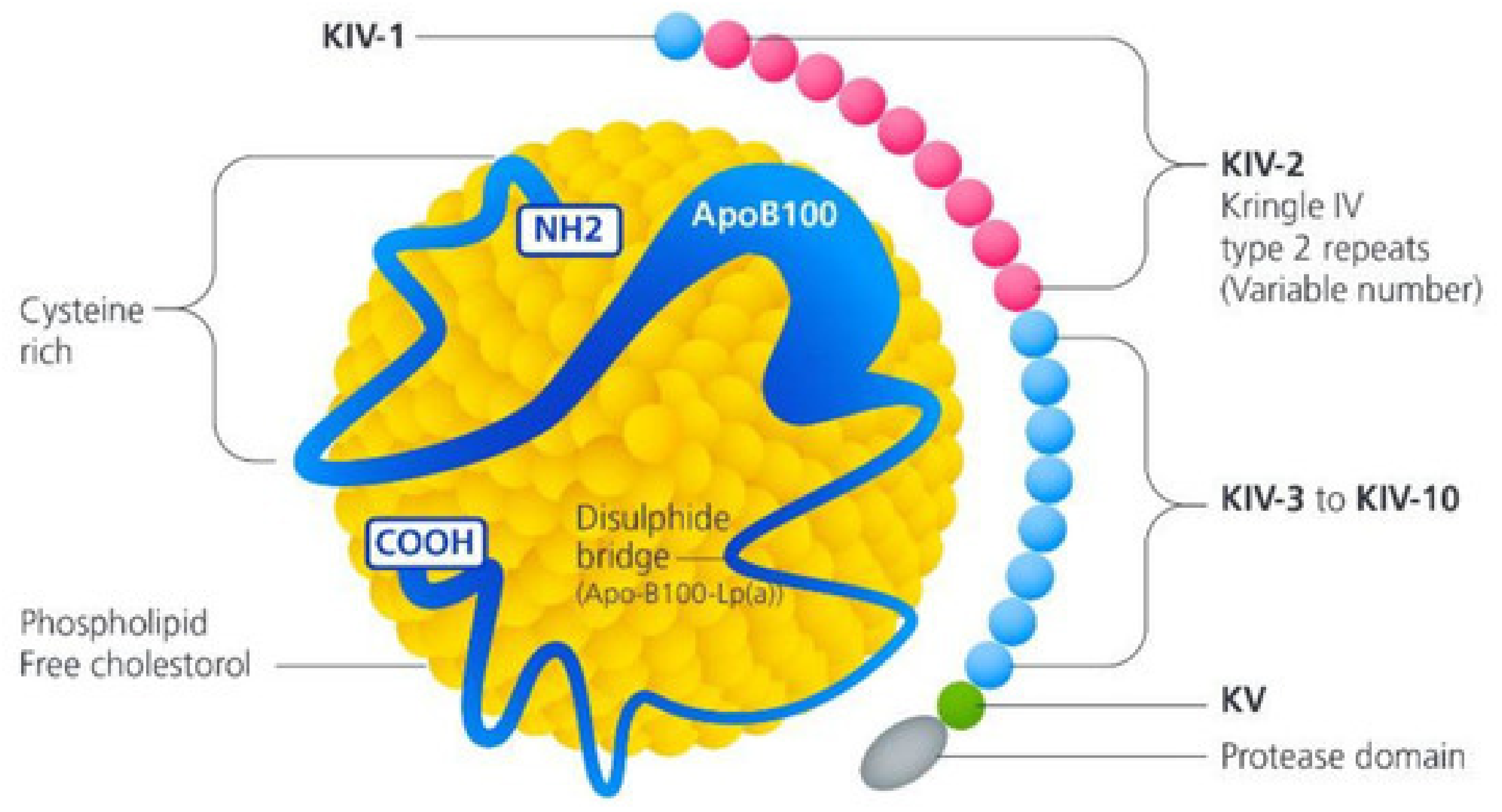
3.1.1. Lipoprotein (A)
3.1.2. Low Density Lipoprotein
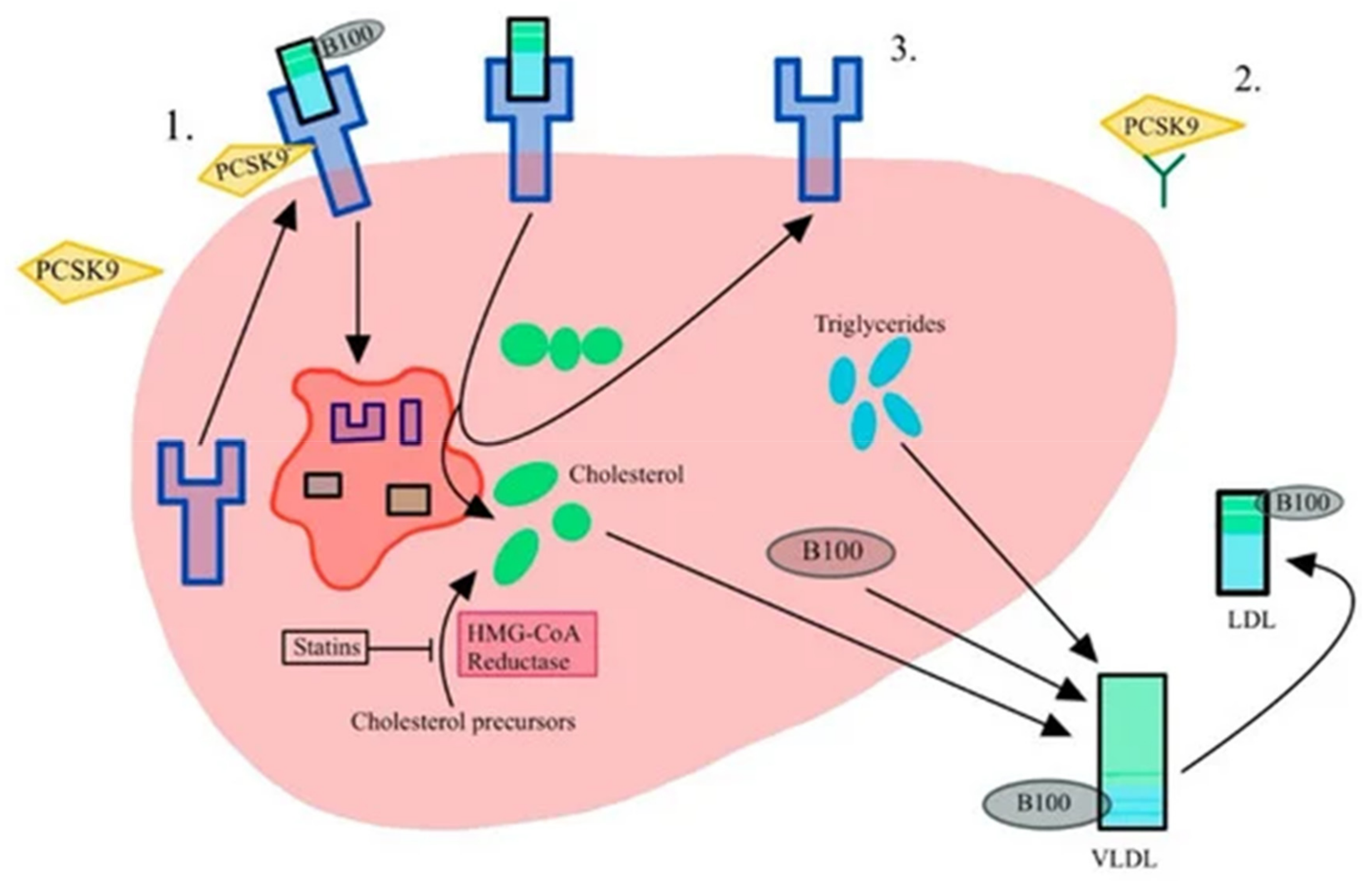
3.2. PCSK9-Proprotein Convertase Subtilisin/Kexin 9
3.3. Apolipoprotein
3.3.1. Apolipoprotein E
3.3.2. Apolipoprotein B
3.4. Lipoprotein-Associated Phospholipase A₂
3.5. Paraoxonase 1
3.6. Inflammatory Pathway Genes and CAS
3.6.1. Interleukin 6
3.6.2. Interleukin 10
3.7. Calcification Pathway Genes and CAS
3.7.1. Palmdelphin
3.7.2. Testis Expressed 41
3.8. Endocrine Pathway Genes and CAS
3.8.1. Parathyroid Hormone
3.8.2. Vitamin D
3.9. Runt-Related Transcription Factor 2 (RUNX2) and Calcium Voltage-Gated Channel Subunit Alpha1 C (CACNA1C)
3.10. Alkaline Phosphatase, Liver/Bone/Kidney
3.11. Miscellaneous Genes and CAS
3.11.1. Neurogenic Locus Notch Homolog Protein 1
3.11.2. Fatty Acid Desaturase 1/2
3.11.3. Neuron Navigator 1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rutkovskiy, A.; Malashicheva, A.; Sullivan, G.; Bogdanova, M.; Kostareva, A.; Stensløkken, K.; Fiane, A.; Vaage, J. Valve Interstitial Cells: The Key to Understanding the Pathophysiology of Heart Valve Calcification. J. Am. Heart Assoc. 2017, 6, e006339. [Google Scholar] [CrossRef] [PubMed]
- Henderson, D.J.; Eley, L.; Chaudhry, B. New Concepts in the Development and Malformation of the Arterial Valves. J. Cardiovasc. Dev. Dis. 2020, 7, 38. [Google Scholar] [CrossRef] [PubMed]
- Di Vito, A.; Donato, A.; Presta, I.; Mancuso, T.; Brunetti, F.S.; Mastroroberto, P.; Amorosi, A.; Malara, N.; Donato, G. Extracellular Matrix in Calcific Aortic Valve Disease: Architecture, Dynamic and Perspectives. Int. J. Mol. Sci. 2021, 22, 913. [Google Scholar] [CrossRef]
- Tsampasian, V.; Merinopoulos, I.; Ravindrarajah, T.; Ring, L.; Heng, E.L.; Prasad, S.; Vassiliou, V.S. Prognostic Value of Cardiac Magnetic Resonance Feature Tracking Strain in Aortic Stenosis. J. Cardiovasc. Dev. Dis. 2024, 11, 30. [Google Scholar] [CrossRef]
- Yadgir, S.; Johnson, C.O.; Aboyans, V.; Adebayo, O.M.; Adedoyin, R.A.; Afarideh, M.; Alahdab, F.; Alashi, A.; Alipour, V.; Arabloo, J.; et al. Global, Regional, and National Burden of Calcific Aortic Valve and Degenerative Mitral Valve Diseases, 1990–2017. Circulation 2020, 141, 1670–1680. [Google Scholar] [CrossRef]
- Tsampasian, V.; Militaru, C.; Parasuraman, S.K.; Loudon, B.L.; Lowery, C.; Rudd, A.; Srinivasan, J.; Singh, S.; Dwivedi, G.; Mahadavan, G.; et al. Prevalence of asymptomatic valvular heart disease in the elderly population: A community-based echocardiographic study. Eur. Heart J. Cardiovasc. Imaging 2024, 25, 1051–1058. [Google Scholar] [CrossRef]
- Kamath, A.R.; Pai, R.G. Risk factors for progression of calcific aortic stenosis and potential therapeutic targets. Int. J. Angiol. Off. Publ. Int. Coll. Angiol. 2008, 17, 63–70. [Google Scholar] [CrossRef]
- Badran, A.; Vohra, H.; Livesey, S. Unoperated severe aortic stenosis: Decision making in an adult UK-based population. Ind. Mark. Manag. 2012, 94, 416–421. [Google Scholar] [CrossRef]
- Tsampasian, V.; Grafton-Clarke, C.; Ramos, A.E.G.; Asimakopoulos, G.; Garg, P.; Prasad, S.; Ring, L.; McCann, G.P.; Rudd, J.; Dweck, M.R.; et al. Management of asymptomatic severe aortic stenosis: A systematic review and meta-analysis. Open Heart 2022, 9, e001982. [Google Scholar] [CrossRef]
- Parikh, P.B.; Kort, S. Racial and Ethnic Disparities in Aortic Stenosis. J. Am. Heart Assoc. Cardiovasc. Cerebrovasc. Dis. 2022, 11, e028131. [Google Scholar] [CrossRef]
- Small, A.M.; Peloso, G.M.; Linefsky, J.; Aragam, J.; Galloway, A.; Tanukonda, V.; Wang, L.-C.; Yu, Z.; Selvaraj, M.S.; Farber-Eger, E.H.; et al. Multiancestry Genome-Wide Association Study of Aortic Stenosis Identifies Multiple Novel Loci in the Million Veteran Program. Circulation 2023, 147, 942–955. [Google Scholar] [CrossRef] [PubMed]
- Benincasa, G.; Suades, R.; Padró, T.; Badimon, L.; Napoli, C. Bioinformatic platforms for clinical stratification of natural history of atherosclerotic cardiovascular diseases. Eur. Heart J. Cardiovasc. Pharmacother. 2023, 9, 758–769. [Google Scholar] [CrossRef] [PubMed]
- Covidence Better Systematic Review Management. Available online: https://www.covidence.org (accessed on 1 August 2024).
- Farrah, K.; Young, K.; Tunis, M.C.; Zhao, L. Risk of bias tools in systematic reviews of health interventions: An analysis of PROSPERO-registered protocols. Syst. Rev. 2019, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Thanassoulis, G.; Campbell, C.Y.; Owens, D.S.; Smith, J.G.; Smith, A.V.; Peloso, G.M.; Kerr, K.F.; Pechlivanis, S.; Budoff, M.J.; Harris, T.B.; et al. Genetic associations with valvular calcification and aortic stenosis. N. Engl. J. Med. 2013, 368, 503–512. [Google Scholar] [CrossRef]
- Arsenault, B.J.; Boekholdt, S.M.; Dubé, M.P.; Rhéaume, E.; Wareham, N.J.; Khaw, K.T.; Sandhu, M.S.; Tardif, J.C. Lipoprotein(a) levels, genotype, and incident aortic valve stenosis: A prospective Mendelian randomization study and replication in a case-control cohort. Circulation. Cardiovasc. Genet. 2014, 7, 304–310. [Google Scholar] [CrossRef]
- Kamstrup, P.R.; Hung, M.Y.; Witztum, J.L.; Tsimikas, S.; Nordestgaard, B.G. Oxidized Phospholipids and Risk of Calcific Aortic Valve Disease: The Copenhagen General Population Study. Arterioscler. Thromb. Vasc. Biol. 2017, 37, 1570–1578. [Google Scholar] [CrossRef]
- Ozkan, U.; Ozcelik, F.; Yildiz, M.; Budak, M. Lipoprotein(a) Gene Polymorphism Increases a Risk Factor for Aortic Valve Calcification. J. Cardiovasc. Dev. Dis. 2019, 6, 31. [Google Scholar] [CrossRef]
- Thériault, S.; Dina, C.; Messika-Zeitoun, D.; Le Scouarnec, S.; Capoulade, R.; Gaudreault, N.; Rigade, S.; Li, Z.; Simonet, F.; Lamontagne, M.; et al. Genetic Association Analyses Highlight IL6, ALPL, and NAV1 As 3 New Susceptibility Genes Underlying Calcific Aortic Valve Stenosis. Circulation. Genom. Precis. Med. 2019, 12, e002617. [Google Scholar] [CrossRef]
- Trenkwalder, T.; Nelson, C.P.; Musameh, M.D.; Mordi, I.R.; Kessler, T.; Pellegrini, C.; Debiec, R.; Rheude, T.; Lazovic, V.; Zeng, L.; et al. Effects of the coronary artery disease associated LPA and 9p21 loci on risk of aortic valve stenosis. Int. J. Cardiol. 2019, 276, 212–217. [Google Scholar] [CrossRef]
- Perrot, N.; Thériault, S.; Dina, C.; Chen, H.Y.; Boekholdt, S.M.; Rigade, S.; Després, A.A.; Poulin, A.; Capoulade, R.; Le Tourneau, T.; et al. Genetic Variation in LPA, Calcific Aortic Valve Stenosis in Patients Undergoing Cardiac Surgery, and Familial Risk of Aortic Valve Microcalcification. JAMA Cardiol. 2019, 4, 620–627. [Google Scholar] [CrossRef]
- Hoekstra, M.; Chen, H.Y.; Rong, J.; Dufresne, L.; Yao, J.; Guo, X.; Tsai, M.Y.; Tsimikas, S.; Post, W.S.; Vasan, R.S.; et al. Genome-Wide Association Study Highlights APOH as a Novel Locus for Lipoprotein(a) Levels—Brief Report. Arter. Thromb. Vasc. Biol. 2021, 41, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Guertin, J.; Kaiser, Y.; Manikpurage, H.; Perrot, N.; Bourgeois, R.; Couture, C.; Wareham, N.J.; Bossé, Y.; Pibarot, P.; Stroes, E.S.; et al. Sex-Specific Associations of Genetically Predicted Circulating Lp(a) (Lipoprotein(a)) and Hepatic LPA Gene Expression Levels With Cardiovascular Outcomes: Mendelian Randomization and Observational Analyses. Circ. Genom. Precis. Med. 2021, 14, e003271. [Google Scholar] [CrossRef]
- Chen, H.Y.; Cairns, B.J.; Small, A.M.; Burr, H.A.; Ambikkumar, A.; Martinsson, A.; Thériault, S.; Munter, H.M.; Steffen, B.; Zhang, R.; et al. Association of FADS1/2 Locus Variants and Polyunsaturated Fatty Acids With Aortic Stenosis. JAMA Cardiol. 2020, 5, 694–702. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.G.; Luk, K.; Schulz, C.-A.; Engert, J.C.; Do, R.; Hindy, G.; Rukh, G.; Dufresne, L.; Almgren, P.; Owens, D.S.; et al. Association of low-density lipoprotein cholesterol-related genetic variants with aortic valve calcium and incident aortic stenosis. JAMA 2014, 312, 1764–1771. [Google Scholar] [CrossRef]
- Allara, E.; Morani, G.; Carter, P.; Gkatzionis, A.; Zuber, V.; Foley, C.N.; Rees, J.M.B.; Mason, A.M.; Bell, S.; Gill, D.; et al. Genetic Determinants of Lipids and Cardiovascular Disease Outcomes: A Wide-Angled Mendelian Randomization Investigation. Circulation. Genom. Precis. Med. 2019, 12, e002711. [Google Scholar] [CrossRef]
- Langsted, A.; Nordestgaard, B.G.; Benn, M.; Tybjærg-Hansen, A.; Kamstrup, P.R. PCSK9 R46L Loss-of-Function Mutation Reduces Lipoprotein(a), LDL Cholesterol, and Risk of Aortic Valve Stenosis. J. Clin. Endocrinol. Metab. 2016, 101, 3281–3287. [Google Scholar] [CrossRef]
- Perrot, N.; Valerio, V.; Moschetta, D.; Boekholdt, S.M.; Dina, C.; Chen, H.Y.; Abner, E.; Martinsson, A.; Manikpurage, H.D.; Rigade, S.; et al. Genetic and In Vitro Inhibition of PCSK9 and Calcific Aortic Valve Stenosis. JACC Basic Transl. Sci. 2020, 5, 649–661. [Google Scholar] [CrossRef]
- Avakian, S.; Annicchino-Bizzacchi, J.; Grinberg, M.; Ramires, J.; Mansur, A. Apolipoproteins AI, B, and E polymorphisms in severe aortic valve stenosis. Clin. Genet. 2001, 60, 381–384. [Google Scholar] [CrossRef]
- Novaro, G.M.; Sachar, R.; Pearce, G.L.; Sprecher, D.L.; Griffin, B.P. Association between apolipoprotein E alleles and calcific valvular heart disease. Circulation 2003, 108, 1804–1808. [Google Scholar] [CrossRef]
- Ortlepp, J.R.; Pillich, M.; Mevissen, V.; Krantz, C.; Kimmel, M.; Autschbach, R.; Langebartels, G.; Erdmann, J.; Hoffmann, R.; Zerres, K. APOE alleles are not associated with calcific aortic stenosis. Heart 2006, 92, 1463–1466. [Google Scholar] [CrossRef]
- Gaudreault, N.; Ducharme, V.; Lamontagne, M.; Guauque-Olarte, S.; Mathieu, P.; Pibarot, P.; Bossé, Y. Replication of genetic association studies in aortic stenosis in adults. Am. J. Cardiol. 2011, 108, 1305–1310. [Google Scholar] [CrossRef] [PubMed]
- Kritharides, L.; Nordestgaard, B.G.; Tybjærg-Hansen, A.; Kamstrup, P.R.; Afzal, S. Effect of APOE ε Genotype on Lipoprotein(a) and the Associated Risk of Myocardial Infarction and Aortic Valve Stenosis. J. Clin. Endocrinol. Metab. 2017, 102, 3390–3399. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-T.; Li, Y.; Ma, Y.-T.; Yang, Y.-N.; Ma, X.; Li, X.-M.; Liu, F.; Chen, B.-D. Association between apolipoprotein B genetic polymorphism and the risk of calcific aortic stenosis in Chinese subjects, in Xinjiang, China. Lipids Health Dis. 2018, 17, 40. [Google Scholar] [CrossRef] [PubMed]
- Perrot, N.; Thériault, S.; Rigade, S.; Chen, H.Y.; Dina, C.; Martinsson, A.; Boekholdt, S.M.; Capoulade, R.; Le Tourneau, T.; Messika-Zeitoun, D.; et al. Lipoprotein-associated phospholipase A2 activity, genetics and calcific aortic valve stenosis in humans. Heart 2020, 106, 1407–1412. [Google Scholar] [CrossRef] [PubMed]
- Moura, L.M.; Faria, S.; Brito, M.; Pinto, F.J.; Kristensen, S.D.; Barros, I.M.; Rajamannan, N.; Rocha-Gonçalves, F. Relationship of PON1 192 and 55 gene polymorphisms to calcific valvular aortic stenosis. Am. J. Cardiovasc. Dis. 2012, 2, 123–132. [Google Scholar]
- Junco-Vicente, A.; Solache-Berrocal, G.; del Río-García, Á.; Rolle-Sóñora, V.; Areces, S.; Morís, C.; Martín, M.; Rodríguez, I. IL6 gene polymorphism association with calcific aortic valve stenosis and influence on serum levels of interleukin-6. Front. Cardiovasc. Med. 2022, 9, 989539. [Google Scholar] [CrossRef]
- Helgadottir, A.; Thorleifsson, G.; Gretarsdottir, S.; Stefansson, O.A.; Tragante, V.; Thorolfsdottir, R.B.; Jonsdottir, I.; Bjornsson, T.; Steinthorsdottir, V.; Verweij, N.; et al. Genome-wide analysis yields new loci associating with aortic valve stenosis. Nat. Commun. 2018, 9, 987. [Google Scholar] [CrossRef]
- Thériault, S.; Gaudreault, N.; Lamontagne, M.; Rosa, M.; Boulanger, M.-C.; Messika-Zeitoun, D.; Clavel, M.-A.; Capoulade, R.; Dagenais, F.; Pibarot, P.; et al. A transcriptome-wide association study identifies PALMD as a susceptibility gene for calcific aortic valve stenosis. Nat. Commun. 2018, 9, 988. [Google Scholar] [CrossRef]
- Li, Z.; Gaudreault, N.; Arsenault, B.J.; Mathieu, P.; Bossé, Y.; Thériault, S. Phenome-wide analyses establish a specific association between aortic valve PALMD expression and calcific aortic valve stenosis. Commun. Biol. 2020, 3, 477. [Google Scholar] [CrossRef]
- Schmitz, F.; Ewering, S.; Zerres, K.; Klomfass, S.; Hoffmann, R.; Ortlepp, J.R. Parathyroid hormone gene variant and calcific aortic stenosis. J. Heart Valve Dis. 2009, 18, 262–267. [Google Scholar]
- Ortlepp, J.R.; Hoffmann, R.; Ohme, F.; Lauscher, J.; Bleckmann, F.; Hanrath, P. The vitamin D receptor genotype predisposes to the development of calcific aortic valve stenosis. Heart 2001, 85, 635–638. [Google Scholar] [CrossRef]
- Guauque-Olarte, S.; Messika-Zeitoun, D.; Droit, A.; Lamontagne, M.; Tremblay-Marchand, J.; Lavoie-Charland, E.; Gaudreault, N.; Arsenault, B.J.; Dubé, M.-P.; Tardif, J.-C.; et al. Calcium Signaling Pathway Genes RUNX2 and CACNA1C Are Associated With Calcific Aortic Valve Disease. Circ. Cardiovasc. Genet. 2015, 8, 812–822. [Google Scholar] [CrossRef]
- Ducharme, V.; Guauque-Olarte, S.; Gaudreault, N.; Pibarot, P.; Mathieu, P.; Bossé, Y. NOTCH1 genetic variants in patients with tricuspid calcific aortic valve stenosis. J. Heart Valve Dis. 2013, 22, 142–149. [Google Scholar] [PubMed]
- Doerfler, A.M.; Park, S.H.; Assini, J.M.; Youssef, A.; Saxena, L.; Yaseen, A.B.; De Giorgi, M.; Chuecos, M.; Hurley, A.E.; Li, A.; et al. LPA disruption with AAV-CRISPR potently lowers plasma apo(a) in transgenic mouse model: A proof-of-concept study. Mol. Ther. Methods Clin. Dev. 2022, 27, 337–351. [Google Scholar] [CrossRef]
- Vassiliou, V.S.; Flynn, P.D.; Raphael, C.E.; Newsome, S.; Khan, T.; Ali, A.; Halliday, B.; Bruengger, A.S.; Malley, T.; Sharma, P.; et al. Lipoprotein(a) in patients with aortic stenosis: Insights from cardiovascular magnetic resonance. PLoS ONE 2017, 12, e0181077. [Google Scholar] [CrossRef]
- Telyuk, P.; Austin, D.; Luvai, A.; Zaman, A. Lipoprotein(a): Insights for the Practicing Clinician. J. Clin. Med. 2022, 11, 3673. [Google Scholar] [CrossRef]
- Dawber, T.R.; Meadors, G.F.; Moore, F.E. Epidemiological approaches to heart disease: The Framingham Study. Am. J. Public Health Nation’s Health 1951, 41, 279–281. [Google Scholar] [CrossRef]
- Harris, T.B.; Launer, L.J.; Eiriksdottir, G.; Kjartansson, O.; Jonsson, P.V.; Sigurdsson, G.; Thorgeirsson, G.; Aspelund, T.; Garcia, M.E.; Cotch, M.F.; et al. Age, Gene/Environment Susceptibility-Reykjavik Study: Multidisciplinary Applied Phenomics. Am. J. Epidemiol. 2007, 165, 1076–1087. [Google Scholar] [CrossRef]
- Bild, D.E.; Bluemke, D.A.; Burke, G.L.; Detrano, R.; Diez Roux, A.V.; Folsom, A.R.; Greenland, P.; Jacobs, D.R., Jr.; Kronmal, R.; Liu, K.; et al. Multi-Ethnic Study of Atherosclerosis: Objectives and design. Am. J. Epidemiol. 2002, 156, 871–881. [Google Scholar] [CrossRef]
- Berglund, G.; Elmstähl, S.; Janzon, L.; Larsson, S.A. The Malmo Diet and Cancer Study. Design and feasibility. J. Intern. Med. 1993, 233, 45–51. [Google Scholar] [CrossRef]
- Yeang, C.; Wilkinson, M.J.; Tsimikas, S. Lipoprotein(a) and Oxidized Phospholipids in Calcific Aortic Valve Stenosis. Curr. Opin. Cardiol. 2016, 31, 440–450. [Google Scholar] [CrossRef]
- Stulnig, T.M.; Morozzi, C.; Reindl-Schwaighofer, R.; Stefanutti, C. Looking at Lp(a) and Related Cardiovascular Risk: From Scientific Evidence and Clinical Practice. Curr. Atheroscler. Rep. 2019, 21, 37. [Google Scholar] [CrossRef]
- Nordestgaard, B.G.; Chapman, M.J.; Ray, K.; Borén, J.; Andreotti, F.; Watts, G.F.; Ginsberg, H.; Amarenco, P.; Catapano, A.; Descamps, O.S.; et al. Lipoprotein(a) as a cardiovascular risk factor: Current status. Eur. Heart J. 2010, 31, 2844–2853. [Google Scholar] [CrossRef] [PubMed]
- Bouchareb, R.; Mahmut, A.; Nsaibia, M.J.; Boulanger, M.C.; Dahou, A.; Lépine, J.L.; Laflamme, M.H.; Hadji, F.; Couture, C.; Trahan, S.; et al. Autotaxin Derived From Lipoprotein(a) and Valve Interstitial Cells Promotes Inflammation and Mineralization of the Aortic ValveCLINICAL PERSPECTIVE. Circulation 2015, 132, 677–690. [Google Scholar] [CrossRef]
- Peeters, F.E.C.M.; Meex, S.J.R.; Dweck, M.R.; Aikawa, E.; Crijns, H.J.G.M.; Schurgers, L.J.; Kietselaer, B.L.J.H. Calcific aortic valve stenosis: Hard disease in the heart: A biomolecular approach towards diagnosis and treatment. Eur. Heart J. 2018, 39, 2618–2624. [Google Scholar] [CrossRef]
- Rawadi, G.; Vayssière, B.; Dunn, F.; Baron, R.; Roman-Roman, S. BMP-2 controls alkaline phosphatase expression and osteoblast mineralization by a Wnt autocrine loop. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 2003, 18, 1842–1853. [Google Scholar] [CrossRef]
- O’Donoghue, M.L.; Fazio, S.; Giugliano, R.P.; Stroes, E.S.G.; Kanevsky, E.; Gouni-Berthold, I.; Im, K.; Lira Pineda, A.; Wasserman, S.M.; Češka, R.; et al. Lipoprotein(a), PCSK9 Inhibition, and Cardiovascular Risk. Circulation 2019, 139, 1483–1492. [Google Scholar] [CrossRef]
- Bergmark, B.A.; O’donoghue, M.L.; Murphy, S.A.; Kuder, J.F.; Ezhov, M.V.; Ceška, R.; Gouni-Berthold, I.; Jensen, H.K.; Tokgozoglu, S.L.; Mach, F.; et al. An Exploratory Analysis of Proprotein Convertase Subtilisin/Kexin Type 9 Inhibition and Aortic Stenosis in the FOURIER Trial. JAMA Cardiol. 2020, 5, 709–713. [Google Scholar] [CrossRef]
- NCT04968509. Effect of PCSK9 Inhibitors on Calcific Aortic Valve Stenosis. Available online: https://clinicaltrials.gov/ct2/show/NCT04968509 (accessed on 1 August 2024).
- Study Details | PCSK9 Inhibitors in the Progression of Aortic Stenosis | ClinicalTrials.gov. Available online: https://www.smartpatients.com/trials/NCT03051360 (accessed on 1 August 2024).
- Ray, K.K.; Wright, R.S.; Kallend, D.; Koenig, W.; Leiter, L.A.; Raal, F.J.; Bisch, J.A.; Richardson, T.; Jaros, M.; Wijngaard, P.L.; et al. Two Phase 3 Trials of Inclisiran in Patients with Elevated LDL Cholesterol. N. Engl. J. Med. 2020, 382, 1507–1519. [Google Scholar] [CrossRef]
- Cowell, S.J.; Newby, D.E.; Prescott, R.J.; Bloomfield, P.; Reid, J.; Northridge, D.B.; Boon, N.A. A randomized trial of intensive lipid-lowering therapy in calcific aortic stenosis. N. Engl. J. Med. 2005, 352, 2389–2397. [Google Scholar] [CrossRef]
- Dichtl, W.; Alber, H.F.; Feuchtner, G.M.; Hintringer, F.; Reinthaler, M.; Bartel, T.; Süssenbacher, A.; Grander, W.; Ulmer, H.; Pachinger, O.; et al. Prognosis and risk factors in patients with asymptomatic aortic stenosis and their modulation by atorvastatin (20 mg). Am. J. Cardiol. 2008, 102, 743–748. [Google Scholar] [CrossRef]
- Rossebø, A.B.; Pedersen, T.R.; Boman, K.; Brudi, P.; Chambers, J.B.; Egstrup, K.; Gerdts, E.; Gohlke-Bärwolf, C.; Holme, I.; Kesäniemi, Y.A.; et al. Intensive lipid lowering with simvastatin and ezetimibe in aortic stenosis. N. Engl. J. Med. 2008, 359, 1343–1356. [Google Scholar] [CrossRef] [PubMed]
- Jassal, D.S.; Tam, J.W.; Bhagirath, K.M.; Gaboury, I.; Sochowski, R.A.; Dumesnil, J.G.; Giannoccaro, P.J.; Jue, J.; Pandey, A.S.; Joyner, C.D.; et al. Association of mitral annular calcification and aortic valve morphology: A substudy of the aortic stenosis progression observation measuring effects of rosuvastatin (ASTRONOMER) study. Eur. Heart J. 2008, 29, 1542–1547. [Google Scholar] [CrossRef]
- Peterson, A.S.; Fong, L.G.; Young, S.G. PCSK9 function and physiology. J. Lipid Res. 2008, 49, 1152–1156. [Google Scholar] [CrossRef]
- Beltran, R.A.; Zemeir, K.J.; Kimberling, C.R.; Kneer, M.S.; Mifflin, M.D.; Broderick, T.L. Is a PCSK9 Inhibitor Right for Your Patient? A Review of Treatment Data for Individualized Therapy. Int. J. Environ. Res. Public Health 2022, 19, 16899. [Google Scholar] [CrossRef]
- Yurtseven, E.; Ural, D.; Baysal, K.; Tokgözoğlu, L. An Update on the Role of PCSK9 in Atherosclerosis. J. Atheroscler. Thromb. 2020, 27, 909–918. [Google Scholar] [CrossRef]
- Tsimikas, S.; Fazio, S.; Ferdinand, K.C.; Ginsberg, H.N.; Koschinsky, M.L.; Marcovina, S.M.; Moriarty, P.M.; Rader, D.J.; Remaley, A.T.; Reyes-Soffer, G.; et al. NHLBI Working Group Recommendations to Reduce Lipoprotein(a)-Mediated Risk of Cardiovascular Disease and Aortic Stenosis. J. Am. Coll. Cardiol. 2018, 71, 177–192. [Google Scholar] [CrossRef]
- Sudlow, C.; Gallacher, J.; Allen, N.; Beral, V.; Burton, P.; Danesh, J.; Downey, P.; Elliott, P.; Green, J.; Landray, M.; et al. UK biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015, 12, e1001779. [Google Scholar] [CrossRef]
- Day, N.; Oakes, S.; Luben, R.; Khaw, K.T.; Bingham, S.; Welch, A.; Wareham, N. EPIC-Norfolk: Study design and characteristics of the cohort. European Prospective Investigation of Cancer. Br. J. Cancer 1999, 80 (Suppl. S1), 95–103. [Google Scholar]
- Watson, A.D.; Berliner, J.A.; Hama, S.Y.; La Du, B.N.; Faull, K.F.; Fogelman, A.M.; Navab, M. Protective effect of high density lipoprotein associated paraoxonase. Inhibition of the biological activity of minimally oxidized low density lipoprotein. J. Clin. Investig. 1995, 96, 2882–2891. [Google Scholar] [CrossRef]
- Artiach, G.; Bäck, M. Omega-3 Polyunsaturated Fatty Acids and the Resolution of Inflammation: Novel Therapeutic Opportunities for Aortic Valve Stenosis? Front. Cell Dev. Biol. 2020, 8, 584128. [Google Scholar] [CrossRef] [PubMed]
- Saxena, A.; Khosraviani, S.; Noel, S.; Mohan, D.; Donner, T.; Hamad, A.R.A. Interleukin-10 paradox: A potent immunoregulatory cytokine that has been difficult to harness for immunotherapy. Cytokine 2015, 74, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Al Musa, T.; Treibel, T.A.; Vassiliou, V.S.; Captur, G.; Chin, C.; Dobson, L.E.; Pica, S.; Loudon, M.; Malley, T.; et al. Sex differences in left ventricular remodelling, myocardial fibrosis and mortality after aortic valve replacement. Heart 2019, 105, 1818–1824. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Zhang, J.; Yang, Z.; Jian, W.; Zhu, Y.; Gao, S.; Liu, Y.; Li, Y.; He, S.; Zhang, C.; et al. Palmdelphin Deficiency Evokes NF-κB Signaling in Valvular Endothelial Cells and Aggravates Aortic Valvular Remodeling. JACC Basic Transl. Sci. 2023, 8, 1457–1472. [Google Scholar] [CrossRef]
- Wang, S.; Yu, H.; Gao, J.; Chen, J.; He, P.; Zhong, H.; Tan, X.; Staines, K.A.; Macrae, V.E.; Fu, X.; et al. PALMD regulates aortic valve calcification via altered glycolysis and NF-κB–mediated inflammation. J. Biol. Chem. 2022, 298, 101887. [Google Scholar] [CrossRef]
- Gade, T.P.; Motley, M.W.; Beattie, B.J.; Bhakta, R.; Boskey, A.L.; Koutcher, J.A.; Mayer-Kuckuk, P. Imaging of alkaline phosphatase activity in bone tissue. PLoS ONE 2011, 6, e22608. [Google Scholar] [CrossRef]
- Vassiliou, V.S.; Pavlou, M.; Malley, T.; Halliday, B.P.; Tsampasian, V.; Raphael, C.E.; Tse, G.; Vieira, M.S.; Auger, D.; Everett, R.; et al. A novel cardiovascular magnetic resonance risk score for predicting mortality following surgical aortic valve replacement. Sci. Rep. 2021, 11, 1–9. [Google Scholar] [CrossRef]
- Vassiliou, V.; Raphael, C.; Ali, A.; Perperoglou, A.; Chin, C.; Nyktari, E.; Alpendurada, F.; Jabbour, A.; Baksi, J.; Murigu, T.; et al. Midwall Fibrosis and Long-Term Outcome in Patients With Aortic Stenosis. J. Am. Coll. Cardiol. 2014, 63, A1199. [Google Scholar] [CrossRef]
- Mathieu, P.; Bouchareb, R.; Boulanger, M.-C. Innate and Adaptive Immunity in Calcific Aortic Valve Disease. J. Immunol. Res. 2015, 2015, 851945. [Google Scholar] [CrossRef]
- Iyer, S.S.; Cheng, G. Role of Interleukin 10 Transcriptional Regulation in Inflammation and Autoimmune Disease. Crit. Rev. Immunol. 2012, 32, 23–63. [Google Scholar] [CrossRef]
- Driscoll, K.; Cruz, A.D.; Butcher, J.T. Inflammatory and Biomechanical Drivers of Endothelial-Interstitial Interactions in Calcific Aortic Valve Disease. Circ. Res. 2021, 128, 1344–1370. [Google Scholar] [CrossRef] [PubMed]
- Monzack, E.L.; Masters, K.S. Can valvular interstitial cells become true osteoblasts? A side-by-side comparison. J. Heart Valve Dis. 2011, 20, 449–463. [Google Scholar]
- Lin, M.-E.; Chen, T.M.; Wallingford, M.C.; Nguyen, N.B.; Yamada, S.; Sawangmake, C.; Zhang, J.; Speer, M.Y.; Giachelli, C.M. Runx2 deletion in smooth muscle cells inhibits vascular osteochondrogenesis and calcification but not atherosclerotic lesion formation. Cardiovasc. Res. 2016, 112, 606–616. [Google Scholar] [CrossRef] [PubMed]
- Mendell, J.R.; Al-Zaidy, S.; Shell, R.; Arnold, W.D.; Rodino-Klapac, L.R.; Prior, T.W.; Lowes, L.; Alfano, L.; Berry, K.; Church, K.; et al. Single-Dose Gene-Replacement Therapy for Spinal Muscular Atrophy. N. Engl. J. Med. 2017, 377, 1713–1722. [Google Scholar] [CrossRef] [PubMed]
- Pipe, S.W.; Leebeek, F.W.; Recht, M.; Key, N.S.; Castaman, G.; Miesbach, W.; Lattimore, S.; Peerlinck, K.; Van der Valk, P.; Coppens, M.; et al. Gene Therapy with Etranacogene Dezaparvovec for Hemophilia B. N. Engl. J. Med. 2023, 388, 706–718. [Google Scholar] [CrossRef]
- D’antiga, L.; Beuers, U.; Ronzitti, G.; Brunetti-Pierri, N.; Baumann, U.; Di Giorgio, A.; Aronson, S.; Hubert, A.; Romano, R.; Junge, N.; et al. Gene Therapy in Patients with the Crigler–Najjar Syndrome. N. Engl. J. Med. 2023, 389, 620–631. [Google Scholar] [CrossRef]
- Alton, E.W.F.W.; Armstrong, D.K.; Ashby, D.; Bayfield, K.J.; Bilton, D.; Bloomfield, E.V.; Boyd, A.C.; Brand, J.; Buchan, R.; Calcedo, R.; et al. Repeated nebulisation of non-viral CFTR gene therapy in patients with cystic fibrosis: A randomised, double-blind, placebo-controlled, phase 2b trial. Lancet Respir. Med. 2015, 3, 684–691. [Google Scholar] [CrossRef]
| Gene(s) | Associated Metabolic Pathway | Related Publications |
|---|---|---|
| LPA | Lipid metabolism | Thanassoulis et al., 2013 [15], Arsenault et al., 2014 [16], Kamstrup et al., 2017 [17], Ozkan et al., 2019 [18], Theriault et al., 2019 [19], Trenkwalder et al., 2019 [20], Perrot et al., 2019 [21], Hoekstra et al., 2021 [22], Guertin et al., 2021 [23], Chen et al., 2020 [24], Small et al., 2023 [11] |
| LDL | Lipid metabolism | Smith et al., 2014 [25], Allara et al., 2019 [26] |
| PCSK9 | Lipid metabolism | Langsted et al., 2016 [27], Perrot et al., 2020 [28] |
| Apolipoprotein | Lipid metabolism | Avakian et al., 2001 [29], Novaro et al., 2003 [30], Ortlepp et al., 2006 [31], Gaudreault et al., 2011 [32], Kritharides et al., 2017 [33], Wang et al., 2018 [34] |
| Lp-PLA2 | Lipid metabolism | Perrot et al., 2020 [35] |
| PONS1 | Lipid metabolism | Moura et al., 2012 [36] |
| IL-6 | Inflammation | Theriault et al., 2019 [19], Junco-Vincente et al., 2022 [37], Small et al., 2023 [11] |
| IL-10 | Inflammation | Gaudreault et al., 2011 [32] |
| PALMD | Calcification pathway | Helgadottir et al., 2018 [38], Theriault et al., 2018 [39], Theriault et al., 2019 [19], Li et al., 2020 [40], Junco-Vincente et al., 2022 [37], Small et al., 2023 [11] |
| TEX41 | Calcification pathway | Helgadottir et al., 2018 [38], Small et al., 2023 [11] |
| PTH | Endocrine metabolism | Schmitz et al., 2009 [41], Gaudreault et al., 2011 [32] |
| Vitamin D Receptor | Endocrine metabolism | Ortlepp et al., 2001 [42], Schmitz et al., 2009 [41] |
| RUNX2 and CACNA1C | Endocrine metabolism | Guauque-Olarte et al., 2015 [43] |
| ALPL | Endocrine metabolism | Theriault et al., 2019 [19] |
| NOTCH1 | Miscellaneous | Ducharme et al., 2013 [44] |
| FADS1/2 | Miscellaneous | Chen et al., 2020 [24] |
| NAV1 | Miscellaneous | Theriault et al., 2019 [19] |
| Lipoprotein(a) ≤ 50 mg/dL | Lipoprotein(a) > 50 mg/dL | |
|---|---|---|
| ε 2 carriers | 1.00 | 1.49 (0.72 to 3.08) |
| Non ε 2 carriers | 1.05 (0.74 to 1.51) | 2.04 (1.46 to 2.26) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vassiliou, V.S.; Johnson, N.; Langlands, K.; Tsampasian, V. Genetics of Calcific Aortic Stenosis: A Systematic Review. Genes 2024, 15, 1309. https://doi.org/10.3390/genes15101309
Vassiliou VS, Johnson N, Langlands K, Tsampasian V. Genetics of Calcific Aortic Stenosis: A Systematic Review. Genes. 2024; 15(10):1309. https://doi.org/10.3390/genes15101309
Chicago/Turabian StyleVassiliou, Vassilios S., Nicholas Johnson, Kenneth Langlands, and Vasiliki Tsampasian. 2024. "Genetics of Calcific Aortic Stenosis: A Systematic Review" Genes 15, no. 10: 1309. https://doi.org/10.3390/genes15101309






